Abstract
It has been reported that nuclei from HeLa cells are responsible for ∼10%–15% of total cellular protein synthesis. We show here that isolated Chinese hamster ovary (CHO) and HeLa cell nuclei are essentially inactive for translation, and that the earlier results were most likely due to cytoplasmic contamination. Moreover, we suggest that the nascent polypeptides observed in nuclei of permeabilized cells may have been due to “overpermeabilization” and consequent damage to the cells. Based on this information, we conclude that nuclear protein synthesis, if it exists, is limited to less than 1% of that in cells.
INTRODUCTION
For many years it was generally accepted that, in eukaryotic cells, transcription and translation are distinct processes of the nuclear and cytoplasmic compartments, respectively. However, a number of recent findings have brought this concept into doubt. First, many components of the protein-synthesizing machinery are present in the nucleus (Sanders et al. 1996; Lund and Dahlberg 1998; Dostie et al. 2000), although for other than the aminoacyl-tRNA synthetases (Nathanson and Deutscher 2000), it is not clear that these components are active. Second, multiple reports support the conclusion that open reading frames need to be recognized within the nucleus for correct transcription and cotranscriptional events (for review, see Brogna 2001), and for the phenomenon of nonsense-mediated decay (NMD) (e.g., Wang et al. 2002). Inasmuch as translating ribosomes are the only known mechanism for inspecting open reading frames, these findings imply that translation can occur in association with nuclei. Finally, Iborra et al. (2001) reported that protein synthesis could be demonstrated in isolated HeLa cell nuclei and that nascent polypeptides could be observed in the nuclei of permeabilized HeLa cells. The level of nuclear translation in these experiments amounted to 9%–15% of that in the whole cell.
In view of the importance of these experiments, we have reexamined protein synthesis in isolated nuclei prepared from Chinese hamster ovary (CHO) cells and from HeLa cells. Although we were generally able to reproduce the data of Iborra et al. (2001) using HeLa cell nuclei prepared by their procedures, we found that these nuclei were significantly contaminated with cytoplasmic adherents. In contrast, purified nuclei from either CHO or HeLa cells, shown to be free of cytoplasmic contamination, were unable to synthesize protein. Thus, on the basis of these data, we cannot confirm the findings of Iborra et al. (2001) that isolated nuclei are able to carry out protein synthesis.
RESULTS
In previous studies of nuclear aminoacyl-tRNA synthetases (Nathanson and Deutscher 2000), we had occasion to prepare purified nuclei from CHO cells. Using such preparations of isolated nuclei, we attempted to reproduce the work of Iborra et al. (2001) to ascertain whether their findings could be extended to nuclei from other mammalian cells. However, in contrast to their results with HeLa cell nuclei, the rate of protein synthesis in isolated CHO cell nuclei, in many experiments, never exceeded 0.8 pmole/20 min/106 nuclei, and averaged ∼0.5 pmole/106 nuclei. This compared with a value of 5 pmole/106 nuclei obtained by Iborra et al. (2001) for HeLa nuclei.
To ensure first that the method of nuclear isolation was not responsible for the difference in results, we repeated the experiments using CHO cell nuclei isolated by the method of Iborra et al. (2001). Protein synthesis also was measured by their procedure (see Materials and Methods). Nevertheless, as shown in Table 1 ▶, the rate of nuclear protein synthesis on an absolute level remained at only one-tenth of what Iborra et al. reported. As a percentage of total cell translation, nuclear protein synthesis in our experiments was even lower, amounting to only 0.3%, compared with the 9%–15% found by Iborra et al. (2001). This was due to the fact that protein synthesis in permeabilized CHO cells prepared and assayed by our methods was about fivefold greater than what they reported for HeLa cells. It should be noted that the CHO cell nuclei prepared by either our method or that of Iborra et al. were highly purified. Thus, based on marker enzymes, contamination of the purified nuclei by either cytoplasm or mitochondria was undetectable (Table 1 ▶). Moreover, staining of the CHO cell nuclei with Azure C, and microscopic examination, revealed no cytoplasmic adherence (data not shown). Based on these data, it appears that isolated CHO cell nuclei either are incapable of translation or do so at an extremely low level.
TABLE 1.
Protein synthesis and marker enzymes in CHO cell nuclei
| Activities (106 cells or nuclei) | |||
| Total cell | Nuclei | Percent nuclear | |
| Protein synthesisa | 210 | 0.5 | 0.3 |
| Lactate dehydrogenaseb | 600 | <0.03 | <0.01 |
| Gly-3-P dehydrogenaseb | 580 | <0.04 | <0.01 |
| Citrate synthaseb | 19.7 | <0.03 | <0.2 |
| Cytochrome C oxidaseb | 2.6 | <0.01 | <0.4 |
Activities were measured in permeabilized cells and in isolated nuclei as described in Materials and Methods.
apmole incorporated per 20 min (average of five amino acids).
bμmole per min.
(CHO) Chinese hamster ovary.
In view of the discrepancy between our results with isolated CHO cell nuclei and those reported by Iborra et al. (2001) with nuclei prepared from HeLa cells, we next reexamined translation in HeLa cell nuclei. The nuclei were prepared as described by Iborra et al. and protein synthesis was measured using their procedure. Interestingly, under these conditions, as shown in Table 2 ▶, protein synthesis proceeded at 5 pmole/20 min/106 nuclei, a value identical to that reported by Iborra et al. (2001). However, on a percentage basis, compared with the whole cell, nuclear translation amounted to only ∼3%, less than the 9%–15% reported by Iborra et al., but again due to the much higher level of protein synthesis in the permeabilized HeLa cells prepared by us.
TABLE 2.
Protein synthesis and marker enzymes in HeLa cell nuclei prepared by two methods
| Activities (106 cells or nuclei) | |||||
| Total cell | Iborra et al. nuclei | Percent nuclear | Purified nuclei | Percent nuclear | |
| Protein synthesisa | 180 | 5.0 | 3.3 | 0.01 | <0.01 |
| Lactate dehydrogenaseb | 900 | 5.1 | 0.6 | <0.02 | <0.01 |
| Gly-3-P dehydrogenaseb | 640 | 5.4 | 0.8 | <0.04 | <0.01 |
| Citrate synthaseb | 19 | 42 | — | <0.01 | — |
| Cytochrome C oxidaseb | 1.4 | 5.3 | — | <0.01 | — |
Activities were measured in permeabilized cells and in isolated nuclei prepared by the method of Iborra et al. (2001) and by a modification of that method as described in Materials and Methods. The higher level of mitochondrial enzymes in nuclei compared with the total cell is due to the inhibition of these activities in cell extracts.
apmole incorporated per 20 min (average of five amino acids).
bμmole per min.
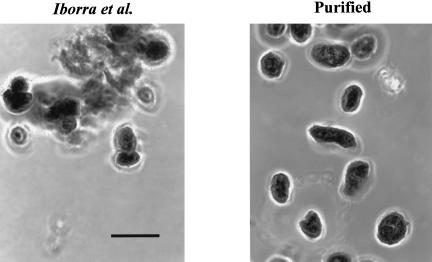
Given these results, we examined the purity of the HeLa cell nuclei, and it was evident that they were significantly contaminated with cytoplasm and mitochondria (Table 2 ▶). All of the marker enzymes assayed were easily detectable in the nuclear preparation. Likewise, microscopy after staining with Azure C demonstrated a considerable level of cytoplasmic adherence and consequent nuclear clumping (Fig. 1 ▶). Iborra et al. (2001) also reported that their preparations of isolated HeLa cell nuclei were only ∼95% pure. Thus, from these observations it appears that HeLa cell nuclei are more difficult to purify than those from CHO cells. Nevertheless, these data raised the obvious question of whether the cytoplasmic contamination was responsible for the apparent nuclear protein synthesis reported by Iborra et al. (2001).
FIGURE 1.
Microscopy of isolated nuclei. Isolated HeLa cell nuclei, prepared either by the method of Iborra et al. (2001) or by the modified procedure, were mixed with an equal volume of 0.5% Azure C in 0.25 M sucrose (Busch 1967) and examined by phase-contrast microscopy. The scale bar is 10 μm.
To address this point, the HeLa cell nuclei were prepared by a slight modification of the procedure of Iborra et al. (2001) to increase their purity (see Materials and Methods). The modifications included the presence of CaCl2 in the homogenization medium to prevent nuclear clumping, one additional homogenization and wash, and centrifugation through 15% rather than 10% glycerol. These minor modifications resulted in a highly purified nuclear preparation that was devoid of cytoplasmic or mitochondrial contamination, but which also displayed essentially no protein synthesis (Table 2 ▶ and Fig. 1 ▶).
To ensure that the alterations of the nuclear purification procedure did not result in damage, we measured transcription in the various nuclear preparations. As shown in Table 3 ▶, there was no significant difference in the level of DNA-dependent RNA synthesis in the purified HeLa cell nuclei compared with the original preparation. Thus, based on this criterion, the highly purified nuclei appear to be functional. These data strongly indicate that the apparent protein synthesis by HeLa cell nuclei was due to cytoplasmic contamination. CHO cell nuclei also display a high level of transcription. Inasmuch as this preparation was made by the unmodified procedure of Iborra et al. (2001), its lack of translation activity could not be due to additional purification.
TABLE 3.
Transcription in various nuclear preparations
| UTP incorporation (pmole/106 nuclei/10 min) | |||
| CHO | HeLa (Iborra et al. 2001) | HeLa (purified) | |
| Untreated | 32.2 | 12.6 | 11.0 |
| Pretreatment with DNase | 3.3 | 2.8 | 0.6 |
| Product treated with RNase | 1.6 | 1.0 | 0.4 |
Incorporation of [α′-32P] UTP into RNA using isolated nuclei preparations was carried out as described in Materials and Methods. DNase treatment was at 28°C for 30 min prior to addition of NTPs using 5 units of RNase-free DNase. RNase treatment was at 28°C for 30 min after completion of synthesis using 100 μg/ml RNase A. (CHO) Chinese hamster ovary.
DISCUSSION
The data presented here raise important questions about the existence and level of nuclear protein synthesis. It should be stated at the outset that we cannot eliminate the possibility that a low level of translation might occur in mammalian nuclei. However, based on our data, nuclear translation, if it exists at all, is unlikely to be at the level of 10%–15% of the whole cell as reported by Iborra et al. (2001). This conclusion also would be in keeping with the low level of aminoacyl-tRNA synthetases previously observed to be present in mammalian nuclei (Nathanson and Deutscher 2000). Thus, given these conflicting reports, it is clear that additional work will be needed to unambiguously demonstrate that nuclear protein synthesis exists.
Iborra et al. (2001) reported that their isolated HeLa cell nuclear preparations were >95% free of cytoplasmic ribosomes, on the basis of standard stereological procedures. However, a ∼5% contamination by cytoplasm is significant, and, as we suggest, was likely responsible for the apparent protein synthesis by isolated nuclei observed by these workers. Iborra et al. (2001) also reported that no mitochondrial signal was observed in their isolated nuclei and that the nuclear periphery was unlabeled after protein synthesis. It is not clear why these microscopic observations differ from our biochemical measurements with regard to mitochondrial and cytoplasmic contamination. One possibility is that the additional washes used in preparing the nuclei for microscopy following translation may have served to remove the contamination. Further work will be needed to reconcile all of these observations.
In addition to their study of isolated nuclei, Iborra et al. (2001) provided evidence for nuclear translation using a permeabilized cell system. However, we have concerns about those studies as well. In earlier work from our laboratory, we showed that saponin permeabilization is a very sensitive procedure and that it is relatively easy to overpermeabilize the cells, resulting in considerable damage to internal membranes and decreased protein synthesis (Stapulionis and Deutscher 1998). Inasmuch as the overall cellular translation reported by Iborra et al. (2001) was approximately one-quarter of that obtained by our procedures, we suspect that their permeabilized cells may have sustained considerable damage. This conclusion is reinforced by the fact that they were also able to use biotin-lysine-tRNA and BODIPY-lysine-tRNA as precursors for protein synthesis. Work from our laboratory showed previously that the mammalian translation system is highly organized (Negrutskii et al. 1994), and that aminoacyl-tRNA is channeled for protein synthesis both in vivo (Negrutskii and Deutscher 1991) and in permeabilized cells (Negrutskii and Deutscher 1992; Negrutskii et al. 1994). Consequently, in undamaged cells, exogenous aminoacyl-tRNA is a very poor substrate for protein synthesis. If, as these data indicate, the permeabilized cells used by Iborra et al. (2001) were damaged, this might result in rapid entry into the nucleus of proteins actually synthesized in the cytoplasm, and might explain the apparent nuclear events observed by these workers.
If nuclear translation does not exist, how then does one explain the presence of nuclear translation components and of nuclear NMD? It may be that assembly of the organized cytoplasmic translation machinery begins in the nucleus, and that components of the apparatus are exported together to the cytoplasm. If so, a small steady-state pool of translation components should always be present in the nucleus (e.g., Fortes et al. 2000; Ishigaki et al. 2001). Also, a low level of certain translation components may be needed for nuclear processes, for example, proofreading of tRNAs (Lund and Dahlberg 1998; Sarkar et al. 1999) or tRNA export (Lund and Dahlberg 1998; Grosshans et al. 2000).
Nuclear-associated NMD may be a consequence of ribosomes associated with the outer nuclear membrane (Kim et al. 2001; Lykke-Anderson et al. 2001; Wang et al. 2002) that are difficult to remove, rather than of intranuclear translation. In fact, as shown in Table 2 ▶, the level of apparent nuclear protein synthesis was four- to fivefold higher than the level of cytoplasmic contamination as determined by marker enzymes. Likewise, the cytoplasmic adherence was revealed by Azure C staining (Fig. 1 ▶), which stains RNA and ribonucleoproteins (Busch 1967). Both of these observations indicate that even a low level of contamination of nuclear fractions by cytoplasm may lead to an enriched contamination by nuclear-bound cytoplasmic ribosomes. These ribosomes may only be removed by more rigorous purification of nuclei. Thus, whereas internal nuclear translation may not exist, nuclear-associated translation is a distinct possibility.
MATERIALS AND METHODS
CHO (CLR-1781) cells and HeLa (CCL-2) cells were obtained from the American Type Culture Collection. A mixture of five 3H-labeled amino acids (leucine, lysine, phenylalanine, proline, and tyrosine) was purchased from Amersham. Rabbit liver tRNA was prepared as described (Negrutskii et al. 1994). Unlabeled amino acids, ATP, GTP, creatine phosphokinase, phosphocreatine, saponin, aminoacyl-tRNA synthetases, trypsin inhibitor, and trypan blue were obtained from Sigma. Cell culture reagents were from HyClone.
CHO cells were cultured as described previously (Nathanson and Deutscher 2000). Hela cells were maintained as a monolayer in Dulbecco’s Modified Eagle’s Medium containing 4.5 g/L glucose and supplemented with 10% fetal bovine serum. Cells were cultured in Nunc flasks at 37°C in air containing 5% CO2 and were transferred every 3 to 4 days. Harvesting and permeabilization of cells were performed as described in Stapulionis et al. (1997), except that HeLa cells were incubated with saponin for 8 min. Nuclei from CHO cells were isolated as described in Iborra et al. (2001) or in Nathanson and Deutscher (2000). Nuclei from HeLa cells also were isolated by the method of Iborra et al. (2001). However, to obtain highly purified nuclei from HeLa cells, we slightly modified this method as follows: After incubation in PB*-diluted at 37°C, cells were homogenized in PB*-diluted containing 2 mM CaCl2 (five to seven strokes). After addition of 0.25% Triton X-100, the homogenate was spun for 2 min at 500 g and the pellet was resuspended in 50 mL of PB*-diluted containing 2 mM CaCl2 and homogenized again (five to seven strokes). After addition of 0.25% Triton X-100, the final homogenate was spun through PB* containing 15% instead of 10% glycerol for 5 min at 250 g.
Transcription was assayed as described in Sambrook and Russell (2001) with minor modifications. The final concentration of all ribonucleotides in the reaction mixture was 0.9 mM. After 10 min of incubation, reactions were stopped by adding 20% trichloracetic acid (TCA) containing 0.9% Na pyrophosphate, and acid-precipitable material was collected on Whatman GF/C filters. Citrate synthase and cytochrome c oxidase were assayed as described in Jarreta et al. (2000). Activities of lactate dehydrogenase and glyceraldehyde-3-phosphate dehydrogenase were determined as described in the Worthington Biochemical Corporation Enzyme Manual (1972). The rate of protein synthesis in isolated nuclei was measured as in Iborra et al. (2001) except that amino acids were present at 250 μM and five were radioactively labeled. Protein synthesis in permeabilized cells was carried out as described in Stapulionis et al. (1997) except that creatine phosphokinase was present at 20 U/mL and GTP was at 0.25 mM.
Acknowledgments
This work was supported by grant GM16317 from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2990203.
REFERENCES
- Brogna, S. 2001. Pre-mRNA processing: Insights from nonsense. Curr. Biol. 11:R838–R841. [DOI] [PubMed] [Google Scholar]
- Busch, H. 1967. Isolation and purification of nucleoli. In: Methods in enzymology (eds. L. Grossman and K. Moldave), Vol. 12, Part A, pp. 448–464. Academic Press, NY.
- Dostie, J., Lejbkowicz, F., and Sonenberg, N. 2000. Nuclear eukaryotic initiation factor 4E (eIF4E) colocalizes with splicing factors in speckles. J. Cell Biol. 148:239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes, P., Inada, T., Preiss, T., Hentze, M.W., Mattaj, I.W., and Sachs, A.B. 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6:191–196. [PubMed] [Google Scholar]
- Grosshans, H., Hurt, E., and Simos, G. 2000. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes & Dev. 14:830–840. [PMC free article] [PubMed] [Google Scholar]
- Iborra, F.J., Jackson, D.A., and Cook, P.R. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293:1139–1142. [DOI] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., and Maquat, L.E. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106:607–617. [DOI] [PubMed] [Google Scholar]
- Jarreta, D., Orus, J., Barrientos, A., Miró, O., Roig, E., Heras, M., Moraes, C.T., Cardellach, F., and Casademont, J. 2000. Mitochondrial function in heart muscle from patients with idiopathic dilated cardiomyopathy. Cardiovasc. Res. 45:860–865. [DOI] [PubMed] [Google Scholar]
- Kim, V.N., Kataoka, N., and Dreyfuss, G. 2001. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science 293:1832–1836. [DOI] [PubMed] [Google Scholar]
- Lund, E. and Dahlberg, J.E. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282:2082–2085. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.-D., and Steitz, J.A. 2001. Communication of the position of exon-exon junction to the mRNA surveillance machinery by the protein RNPS1. Science 293:1836–1839. [DOI] [PubMed] [Google Scholar]
- Nathanson, L. and Deutscher, M.P. 2000. Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J. Biol. Chem. 275:31559–31562. [DOI] [PubMed] [Google Scholar]
- Negrutskii B.S., Deutscher M.P. 1991. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. 88:4991–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———1992. A sequestered pool of aminoacyl-tRNA in mammalian cells. Proc. Natl. Acad. Sci. 89:3601–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii, B.S., Stapulionis, R., and Deutscher, M.P. 1994. Supramolecular organization of the mammalian translation system. Proc. Natl. Acad. Sci. 91:964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russel, D.W. 2001. Molecular cloning. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sanders, J., Brandsma, M., Janssen, G.M.C., Dijk, J., and Möller, V. 1996. Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of elongation factor-1βδγ in the endoplasmic reticulum. J. Cell Sci. 109:1113–1117. [DOI] [PubMed] [Google Scholar]
- Sarkar, S., Azad, A.K., and Hopper, A.K. 1999. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 96:14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapulionis, R. and Deutscher, M.P. 1998. Permeabilized mammalian cells as a system for protein synthesis. In: Protein synthesis. Methods and protocols (ed. R. Martin), pp. 23–33. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Stapulionis, R., Kolli, S., and Deutscher, M.P. 1997. Efficient mammalian protein synthesis requires an intact F-actin system. J. Biol. Chem. 272:24980–24986. [DOI] [PubMed] [Google Scholar]
- Wang, J., Vock, V.M., Li, S., Olivas, O.R., and Wilkinson, M.F. 2002. A quality control pathway that down-regulates aberrant T-cell receptor (TCR) transcripts by a mechanism requiring UPF2 and translation. J. Biol. Chem. 277:18489–18493. [DOI] [PubMed] [Google Scholar]
- Worthington Biochemical Corporation. 1972. Worthington enzyme manual. Freehold, NJ.