Abstract
A stable 100-kD complex from mitochondria of Leishmania tarentolae containing two RNA-binding proteins, Ltp26 and Ltp28, was identified by cross-linking to unpaired 4-thiouridine nucleotides in a partially duplex RNA substrate. The genes were cloned and expressed and the complex was reconstituted from recombinant proteins in the absence of RNA or additional factors. The Ltp26 and Ltp28 proteins are homologs of gBP27 and gBP29 from Crithidia fasciculata and gBP25 and gBP21 from Trypanosoma brucei, respectively. The purified Ltp26/Ltp28 complex, the individual recombinant proteins, and the reconstituted complex are each capable of catalyzing the annealing of complementary RNAs, as was previously shown for gBP21 from T. brucei. A high-molecular-weight RNP complex consisting of the Ltp26/Ltp28 complex and several 55–60-kD proteins together with guide RNA could be purified from mitochondrial extract of L. tarentolae transfected with Ltp28-TAP. This complex also interacted in a less stable manner with the RNA ligase-containing L-complex and with the 3′ TUTase. The Ltp26/Ltp28 RNP complex is a candidate for catalyzing the annealing of guide RNA and pre-edited mRNA in the initial step of RNA editing.
Keywords: RNA editing, RNA annealing, RNA binding proteins, trypanosomes
INTRODUCTION
The “enzyme cascade” model (Blum et al. 1990) of U-insertion/deletion RNA editing that occurs in mitochondria of kinetoplastid protozoa predicts the involvement of at least four enzymatic activities (endoribonuclease, 3′ TUTase, RNA ligase, 3′-5′ exonuclease) and most likely several structural or RNA-binding proteins within large ribonucleoprotein complexes (Pollard et al. 1992; Peris et al. 1994;Corell et al. 1996). The only enzymatic components isolated to date and shown to be involved in editing by gene knockout or RNAi are the larger RNA ligase (REL1; Schnaufer et al. 2001), a putative RNA helicase which may be required for the 3′ to 5′ processivity of editing or for gRNA-mRNA hybrid unwinding (Missel et al. 1997), and a 3′ terminal uridylyltransferase (TUTase; Aphasizhev et al. 2002). TbMP81 (Drozdz et al. 2002) and band III protein (Huang et al. 2002) are likely to be structural components of core RNA editing complexes in Trypanosoma brucei, as also shown by gene knockout studies. A smaller RNA ligase (REL2) and several unidentified proteins have also been isolated as components of a large “editing complex” (Panigrahi et al. 2001). A gRNA-directed mitochondrial endoribonuclease has been partially purified from T. brucei (Seiwert et al. 1996; Rusche et al. 1997) and a 3′-5′ U-specific mitochondrial exonuclease has been partially purified from Leishmania tarentolae (Aphasizhev and Simpson 2001). The REAP-1 mitochondrial protein from T. brucei was isolated as a component of high-molecular-weight complexes, and an inhibition of in vitro editing by anti-REAP-1 antibodies was reported (Madison-Antenucci et al. 1998), but the specific role of this protein remains to be identified.
One approach for the detection of components of the editing machinery has been to search for gRNA-binding or mRNA-binding proteins. This approach has had varied success. For example, in L. tarentolae, three proteins, p18, p51, and p110, were identified by RNA binding and then discovered to be the mitochondrial metabolic enzymes ATPase subunit b, aldehyde dehydrogenase, and glutamate dehydrogenase, respectively (Bringaud et al. 1995, 1997), and a double knockout of the glutamate dehydrogenase gene had no effect on editing (Estévez et al. 1999). TBRGG1, a mitochondrial protein from T. brucei that binds poly[U] and gRNAs, was cloned by homology to nucleolin and shown to cosediment with in vitro editing activity, but no role in editing has been ascribed to this protein (Leegwater et al. 1995; Vanhamme et al. 1998). RBP16 from T. brucei which binds in vitro to the gRNA 3′ oligo[U] tail contains a cold-shock domain, but its role is uncertain (Hayman and Read 1999; Pelletier et al. 2000).
The RNA-binding mitochondrial protein gBP21, from T. brucei, has been extensively studied (Köller et al. 1994; Allen et al. 1998; Lambert et al. 1999; Muller et al. 2001) and has several properties that make it a very likely candidate for indirect involvement in editing. These involve high-affinity binding to gRNA and coimmunoprecipitation with in vitro editing activity. An RNA annealing activity was also demonstrated, and it was proposed that this protein played a role in the initial interaction of mRNA and the cognate gRNA (Muller et al. 2001). However, a targeted knockout of the corresponding gene had no apparent affect on editing in vivo (Lambert et al. 1999). In Crithidia fasciculata, gBP29, an ortholog of gBP21, and a second related RNA-binding protein, gBP27, were cloned. In addition, an ortholog of gPB27 from T. brucei, gBP25, was identified (Leegwater et al. 1995; Blom et al. 2001). These proteins from C. fasciculata coimmunoprecipitated and were present in the form of variable-sized high-molecular-weight complexes in the mitochondrial extract.
Here we present the isolation and characterization of a stable heterotetrameric 100-kD complex of two RNA-binding mitochondrial proteins, Ltp28 and Ltp26 from L. tarentolae. The Ltp28 protein is an ortholog of the T. brucei gBP21, and the Ltp26 protein is an ortholog of the T. brucei gBP25. Both the complex and each recombinant protein catalyze the annealing of RNA. The complex is present in the extract as a high-molecular-weight RNP complex, which interacts with the two mitochondrial RNA ligases and the 3′ TUTase.
RESULTS
Crosslinking of protein to unpaired 4-thiouridine nucleotides in a model substrate RNA
As shown in 1986 (Favre et al.), 4-thiouridine forms efficient and stable crosslinks when RNA and protein molecules are positioned in close proximity and the crosslinking is activated by long-wavelength UV. RNA-I, shown in Figure 1A ▶, was radioactively labeled and 4-thioU residues site-specifically inserted during T7 transcription by incorporation of a mixture of [α32P]UTP and 4-thiouridine. The partially complementary RNA-II which hybridized downstream and upstream of the 4-thio Us was also produced by T7 transcription and subsequent annealing of the two RNAs, yielding the substrate RNA shown in Figure 1A ▶, which emulates an mRNA-gRNA substrate with four unpaired U (and thio-U) residues at the initial U-deletion site. It should be noted that we have not tested this substrate for activity in an RT-PCR editing assay (Byrne et al. 1996) or a precleaved editing assay (Igo et al. 2000).
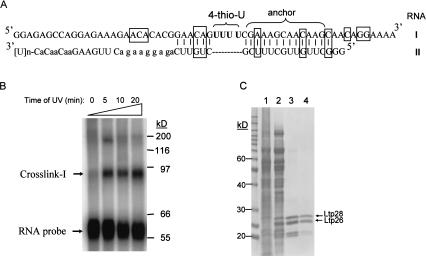
FIGURE 1.
Isolation of Ltp26/Ltp28 100-kD complex from L. tarentolae mitochondria. (A) The model mRNA editing substrate contains four 4-thiouridines (in bold) in the first editing site. The anchor region is labeled, and the locations of the original U nucleotides which were mutated to non-U nucleotides outside the editing site are indicated by boxes. In five cases, the complementary nucleotides were also mutated to preserve base pairing. (B) The annealed labeled mRNA-gRNA substrate was incubated with mitochondrial extract and UV-irradiated for 0–20 min. The extract was fractionated on a 12% SDS gel and the gel was exposed to a Phosphoimager plate. RNA-protein crosslinks I and II are indicated. (C) Crosslink I in the S100 mitochondrial extract (lane 1) was followed through ammonium sulfate precipitation (lane 2), heparin affinity chromatography (lane 3), and Superose 12 size-fractionation (lane 4; see Materials and Methods). Active fractions were analyzed on a 12% SDS gel and stained with Coomassie Blue.
The labeled RNA substrate was incubated with mitochondrial extract which had been preincubated with 100-fold mass excess of nonspecific RNA, and then subjected to 300-nm UV irradiation. One major crosslink was observed on an SDS gel (Fig. 1B ▶). The formation of crosslink I was dependent on the annealing of RNA-II and was inhibited more than 90% by a twofold mass excess of heparin or a 3–5-fold excess of a poly[U] homopolymer. Poly[C] only achieved this level of inhibition at 10-fold excess, and poly[A] and nonspecific RNA did not inhibit the formation of the crosslink until more than a 500-fold excess (not shown).
Isolation of Ltp26 and Ltp28 proteins and cloning of genes
Crosslink I was preparatively gel-isolated and digested with RNase, and the labeled proteins separated in an SDS gel (data not shown). Label transfer after RNase digestion was observed mostly to the upper 28 kD band (data not shown).
Based on this information, a purification method was developed using the generation of crosslink I as an analytical assay. As described in Materials and Methods, a fractionation method employing a 40%–55% ammonium sulfate cut of the mitochondrial extract, followed by heparin affinity chromatography, and finally by size fractionation in a Superose 12 column led to the isolation of a fraction that was active in the generation of crosslink I. A single 100-kD protein peak was seen in the final Superose 12 gel fractionation (see Fig. 5 ▶ below). Interestingly, an enrichment for two major bands of 26 and 28 kD and two minor bands of 21 and 23 kD occurred during this fractionation, especially after the heparin affinity chromatography (Fig. 1C ▶).
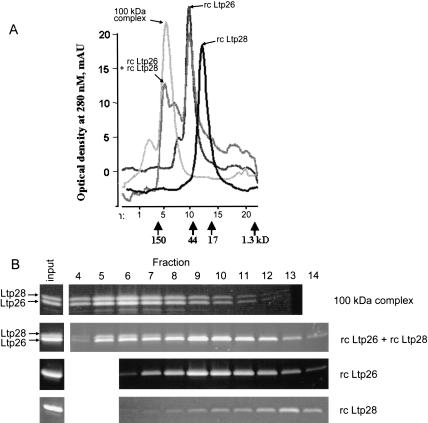
FIGURE 5.
Reconstitution of Ltp26/Ltp28 100-kD complex. (A) Superose 12 gel filtration profiles of purified Ltp26/Ltp28 complex, of the material reassembled from the purified recombinant Ltp26 and Ltp28 proteins, and of the individual proteins. Positions of molecular weight markers are indicated. (B) Protein composition of native and reconstructed complexes. Fractions were collected from the same starting point in chromatographic runs (above) and separated on 10%–20% SDS gradient gels which were stained with Sypro Ruby.
Both major bands were subjected to N-terminal and internal protein microsequencing, and N-termini were also determined for the minor 21- and 23-kD bands. The Ltp28 protein was blocked at the N-terminus, but the Ltp26 protein did yield an N-terminal sequence, which was used to establish the site of cleavage of the mitochondrial signal sequence (see below). Degenerate oligonucleotides based on the peptide sequences were used to amplify genomic fragments, which were employed as probes to clone the genes from a subgenomic library. The genes were expressed in E. coli, and the recombinant proteins were used to generate polyclonal antisera.
The amino acid sequences of the Ltp28 and Ltp26 genes (Fig. 2 ▶, peptides indicated by lines) contained the peptides obtained from the microsequencing of p23 and p21, respectively, confirming that these represented proteolytic fragments. An N-terminal mitochondrial targeting sequence in the Ltp28 gene was recognized by the SignalP and Mitoprot programs with high confidence levels. The cleavage site of the N-terminal targeting sequence in Ltp26 was determined by the N-terminal amino acid sequence of the mature protein (Fig. 2A, arrow ▶). The 46-aa signal peptide in Ltp26 probably accounts for the observed difference in the electrophoretic mobility of Ltp28 and Ltp26, given that the calculated molecular weight of the Ltp28 preprotein is only 0.25 kD larger that Ltp26.
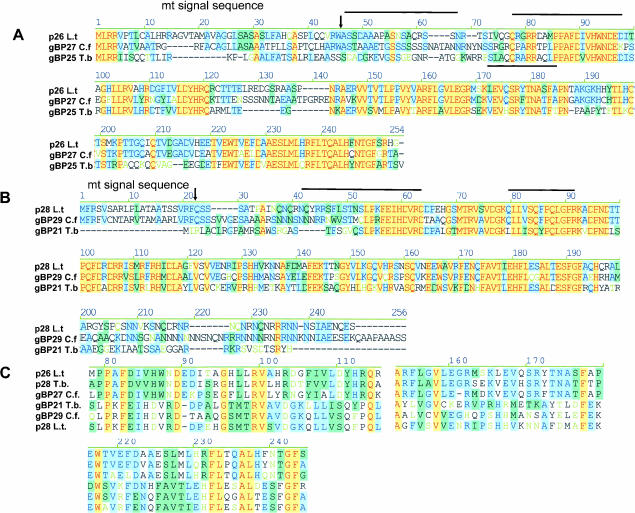
FIGURE 2.
Protein sequence alignments of gRNA-binding proteins. (A) The L. tarentolae Ltp26 amino acid sequence aligned with gBP25 from T. brucei and gBP27 from C. fasciculata (Blom et al. 2001). (B) The L. tarentolae Ltp28 amino acid sequence aligned with gBP29 from C. fasciculata (Blom et al. 2001) and gBP21 from T. brucei (Koller et al. 1997). (C) Motifs conserved between the p26 and p28 homologs from all three species. The alignments above were obtained using AlignX in the Vector NTI Suite (Informax). The extent of nucleotide similarity is color-coded: blue on cyan denotes a consensus residue from a block of similar residues at a given location; black denotes nonhomologous residues; black on green denotes a consensus residue from a single conservative residue at a given location; red on yellow denotes a consensus residue from completely conservative residues at a given position. The predicted mitochondrial signal sequence cleavage sites in A and B are indicated by arrows, and the internal peptides obtained by microsequencing are indicated with lines.
A BLAST search revealed a strong homology of Ltp28 with T. brucei gBP21 (Koller et al. 1997) and C. fasciculata gBP29 (Blom et al. 2001). The L. tarentolae Ltp26 sequence showed strong homology with C. fasciculata gBP27 (Blom et al. 2001). The L. tarentolae Ltp26 protein also produced a significant hit with an EST clone (AI881040) in the T. brucei Genome Database. This sequence was PCR-amplified and used to clone the ortholog from T. brucei, which proved to be gBP25, a gene previously cloned from T. brucei (Blom et al. 2001).
An alignment of these RNA-binding proteins from T. brucei, C. fasciculata, and the Ltp26 and Ltp28 proteins from L. tarentolae is shown in Figure 2A,B ▶. This analysis extends the findings of Blom et al. (2001) with sequences from three organisms and indicates several highly conserved motifs in each ortholog group. A limited conservation of three motifs can be seen in the alignments of all six sequences in Figure 2C ▶. No functional motifs were recognized by Prosite or Profile analysis, including any known RNA-binding motifs.
Mitochondrial localization of the Ltp26/Ltp28 complex
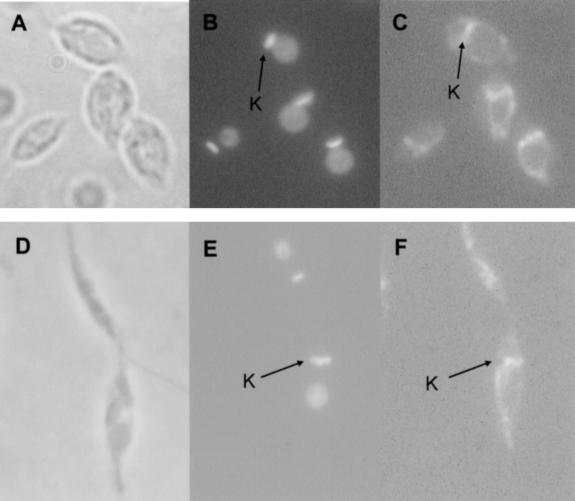
In situ immunofluorescence analysis using the antiserum against the purified 100-kD complex, which recognizes both proteins, showed that the proteins are confined to the circular mitochondrion in log-phase cells (Fig. 3A–C ▶) and the asymmetric mitochondrion in stationary-phase cells (Fig. 3D–F ▶; Simpson and Kretzer 1997). Interestingly, there was a predominant localization in the kinetoplast region (K) of the mitochondrion in addition to a diffuse localization throughout the tubular mitochondrial structure (Fig. 3C,F ▶).
FIGURE 3.
Mitochondrial localization of Ltp26 and Ltp28 proteins. Log-phase (A–C) or stationary-phase (D,E) cells were subjected to indirect immunofluorescence using an antiserum against the purified Ltp26/Ltp28 complex. A,D: phase contrast, 1000×. B,E: Same cells stained with DAPI. Location of kinetoplast (K) indicated by arrows. C,F: Same cells stained with Ltp26/Ltp28 antiserum. Location of K indicated by arrows.
Ltp26 and Ltp28 are components of a stable 100-kD complex
As mentioned above and shown in Figure 5A ▶ below, the purification procedure yielded a single peak in a Superose 12 fractionation with an apparent molecular weight of 100 kD. This peak contained both Ltp26 and Ltp28 in approximately equimolar ratio. This purified complex was used to generate polyclonal antiserum, which showed reactivity against both Ltp26 and Ltp28 with approximately equal affinities. The equal intensity of the Ltp26 and Ltp28 bands in gels stained with Coomassie Blue (Fig. 1C ▶, lane 4) and Sypro Ruby (see Fig. 5B ▶ below) suggested that the proteins are present at a 1:1 ratio. The yields of tryptic peptides from the two bands for protein sequencing were also close to this ratio (Harvard Microchemical Facility, pers. comm.). The 1:1 ratio and the 100-kD molecular weight of the complex argue for an α2β2 organization.
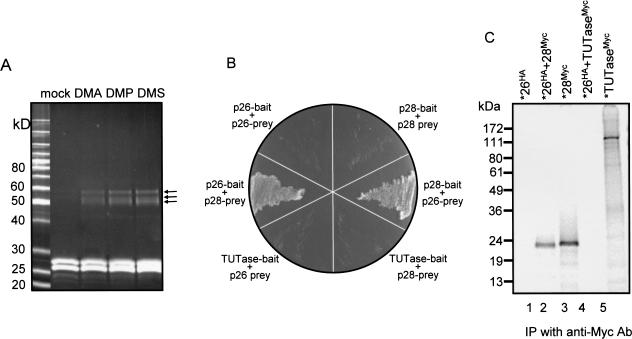
To investigate the topography of protein–protein interactions within the Ltp26/Ltp28 complex, chemical crosslinking was performed, a fraction of which had undergone an additional purification step on Phenyl Superose (see Materials and Methods). Incubation of the complex with homobifunctional crosslinking reagents with different chain lengths produced three closely migrating crosslinks with molecular weights that are commensurate with all pairwise combinations, Ltp26-Ltp28, Ltp26-Ltp26, and Ltp28-Ltp28 (Fig. 4A ▶). The low efficiency and the absence of a crosslink which would correspond to three or four subunits together may be due to the low lysine content in these proteins (four in Ltp26 and seven in Ltp28).
FIGURE 4.
Interaction of Ltp26 and Ltp28 proteins. (A) Chemical crosslinking of Ltp26/Ltp28 complex. Purified Ltp26/Ltp28 complex was incubated with homobifunctional protein–protein crosslinking reagents, and the products were separated on an SDS gel which was stained with Sypro Ruby. (B) Yeast two-hybrid analysis of the interaction between Ltp26 and Ltp28. Samples of cotransformed yeast grown on medium lacking tryptophan, leucine, and histidine. (C) Coimmunoprecipitation of epitope-tagged proteins synthesized in vitro in a coupled transcription/translation system. All IPs were with a monoclonal anti-Myc antibody. Lane 1: 35S-labeled Ltp26 tagged with HA. Lane 2: 35S-labeled Ltp26 tagged with HA mixed with unlabeled Ltp28 tagged with Myc. Lane 3: 35S-labeled Ltp28 tagged with Myc. Lane 4: 35S-labeled Ltp26 tagged with HA mixed with unlabeled TUTase tagged with Myc. Lane 5: 35S-labeled TUTase tagged with Myc.
No endogenous RNA component could be detected by phenol-extracting 50 μg of the purified Ltp26/Ltp28 complex, ethanol-precipitating, and incubating the pellet with RNA ligase and [α32P]pCp or with poly[A] polymerase and [α32P]ATP (not shown). This is not surprising however, because the complex was subjected to heparin affinity chromatography and high salt conditions during the isolation.
Yeast two-hybrid and coimmunoprecipitation evidence of an interaction of Ltp26 and Ltp28
To assess the relative affinity of protein–protein interactions within the Ltp26/Ltp28 complex, yeast two-hybrid analysis (Chien et al. 1991) was performed by cotransfecting a yeast strain deficient for several nutritional markers with Ltp28 and Ltp26 fusion constructs followed by selection on the appropriate media (see Materials and Methods). The survival of the cells on selective media depended on the presence of both Ltp26 fused with the gal4 binding domain (= bait) and Ltp28 fused with the gal4 activation domain (= prey), or vice-versa (Fig. 4B ▶). No interaction of Ltp26 with Ltp26 or Ltp28 with Ltp28 could be detected under the same stringency conditions. In addition, cotransformants of yeast with a fusion construct of the L. tarentolae 3′ TUTase and a fusion construct of Ltp26 or Ltp28 did not grow on selective medium, possibly suggesting a lack of direct interaction between Ltp26 or Ltp28 and TUTase, although other explanations are of course possible for this negative result in a heterologous system.
Interactions were also assayed by coimmunoprecipitation. S35-labeled HA-tagged or Myc-tagged proteins were synthesized by in vitro transcription-translation and then mixed, and physical interactions were assayed by coimmunoprecipitation using an anti-Myc monoclonal antibody (Fig. 4C ▶). The anti-Myc antibody pulled down Myc-tagged S35-labeled Ltp28 (lane 3) but only pulled down HA-tagged S35-labeled Ltp26 in the presence of unlabeled Myc-tagged Ltp28 (lanes 1 and 2), thus confirming the in vivo yeast two-hybrid results. In addition, this procedure could not demonstrate an association between TUTase and Ltp26 (lanes 4 and 5), which is consistent with the negative yeast two-hybrid results (Fig. 4B ▶). The reciprocal pull down of Ltp28 by Ltp26 was not technically possible as in vitro-synthesized Ltp28 (unlike TUTase or Ltp26) partially precipitated out of solution during the immunoprecipitation. Together these data provide evidence of a strong interaction of Ltp26 and Ltp28.
In vitro reconstitution of Ltp26/Ltp28 complex with recombinant Ltp26 and Ltp28 proteins
The Ltp26 and Ltp28 proteins were expressed in E. coli with His-tags at the N-termini of the mature proteins. The Ltp26 protein was purified under native conditions by metal and heparin affinity chromatography to more than 95% purity, as estimated from Sypro Ruby staining of an SDS gel. The Ltp28 protein formed inclusion bodies in E. coli and was therefore purified by metal affinity chromatography under denaturing conditions; the denatured Ltp28 protein was then refolded while attached to the matrix and eluted (see Materials and Methods). Subsequent heparin column purification of Ltp28 resulted in a fully soluble preparation of more than 95% purity, as estimated by SDS gel analysis.
The reconstitution of high-molecular-weight complexes by the recombinant proteins after incubation for 16 h at 2 mg/mL protein concentration was monitored by Superose 12 chromatography (Fig. 5A ▶). As mentioned above, the Ltp26/Ltp28 complex purified from the mitochondrial extract migrated as a sharp 100-kD peak. Recombinant Ltp28 migrated as an apparent monomer, which is consistent with previously described properties of the T. brucei ortholog, gBP21 (Koller et al. 1997). Recombinant Ltp26 migrated as an apparent homodimer, with a small shoulder migrating at a possible homotrimer position. The interaction of Ltp26 with itself was not detected in the above yeast two-hybrid assay, suggesting that this interaction may be less stable than the Ltp26/Ltp28 interaction. Incubation of Ltp28 and a two-fold molar excess of Ltp26 yielded a substantial peak comigrating with the purified 100-kD Ltp26/Ltp28 complex in addition to a major dimer peak and a minor trimer peak. The molar excess of Ltp26 was necessary to achieve incorporation of Ltp28 into the tetrameric complex (data not shown). Figure 5B ▶ shows the SDS gel profiles of the proteins from chromatographic fractions 4–14. The stoichiometric ratio of Ltp28:Ltp26 in the reconstituted tetramer peak in fractions 5–8 was approximately 1:2. We attribute this deviation from a 1:1 ratio to cross contamination of the tetramer peak with the overlapping Ltp26 trimer peak. Consistent with this interpretation, the ratio of Ltp26 to Ltp28 in the leading edge of the tetramer peak (fraction 4) is close to 1:1.
RNA-binding properties of the purified and reconstituted Ltp26/Ltp28 complex and the recombinant Ltp26 and Ltp28 proteins
The equilibrium dissociation constants (Kd) for binding single-stranded (denatured RNA-III, Fig. 7 ▶ below) and double-stranded RNA (annealed RNA-III+RNA-V) were determined for the purified native Ltp26/Ltp28 complex, the individual recombinant proteins, and the reconstructed Ltp26/Ltp28 complex. As shown in Table 1 ▶, binding to denatured RNA-III and to annealed RNA-III+RNA-V was approximately the same, with a possible slight preference of the native and reconstituted complexes and the Ltp28 for the double-stranded RNA. However, the Kd values were significantly lower for both the native and the reconstructed complexes, indicating a higher affinity of binding of the complex than either of the component proteins. The Kd for the binding of Ltp28 with single-stranded RNA was approximately 10-fold higher than that reported for the binding of T. brucei gBP21 to single-stranded RNA (Muller et al. 2001), which may reflect a species variation or differences in the recombinant protein preparation. However, the Kd of binding of the Ltp26/Ltp28 complex to single-stranded RNA (9.6 nM) is nearly identical to that of gBP21 (Muller et al. 2001).
FIGURE 7.
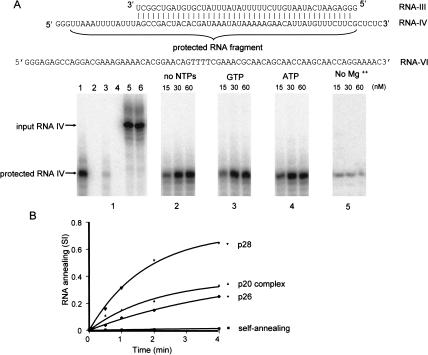
RNA annealing activity of Ltp26/Ltp28 100-kD complex. (A) Specificity and cofactor requirements. Panel 1: lane 1, RNA-III at 2 nM was annealed with 50-fold excess of radioactively labeled RNA-IV by heating and slow cooling prior to addition of T1 nuclease in the absence of added protein. Lane 2, labeled RNA-IV + Ltp26/Ltp28 complex in the absence of complementary RNA followed by T1 nuclease digestion. Lane 3, RNA-III + labeled RNA-IV (2 nM each) in absence of protein, followed by T1 nuclease digestion. Lane 4, labeled RNA-IV + noncomplementary RNA-VI of approximately the same length + Ltp26/Ltp28 complex, followed by T1 nuclease digestion. Lane 5, RNA-III + labeled RNA-IV (2 nM) + Ltp26/Ltp28 complex, no T1 nuclease. Lane 6, labeled RNA-IV, no T1 nuclease. Panel 2: Ltp26/Ltp28 complex was added at increasing concentrations to RNA-III + labeled RNA-IV in absence of added NTPs, followed by T1 nuclease digestion. Panel 3: Same as Panel 2 but with 1mM GTP. Panel 4: Same as Panel 2 but with 1 mM ATP. Panel 5: Same as Panel 2 but without Mg2+. (B) Kinetics of annealing catalyzed by the recombinant p26 and p28 proteins and by the native 100-kD complex at 50 nM . SI, signal intensity of the T1-protected fragment. A control for the kinetics of annealing in the absence of protein is also shown.
TABLE 1.
RNA binding and RNA annealing properties of the Ltp26/Ltp28 100 kDa complex and individual recombinant proteins
| Single-stranded RNA | Double-stranded RNA | ||
| Protein | Kd (nM) | Kd (nM) | Fold annealing stimulation |
| Native complex | 9.6 ± 2 | 2.6 ± 0.3 | 38 |
| Ltp28 | 128 ± 21 | 96 ± 6 | 67 |
| Lpt26 | 225 ± 52 | 259 ± 65 | 21 |
| Reconstituted complex | 24 ± 2 | 4.5 ± 1.2 | N/D |
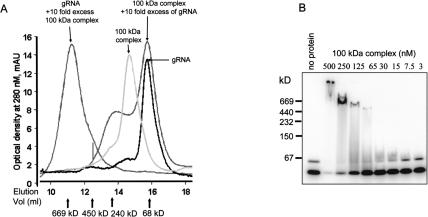
The interaction of the purified native Ltp26/Ltp28 complex with in vitro transcribed gRPS12-I gRNA (see Materials and Methods) was assayed by gel filtration chromatography. At a 10-fold excess of protein, the formation of an ∼700-kD complex was observed, which corresponds to approximately 5–6 100-kD complexes bound to one RNA molecule (Fig. 6A ▶). A 10-fold molar excess of RNA resulted in the formation of an ∼200-kD complex, which may correspond to a single complex bound per RNA molecule. The 100-kD complex alone and the labeled RNA alone are also shown. It should be noted that the use of the T7 transcribed gRNA as substrate in this assay does not imply any specificity of binding for gRNA, but was used since it represents a natural ligand for the complex (see below).
FIGURE 6.
Binding of Ltp26/Ltp28 complex to T7 transcribed gRNA. (A) Binding of Ltp26/Ltp28 100-kD complex to RNA. In vitro transcribed labeled gRPS12-I gRNA was incubated with purified Ltp26/Ltp28 complex in 100 μL of 20 mM HEPES, pH 7.5, 5 mM MgCl2, 150 mM KCl, and separated on a Superose 6 column. The peak on left shows 50 pmole of gRNA and 500 pmole of protein, and the large peak on right 50 pmoles Ltp26/Ltp28 and 500 pmoles gRNA. (B) Radiolabeled gRPS12-I gRNA (20 nM) was incubated with increasing concentrations of Ltp26/Ltp28 complex under the same conditions in 5 μL aliquots and separated on 8%–16% native Tris-glycine gel.
Native gel analysis of increasing amounts of Ltp26/Ltp28 complex mixed with 20 nM labeled gRNA showed a series of discrete higher-molecular-weight protein/RNA complexes (Fig. 6B ▶). This method, however, does not allow determination of accurate molecular weights due to the high isoelectric point (pH 10.1) of the Ltp26/Ltp28 complex, because in the absence of RNA these proteins do not enter the native gel at pH 8.4. The two RNA bands of unequal intensity in the “no protein” control lane probably represent folded and unfolded RNA molecules.
The Ltp26/Ltp28 complex and the recombinant Ltp26 and Ltp28 proteins catalyze the annealing of complementary RNAs
A T1 nuclease protection assay (Portman and Dreyfuss 1994) was used to determine whether the Ltp26/Ltp28 complex catalyzes the annealing of complementary RNAs as has been previously shown for the T. brucei gBP21 homolog of the Ltp28 protein (Muller et al. 2001). The substrate consisted of two partially complementary RNAs (RNA-III and RNA-IV in Fig. 7 ▶). RNA-IV was uniformly labeled during transcription. T1 nuclease cleaves 3′ of G residues only in single-stranded RNA. As shown in Figure 7 ▶, the T1 nuclease-protected fragment of labeled RNA-IV which is annealed to RNA-III is eight nucleotides shorter than the input RNA.
The increase in rate of annealing in the presence of the Ltp26/Ltp28 complex is shown by comparing the protected fragments produced in the presence of added protein to those produced in the absence of protein (Fig. 7, cf. panels 2–4 to panel 1, lane 3 ▶). The annealing of labeled RNA-IV and the partially complementary RNA-III under these conditions in the absence of Ltp26/Ltp28 complex represented approximately 5 % (panel 1, lane 3) of that achieved by incubation in the presence of Ltp26/Ltp28 protein or by heating and slow cooling with a 50-fold excess of RNA-IV (panel 1, lane 1). A control experiment showed that annealing of the labeled RNA-IV to the noncomplementary RNA-VI did not produce a T1-protected fragment (Fig. 7, panel 1, lane 4 ▶).
Nucleotides were not required for the catalysis of RNA annealing by the Ltp26/Ltp28 complex (Fig. 7, panels 2–4 ▶), but omitting MgCl2 greatly reduced the efficiency of annealing (panel 5).
Similar results were obtained with the RNA constructs which were used for the crosslinking experiments in Figure 1 ▶, suggesting a lack of RNA sequence specificity of the RNA annealing activity (data not shown).
The second-order association rate constants (k2) for RNA annealing in the absence and presence of the proteins were obtained as described in Materials and Methods. The stimulation of annealing, which was defined as the ratio of k2 (with protein)/k2 (no protein) where k2 in the absence of protein = 1.6 × 104 M−1 s−1, varied from 20- to almost 70-fold. The recombinant Ltp28 protein showed a threefold higher stimulation of annealing compared to the Ltp26 protein and showed a higher value than the purified Ltp26/Ltp28 complex (Table 1 ▶). Recombinant T. brucei gBP25 protein showed annealing activity similar to that of Ltp26 (Table 1 ▶).
Interaction of the Ltp26/Ltp28 complex with components of editing machinery in mitochondrial extract
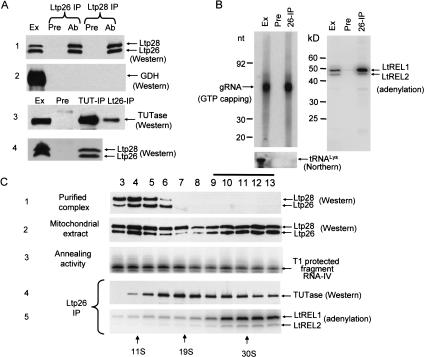
Affinity-purified polyclonal antibodies raised against the purified Ltp26/Ltp28 complex and the recombinant proteins were used for immunoprecipitation (IP) of mitochondrial extract. The anti-Ltp26 antibody coprecipitated Ltp26 and Ltp28 and the anti-Ltp28 antiserum did the same, confirming that the complex is composed of both proteins (Fig. 8A ▶, panel 1). The specificity of these antisera and the absence of crossreactivity between Ltp26 and Ltp28 was confirmed by Western blotting analysis of the mitochondrial extract (data not shown).
FIGURE 8.
Association of Ltp26/Ltp28 complex with components of RNA editing machinery. (A) Co-IP of Ltp26/Ltp28 complex and 3′TUTase. IP from mitochondrial extract was performed using Sepharose G beads coated with affinity-purified mouse polyclonal antibodies against recombinant Ltp26, recombinant Ltp28, and L. tarentolae 3′ TUTase (Ltp26-IP, Ltp28-IP, and TUT-IP lanes). Immunodetection was performed with rabbit antibodies raised against purified Ltp26/Ltp28 complex, recombinant glutamate dehydrogenase (GDH) (Bringaud et al. 1997), and TUTase. Ext, mitochondrial extract; Pre, proteins bound to G-beads coated with preimmune serum; Ab, proteins bound to G-beads coated with specific antibodies. 10%–20% SDS gels blotted to nitrocellulose membranes and probed with respective antibodies. (B) Co-IP of Ltp26/Ltp28 complex, gRNA, and RNA ligase. IP was performed with preimmune (Pre) or anti-Ltp26 (IP) antibody. Left panel: Total RNA was isolated from mitochondrial extract and the IP material, labeled with [α32P]GTP in the presence of guanylyltransferase and separated on a 12% acrylamide/urea gel. Lower panel: The presence of tRNALys was assayed in the same material by Northern blotting with an oligonucleotide probe. Right panel: The same samples were incubated with [α32P]ATP to visualize the LtREL1 and LtREL2 proteins and analyzed on a 10%–20% SDS gel. (C) Association of Ltp26/Ltp28 complex with TUTase and with RNA ligase. Purified Ltp26/Ltp28 complex (10 μg; panel 1) and mitochondrial extract (250 μL, 10 mg/mL; panels 2,3) were fractionated on a 10%–30% glycerol gradient in the SW41 rotor for 20 h at 35,000 rpm. Panel 2, Western analysis of Ltp26 and Ltp28. Panel 3, T1 nuclease protection assay for RNA annealing activity. Panels 4 and 5, co-IP of TUTase and LtREL1 and LtREL2 by anti-Ltp26 antibody.
The anti-Ltp26 antiserum also coprecipitated a fraction of the TUTase from the extract (Fig. 8A ▶, panel 3). A control for specificity of the IP was provided by using antibody against glutamate dehydrogenase (GDH), an abundant mitochondrial matrix protein with a high affinity for RNA but which is apparently not involved in RNA editing (Estévez et al. 1999), and by showing that this protein was present in the extract but not in the anti-Ltp26 IP (Fig. 8A ▶, panel 2). A reciprocal IP with anti-TUTase antibody confirmed these results (Fig. 8A ▶, panel 4). These data suggest an interaction of the Ltp26/Ltp28 complex with TUTase or with a TUTase-containing complex. The latter is suggested by the experiments described in Figure 4C ▶, in which recombinant TUTase was shown not to associate with Ltp26, Ltp28, or the Ltp26/Ltp28 complex in vitro, and by the yeast two-hybrid experiments in Figure 4B ▶ in which no direct interaction could be detected in vivo between TUTase and Ltp26 or Ltp28.
The LtREL1 and LtREL2 RNA ligase proteins can be detected by adenylation with [α32P]ATP (Sabatini and Hajduk 1995,Peris et al. 1994; 1997). Evidence of an interaction of ATP-labeled LtREL1 and LtREL2 in mitochondrial extract with Ltp26 was provided by the co-IP with anti-Ltp26 antiserum shown in Figure 8B ▶, right panel. The co-IP of LtREL1 and LtREL2 (and TUTase) was stable enough to withstand high-stringency washes with 0.5 M NaCl and 0.5% Triton X100 (not shown).
We also investigated the possibility that the Ltp26/Ltp28 complex interacts with endogenous gRNA. Guide RNA in the mitochondrial extract can be specifically 5′ end-labeled with [α32P]GTP and guanylyltransferase, as shown for total mitochondrial extract in Figure 8B ▶ (left panel, Ex lane). IP of the extract with anti-Ltp26 antiserum was performed and RNA extracted from the washed beads and labeled with GTP. A substantial amount of the gRNA in the extract co-immunoprecipitated with Ltp26 and Ltp28 (Fig. 8B ▶, left panel, IP lane). A control for the specificity of the co-IP of gRNA was shown by the lack of co-IP of the abundant mitochondrial tRNALys by the anti-Ltp26 antiserum (Fig. 8B ▶, lower panel).
To investigate these interactions in more detail, mitochondrial extract was fractionated in a glycerol gradient and the distribution of the Ltp26/Ltp28 complex assayed. The purified Ltp26/Ltp28 complex sedimented as expected in one band, peaking in fractions 4–5 (Fig. 8C ▶, panel 1). However, the Ltp26/Ltp28 complex in the extract showed, in addition to a peak comigrating with the purified complex, another peak sedimenting in approximately the 30S region, suggesting an association of the Ltp26/Ltp28 complex in the extract with RNA and/or protein components. In agreement with this distribution, RNA annealing activity was spread throughout the gradient, with the majority in the ∼10–15S region (Fig. 8C ▶, panel 3). The RNA annealing activity in the 10–15S region appears to be higher than the ∼30S region, although a similar amount of Ltp26/Ltp28 complex is present, which may indicate the saturation of higher-molecular-weight forms of the complex with substrate RNA. Each gradient fraction was also subjected to IP with anti-Ltp26 antiserum and analyzed for co-IP of TUTase and LtREL1 and LtREL2. Co-IP of TUTase (Fig. 8C ▶, panel 4) occurred throughout the gradient, with the relative amount precipitating in different regions, coinciding with the previously reported distribution of TUTase in glycerol gradients (Peris et al. 1997; Aphasizhev et al. 2002). Co-IP of the two RNA ligases was mainly limited to the higher-molecular-weight region of the gradient, coinciding with the second peak of Ltp26/Ltp28 (Fig. 8C ▶, panel 5, indicated by line above fractions 9–13).
Isolation of Ltp26/Ltp28-associated complex
In order to more directly assess the Ltp26/Ltp28 interactions, Ltp28 protein with the mitochondrial targeting sequence was ectopically expressed in L. tarentolae from the pX vector (LeBowitz et al. 1990) with a C-terminal fusion of a dual TAP epitope tag, which consists of an IgG-binding Protein A domain and a calmodulin-binding domain (Puig et al. 2001). Mitochondria were isolated from transfected cells, and clarified mitochondrial extract was sedimented through a glycerol gradient. Each gradient fraction was subjected to affinity binding with IgG Sepharose. As shown in Figure 9A ▶, both the wild-type Ltp28 protein and the Ltp28-TAP protein were present in the IgG “pull down,” indicating that the tagged protein was incorporated into the Ltp26/Ltp28 complex. In addition, as also found above in the co-IP from mitochondrial extract, the IgG “pull down” from the higher-molecular-weight gradient fractions (indicated by line above fractions) contained TUTase and LtREL1 and LtREL2 (Fig. 9A ▶). The consistency of these results with the IP data from mitochondrial extract of nontransfected cells validates the use of the tandem affinity purification (TAP) system for studying higher-order interactions of the Ltp26/Ltp28 complex.
FIGURE 9.
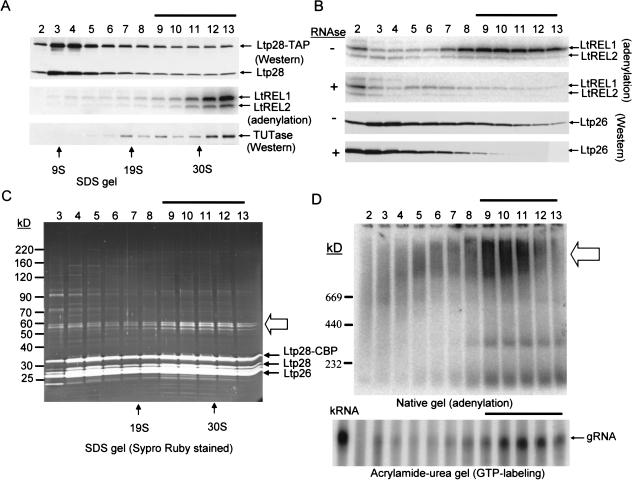
Purification of high-molecular Ltp26/Ltp28 RNP complex weight by TAP chromatography. (A) Mitochondrial extract from Ltp28-TAP transfected cells was sedimented through a glycerol gradient. Each fraction (0.5 mL) was incubated with 20 μL of IgG Sepharose beads and, after extensive washing in 20 mM Tris-HCl, pH 7.6, 60 mM KCl, 10 mM MgCl2, 0.1% NP40, bound material was released from the beads and analyzed for the presence of the endogenous Ltp28 and the Ltp28 TAP fusion protein by Western analysis using anti-p28 antiserum, for the LtREL1 and LtREL2 RNA ligases by adenylation, and for TUTase by Western analysis using anti-TUTase antiserum. The gradient fractions containing the cosedimenting TUTase and RNA ligases are indicated by a line above. (B) IgG purification of Ltp28-associated RNP complex. Mitochondrial extract as in A was subjected to binding to IgG Sepharose and release by cleavage with TEV protease. In a separate experiment, RNase A (0.1 mg/mL) was added during the binding to IgG. The purified “pull down” was fractionated on a glycerol gradient, and the fractions assayed for the RNA ligases and the Ltp26 protein. The fractions showing a depletion of the ligases and Ltp26 are indicated by a line above. (C) Complete TAP purification of mitochondrial extract. This was carried out through the IgG binding with TEV protease release, and calmodulin binding with EGTA release. The purified fraction was subjected to glycerol gradient sedimentation, and each fraction was separated on an SDS gel, which was stained with Sypro Ruby. Arrows indicate the location of the endogenous Ltp26, Ltp28, and the ectopically expressed Ltp28-TAP bands. The three 55–60-kD associated bands are indicated by an open arrow. (D) Upper panel, the fractions in C were subjected to adenylation and fractionated in a 4%–12% native gel. The location of the adenylated complexes is indicated by an open arrow. Lower panel, RNA was extracted from each fraction, the gRNA labeled with [αP32] GTP in the presence of guanylyltransferase and separated on 12% acrylamide/urea. The gRNA band is indicated. RNA from total mitochondrial extract (Ex) was used as a control.
The Ltp26/Ltp28-associated RNP complex was isolated from total mitochondrial extract by first binding the extract to IgG, releasing the bound material by cleavage with TEV protease, and then binding to a calmodulin matrix and eluting with 2 mM EGTA (see Materials and Methods). The eluted material was subjected to glycerol gradient sedimentation. As shown in the stained SDS gel of each fraction in Figure 9C ▶, a complex composed of Ltp28-CBP, Ltp28, Ltp26, and three major 55–60-kD proteins was detected sedimenting in fractions 9–13 (indicated by line above fractions), in addition to a substantial amount of the free 100-kD complex sedimenting in the upper region of the gradient. The identical protein profile was obtained when Ltp26 was used as an entry for the TAP purification system (not shown). GTP-labeled gRNA also sedimented in this region of the gradient (Fig. 9D ▶, lower panel), in agreement with the co-IP results described above.
Adenylation of each fraction and SDS (Fig. 9B ▶) and native gel analysis (Fig. 9D ▶, upper panel) showed the presence of the LtREL1 and LtREL2 RNA ligases sedimenting in the region of the gradient of the purified IgG + calmodulin “pull down” material. The heterogeneous appearance of the adenylated complexes in the native gel is probably a result of breakdown during the electrophoresis and/or to the presence of variable amounts of bound RNA.
Treatment of the mitochondrial extract with RNase A during binding to the IgG matrix led to a disappearance of the high-molecular-weight RNP complex, as shown in Figure 9B ▶ for LtREL1 and LtREL2 and Ltp26/p28 complex, as detected by Western analysis with anti-Ltp26 antibody (Fig. 9B ▶), suggesting an RNA requirement for the interaction of the p26-p28 complex with the L-complex which contains LtREL1 and LtREL2 (Peris et al. 1997). It should be noted that the LtREL1 and LtREL2 RNA ligases and the 120-kD TUTase were not stoichometric with the Ltp26-28-28-CBP and the 55–60-kD proteins (Fig. 9C ▶) and could only be detected by Western analysis or adenylation. This suggests that the Ltp26/Ltp28 RNP complex interacts less stably with TUTase and the L-complex (Peris et al. 1997; Aphasizhev et al. 2002).
DISCUSSION
We have confirmed and extended previous results from T. brucei (Koller et al. 1997; Allen et al. 1998; Lambert et al. 1999; Blom et al. 2001; Muller et al. 2001) and C. fasciculata (Blom et al. 2001) for the presence of two small mitochondrial RNA-binding proteins, with L. tarentolae. Several features of the L. tarentolae proteins are similar to those reported previously, but there are several novel findings. One was the discovery of a stable 100-kD complex that contained two copies each of Ltp26 and Ltp28. An association as a part of a high-molecular-weight complex was suggested previously by a co-IP of the homologs from C. fasciculata (Blom et al. 2001), but this is the first report of a stable tetrameric complex. In addition, the Ltp26/Ltp28 complex was isolated from mitochondrial extract by sequential enrichment by ammonium sulfate fractionation, heparin affinity, and gel-filtration chromatography of a protein fraction capable of forming a crosslink with a model 4-thiouridine-containing RNA that emulated an mRNA-gRNA substrate. Although this interaction was found to be specific for unpaired U residues present as a bulge in a duplex RNA and poly[U] was the most efficient homopolymer competitor (data not shown), a more detailed analysis with several different RNAs indicated no great difference in binding affinity between single-stranded and double-stranded RNA.
The 100-kD complex could be reconstituted with recombinant proteins in the absence of RNA or additional cofactors. An interaction between Ltp26 and Ltp28 recombinant proteins was established by the in vivo yeast two-hybrid results and in vitro co-IP of transcribed and translated epitope-tagged proteins. Although recombinant Ltp26 can form a dimer and possibly a trimer after extended incubation in high concentration, an interaction of Ltp26 with itself was not detected in the yeast two-hybrid and co-IP experiments, suggesting that the observed Ltp26-Ltp26 interaction is low-affinity. However, since we found that a twofold molar concentration of the Ltp26 protein over the Ltp28 protein was necessary for efficient reconstitution of the tetramer, the most likely scenario for the reconstitution of the Ltp26/Ltp28 complex in vitro consists of Ltp26 first forming a homodimer, one component of which is then displaced by Ltp28. Two heterodimers could then interact to form a tetramer, which is consistent with the detection of three protein–protein crosslinks in the purified Ltp26/Ltp28 complex. This model must of course be confirmed by further experimentation.
It was shown previously that the T. brucei gPB21 showed RNA-binding properties and also catalyzed RNA annealing, although only a minority of the annealing activity of the mitochondrial extract could be attributed to this protein (Muller et al. 2001). We confirmed an annealing activity for the gPB21 homolog from L. tarentolae (Ltp28) and showed that Ltp26 and the T. brucei gPB25 homolog of Ltp26 also had RNA annealing activities, although their Kd values (Table 1 ▶) indicate a lower affinity for both single- and double-stranded RNAs. Furthermore, both the purified and reconstituted Ltp26/Ltp28 complex showed higher-affinity RNA binding than either of the component proteins and stimulated RNA annealing approximately 40-fold. The binding of purified Ltp26/Ltp28 100-kD complex to a synthetic single-stranded RNA was shown to occur stepwise essentially to saturation, with one protein complex for approximately 5–7 nucleotide bases. Although an RNA component was not detected in the purified Ltp26/Ltp28 100-kD complex, the co-IP analysis and the TAP pull-down analysis indicated an interaction of the Ltp26/Ltp28 complex with gRNA and with several 55–60-kD proteins to form a stable high-molecular-weight RNP complex.
T. brucei gPB21 has been shown to coimmunoprecipitate with in vitro RNA editing activity, ATP-labeled RNA ligase proteins, and TUTase activity, and the interaction was sensitive to micrococcal nuclease treatment, suggesting involvement of an RNA component (Allen et al. 1998). The C. fasciculata gPB27 and gPB29 proteins, however, were reported not to interact with any identified editing components or RNA (Blom et al. 2001). We showed by TAP affinity chromatography of tagged Ltp28 targeted to the mitochondrion and incorporated into the 100-kD complex that the 100-kD Ltp26/Ltp28 complex interacts via an RNA component with both the 3′ TUTase and the RNA ligase-containing L-complex to form a heterogeneous high-molecular-weight RNP complex sedimenting at approximately 30S in glycerol gradients. We recently found evidence for the existence of an ∼20–25S RNP complex that shows in vitro editing activity and consists of the L-complex interacting via an RNA component with the 3′ TUTase and with the 100-kD Ltp26/Ltp28 complex (unpubl. results). We speculate that the differences in sedimentation may be due to variable extents of interaction and/or bound RNA.
We speculate that the Ltp26/Ltp28 RNP complex plays a specific role in the initiation of RNA editing in which a gRNA forms a duplex anchor with the cognate preedited mRNA, thereby creating a substrate for an initial site-directed cleavage. However, as mentioned above, previous evidence for involvement of the T. brucei gBP21 protein in RNA editing is somewhat equivocal. gBP21 was reported to coprecipitate with a 20S in vitro editing activity (Allen et al. 1998), whereas gBP21 null mutants of bloodstream T. brucei showed an overall decrease in the abundance of mitochondrial transcripts but no effect on editing in vivo (Lambert et al. 1999). One explanation of these earlier results could be the observed redundancy of the two proteins of the Ltp26/Ltp28 complex. In this regard, we recently showed that inhibition of expression of T. brucei gPB21 by RNAi had no effect on viability, whereas inhibition of expression of both gBP21 and gBP25 produced a growth defect (A. Simpson, R. Aphasizhev, L. Simpson, unpubl.). Further work is required to determine the precise biological role of the Ltp26 and Ltp28 proteins.
MATERIALS AND METHODS
Cell growth and isolation of mitochondria
L. tarentolae cells were grown in 15-L batches in brain-heart infusion medium with a supplement of hemin to late log phase (∼180 × 106 cells/mL) in a BioFlow IV Fermentor (New Brunswick). Mitochondria were isolated by the hypotonic lysis Renografin density gradient method (Braly et al. 1974).
RNA synthesis and RNA-protein crosslinks
The sequences of RNA-I and RNA-II (Fig. 1A ▶) were initially derived from ND7 mRNA and gND7 gRNA (Kapushoc and Simpson 1999) and were modified to remove all the encoded U residues from RNA-I except the four Us at the editing site and to make compensatory changes in RNA-II for base-pairing. RNA-I was synthesized by T7 transcription from linearized plasmid DNA in the presence of 4-thiouridine, thereby inserting 4-thio U residues at a specific site in the RNA molecule as shown in Figure 1A ▶ (Kim and Abelson 1996). Typically, 5 μg plasmid was transcribed in the buffer supplied by the manufacturer (Life Technologies) in the presence of 0.5 mM of 4-thio UTP, 50 μCi [α32P]UTP, 2 mM of ATP, GTP, and CTP, and 100 U of T7 RNA polymerase. RNA was purified by electrophoresis in 15% acrylamide/8M urea, eluted in 0.1 M sodium acetate, pH 5.5/0.1%SDS, extracted with phenol/chloroform, and ethanol-precipitated. All other RNAs were synthesized under the same conditions with standard NTPs.
For analytical crosslink analysis, 1 pmole of 4-thioU RNA-I (Fig. 1A ▶) was annealed with 2 pmole of ligand RNA-II in 10 μL of 10 mM HEPES, pH 7.8 by heating to 80°C and cooling to 25°C in 15 min. Then, 1 μL of 10X crosslinking buffer (200 mM HEPES, pH 7.8, 1 M KCl, 10 mM DTT, 50 mM MgCl2) was added and incubated for 10 min. The mitochondria were resuspended in 1× crosslinking buffer at 50 mg of total protein/mL in the presence of protein inhibitors (Complete, EDTA free, Roche), lysed on ice by adding Triton X100 to 0.3% and centrifuged for 10 min at 200,000 g. Fragmented total yeast tRNA (Sigma) was added to the supernatant to 0.1 mg/mL. The annealed RNA was incubated with an equal volume of supernate for 30 min at 27°C and irradiated on ice with a UV-lamp at 5 cm distance with a 300 nM cut-off filter for 5 min, unless otherwise indicated. The products were analyzed on an SDS gel followed by autoradiography.
Purification of UV-crosslinked proteins
The generation of crosslink I was followed as an analytical assay for purification of the associated proteins. Chromatography was performed on an Akta Explorer 100 (AP Biotech) at 4°C unless otherwise indicated. Purified mitochondria (1200 mg total protein) were extracted in 100 mL (final volume) of 50 mM HEPES, 300 mM KCl, 1 mM DTT, three tablets of Complete Inhibitors (Roche), 2 mM CHAPS with sonication. The extract was centrifuged at 50,000 rpm in the Ti 60 rotor (Beckmann) for 1 h. Approximately 250 mg of soluble protein was recovered. Ammonium sulfate (AmS, fine powder) was added to 40% and the centrifugation repeated. The supernatant was recovered and AmS added to 55%. The precipitate, which contained the crosslink activity, was collected by centrifugation, dissolved in 50 mM HEPES, 10 mM MgCl2, 1 mM DTT, 5% glycerol, and the conductivity adjusted to 15 mSi/cm2 (80 mg of protein) and loaded on a 5-mL HiTrap heparin column (AP Biotech). The column was washed with 200 mM of KCl in the same buffer and developed with a 100-mL gradient from 200 to 1000 mM of KCl at 1 mL/min. The fractions which were active in generation of crosslink I (6 mL, 0.8 mg of protein, which eluted at 500–600 mM) were concentrated to 200 μL and loaded on a Superose 12 column equilibrated with 50 mM HEPES, pH 7.8, 300 mM KCl, 0.1 mM EDTA, 1 mM DTT, 10% glycerol. A single A280 peak with an apparent molecular weight of 100 kD was observed in the Superose 12 chromatography (see Fig. 5 ▶). The final yield was consistently 0.5–0.3 mg of protein. Proteins were either transferred onto a PVDF membrane for N-terminal sequencing (UCLA Microsequencing Facility) or excised from Coomassie-stained gels and submitted to the Harvard Microsequencing Facility for tryptic digestion and internal peptide sequencing.
For the analysis of protein–protein crosslinks, a Phenyl Resource (AP Biotech) column was introduced prior to gel-filtration. AmS was added to heparin fractions to1.2 M and centrifuged for 10 min at 100,000 g. The supernatant was loaded onto a 1-mL Phenyl Resource column preequilibrated with 1.5M of AmS in 50 mM HEPES, pH 7.8, and the column was developed with a 20-mL gradient from 1.5 M to no salt. The complex eluted at 1–1.2 M salt as a single sharp peak.
Gene cloning and protein expression
N-terminal sequences were obtained from the 26-kD and 28-kD bands and two minor bands of 20 kD and 22 kD. Two internal peptide sequences from the 26-kD band and three peptides from the 28-kD band were used to construct degenerate oligonucleotides with the Codehop program. All possible pairwise combinations of oligonucleotides were tested for PCR using genomic DNA. A 262-bp product was obtained for the 26-kD protein and a 378-bp product for the 28-kD protein. The PCR products were used as probes to clone 4-kb KpnI and 3.5-kb PstI genomic fragments that contained the entire genes for Ltp26 and Ltp28, respectively. The Ltp26 ortholog from T. brucei was cloned using a PCR-derived probe based on the EST sequence AI881040 (Parasite Genomes Database) from a KpnI subgenomic library. The 5.3-kb genomic clone contained the full-length gene. For expression in E. coli, cloned genes were PCR-amplified, with NdeI and BamHI sites introduced at the 5′ and 3′ ends, respectively, and cloned into the pET 15b vector, which provides an N-terminal 6 His affinity tag. The experimentally determined mitochondrial localization signal (position −46) for Ltp26 from L. tarentolae, and the predicted signals (position −22) for Ltp28 and (position −30) for the Ltp26 ortholog from T. brucei were removed. Both Ltp26 proteins were partially soluble (30%–50%) when expressed at 25°C and were purified by Talon metal affinity chromatography as suggested by the manufacturer, followed by chromatography on a 1-mL heparin HiTrap column as described above which was downscaled fivefold. The Ltp28 formed inclusion bodies and was purified under denaturing conditions. The protein was refolded while still attached to the Talon resin at 20°C by gradually decreasing the urea concentration from 8 to 0 M in 20 mM sodium phosphate pH 7.0, 300 mM NaCl. The gradient was run for 15 h at 1 column volume per h. The protein was eluted with 200 mM of imidazole and further purified on a heparin column. All proteins were at least 95% pure as determined by Sypro Ruby (Molecular Dyes) staining of SDS gels.
Protein–protein interactions
DMA, DMP, and DMS are homobifunctional imidoester crosslinking agents (Pierce) that react with primary amines and differ in spacer arm length (8.6, 9.2, and 11 A, respectively). Purified Ltp26/28 complex (0.1 mg/mL) in 50 mM sodium phosphate buffer, pH 8.0, 150 mM KCl, was incubated for 40 min at 5°C in the presence of 10 mM of the crosslinking reagent. The reaction was stopped by adding 1 M Tris-HCl, pH 6.8 to 1/10 of the volume, and the products were analyzed on a 10%–20% SDS gel followed by Sypro Ruby staining.
The full-length coding regions for the Ltp26 and Ltp28 L. tarentolae proteins, and the L. tarentolae 3′ TUTase were cloned in the proper reading frames into both the ‘bait” (pGBKT7) and “prey” (pGADT7) vectors used in the Matchmaker 3 Yeast Two-Hybrid Screening Kit™ (Clontech). Each construct alone or in various pairwise combinations with other constructs were used to transform yeast strain AH109 (MATa, trp1–109, leu2–3, 112, ura3–5, his3–200, ade2–101, gal4Δ, gal80Δ, LYS2•GAL1UAS-GAL1TATA-HIS3, GAL2 UAS-GAL2 TATA-ADE2, URA3•MEL1 UAS-MEL1TATA-lacZ) using appropriate nutritional selection; the resulting yeast transformants were checked for fusion protein expression by Western analysis. In this manner, each of the three L. tarentolae proteins were shown to be expressed fused at their amino-terminal end to either the DNA-binding domain of the Gal4 protein epitope-tagged with Myc or the activation domain of the Gal4 protein epitope-tagged with HA. Each of the plasmid constructs was also used for coupled transcription/translation (TnT T7 System, Promega). Because of the location of the T7 promoter in the parent pGBKT7 and pGADT7 vectors, in vitro translation products retain the amino-terminal epitope tags, Myc and HA, respectively, but do not contain any portion of the Gal 4 protein. Pairwise mixtures of these were analyzed for association in vitro using a co-IP protocol (Matchmaker Co-IP Kit) that included clarification of reactions to remove any nonspecific precipitation and/or nonspecific adsorption.
Reconstruction of Ltp26/Ltp28 complex
Purified recombinant proteins were mixed at a 1:2 (Ltp28:Ltp26) molar ratio and a final protein concentration of 2 mg/mL, and dialyzed against 20 mM sodium phosphate, 300 mM NaCl, for 16 h at 25°C. We found that an excess of the Ltp26 protein was necessary for in vitro complex formation. After 10 min centrifugation at 200,000 g, the samples were loaded on a Superose 12 column and chromatographed in the same buffer at 0.3 mL/min. Fractions of 0.4 mL were collected and analyzed on a 10%–20% acrylamide-SDS gel and stained with Sypro Ruby. Molecular weight markers were bovine gamma globulin, ovalbumin, myoglobin, and Vitamin B12 (Biorad). For gRNA-protein reconstruction, purified Ltp26/28 complex and gRNA were incubated with in vitro transcribed RPS12-I gRNA (see Results) and separated on Superose 6 columns in 20 mM HEPES, pH 7.5, 5 mM MgCl2, 150 mM KCl buffer. Molecular weight markers were thyroglobulin, ferritin, catalase, and BSA. Native precast 8%–16% gels (Invitrogen) were run for 12 h at 40 V, fixed, dried, and exposed to X-ray film. Premixed high-molecular-weight markers for native gel electrophoresis were from AP Biotech.
Catalysis of RNA annealing and RNA binding
RNA molecules (RNA-III and RNA-IV in Fig. 7 ▶) were synthesized by run-off transcription from a linear DNA template (Kapushoc et al. 1999) using T7 RNA polymerase. RNA-IV was labeled with [α-32P]UTP during transcription and purified on an 8M urea- 5% polyacrylamide gel. The RNA annealing reaction contained 0.5 nM of mRNA and 0.5–50 nM of gRNA, and the protein concentration varied from 0 to 5 μM. All reactions were performed in 20 μL of 20 mM HEPES, pH 7.6, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT for 0.5, 1, 2, 4, and 10 min at 27°C. The reaction was stopped by the addition of 1 μL T1 RNase (18u/μL) and incubated for 10 min at 27°C, followed by phenol/chloroform extraction and ethanol precipitation. Products were analyzed in 15% polyacrylamide/urea gels, exposed to PhosphoImager plates, and quantified with ImageQuant software.
The first- and second-order association constants were calculated as described (Muller et al. 2001). The affinity of RNA binding was measured as the equilibrium dissociation constant, Kd, by fitting nitrocellulose filter binding data from three independent experiments into an equilibrium binding model. RNA-III (Fig. 7 ▶) was used as the single-stranded RNA substrate. The double-stranded RNA substrate was obtained by annealing of RNA-III to RNA-V, a complementary RNA of the same size: 3′-GGCCTCT TAGTATTACAAGAAAAATATAAATAGCACATCAGCCGA-5′.
For annealing, the RNAs at 5 μM were denatured by incubating at 95°C for 2 min in 20 mM HEPES, pH 7.1, and cooled to 20°C over 30 min followed by addition of MgCl2 to 10 mM. RNA-III was denatured and quick-cooled to melt out a possible hairpin structure produced by self-folding. For Kd determinations, the RNA concentration was kept constant at 5 nM, and increasing amounts of protein were used. The percentage of active protein was determined in reciprocal experiments with 2 nM of protein and increasing concentrations of RNA and used to adjust concentration values in the Kd calculations. The purified complex was determined to be 60% active, and the recombinant Ltp28 and Ltp26 were 25%–35% active, depending on the preparation. The Prizm3 (GraphPad) software package was used for all nonlinear regression curve fitting and statistical analyses.
Western blotting and immunoprecipitation
Rabbit and mouse polyclonal antibodies were raised against the Ltp26/28 complex from L. tarentolae, which was purified by heparin fraction and preparative 10%–20% SDS gel electrophoresis, and also against recombinant proteins (Covance Research Products). Antibodies were affinity-purified on immobilized antigen columns by standard techniques. IPs were carried out in 300–400 μL of extract (2 mg/mL) or glycerol gradient fractions (supplemented with BSA to 1 mg/mL) with 10–15 μL of GammaBind protein G beads (AP Biotech) coated with 1–5 μg of affinity-purified antibodies or 100 μg of total IgG fraction for 2 h at 4°C with constant mixing. Beads were pelleted by brief centrifugation, washed 3 times with 1 mL of 20 mM Tris HCl, pH 7.5, 500 mM NaCl, 0.5% TritonX100 and resuspended in 50 μL of 1×SDS loading buffer. Alternatively, total RNA was isolated from mitochondrial extract or IP material by adding SDS to 1%, followed by phenol/chloroform extraction and ethanol precipitation. Electrophoretic transfer on nitrocellulose membrane from SDS gels was performed for 2 h at 80 V in Mini Trans-Blot cells (Bio-Rad) in 25 mM Tris, 190 mM glycine, 10% methanol. Immunodetection was carried out with SuperSignal West Pico chemiluminescent substrate (Pierce) by standard techniques.
Glycerol gradient sedimentation
Mitochondrial pellets were resuspended in 10 mM HEPES, pH 7.6, 10 mM MgCl2, 60 mM KCl, 125 mM sucrose at 25 mg total protein per mL, and lysed on ice by adding Triton×100 to 0.3%. The extract was clarified by centrifugation at 200,000 g for 15 min and 200 μL of supernatant loaded on 10%–30% glycerol gradient formed in a centrifuge tube for the SW 41 Beckman rotor. After centrifugation for 20 h at 30,000 rpm, 750-μL fractions were collected from the top of the gradient. Adenylation reactions were performed on each fraction as described (Peris et al. 1997). RNA was isolated from each fraction and the gRNA specifically 5′-labeled with [α32P]GTP as described (Blum et al. 2000).
Tandem affinity purification (TAP) of Ltp28-associated proteins
The entire Ltp28 gene was fused at the C-terminus with the TAP cassette and inserted into the pX vector (LeBowitz et al. 1990) at the BamHI site. L. tarentolae cells were transfected with this plasmid and selected for resistance to G418 by agar plating. The expression of the Ltp28-TAP and targeting of the protein to the mitochondrion was confirmed by Western analysis of subcellular fractions using the PAP reagent which recognizes the protein A moiety of the fusion protein. Cells were grown to late log phase, and the mitochondrial fraction was isolated.
The TAP protocol was adapted from the published method (Puig et al. 2001). Mitochondria (200 mg of total protein) were lysed on ice for 15 min in 12 mL of TMK buffer (20 mM Tris-HCl, pH 7.6, 60 mM KCl, 10 mM MgCl2) plus 0.5% NP40. One tablet of Complete proteinase inhibitors (Roche) was added directly to the extract. After centrifugation at 200,000 g for 10 min, the pellet was sonicated 3 times for 10 sec in 12 mL of the same buffer without detergent and the centrifugation was repeated. Supernatants from both extraction were pooled and incubated with 0.6 mL of IgG Sepharose FF (AP Biotech) for 2 h in batch, transferred to three disposable 5-mL columns (Pierce) and washed sequentially with 20 mL of TMK buffer plus 0.1% of NP40 and 10 mL of TEV proteinase cleavage buffer (20 mM Tris-HCl, pH 7.6, 60 mM KCl, 10 mM MgCl2, 0.1% NP40, 1 mM DTT). One mL of the same buffer with 100 U of TEV proteinase was added to each column, and the columns were incubated overnight with constant mixing. The IgG resin was drained and washed with 2 mL of CB buffer (20 mM Tris-HCl, pH 7.6, 60 mM KCl, 10 mM MgCl2, 1 mM imidasole, 10 mM β-mercaptoethanol, 2 mM CaCl2, 0.1% NP40). Eluted material was incubated with 0.6 mL of calmodulin agarose (Stratagene) for 1 h, transferred to two disposable 5-mL columns (Pierce), washed with 20 mL of CB buffer, and eluted with 3 mL of 20 mM Tris-HCl, pH 7.6, 60 mM KCl, 10 mM MgCl2, 2 mM EGTA, 2 mM CHAPS. Eluted proteins were concentrated with Slide-A-Lyzer solution (Pierce) to 300 μL and loaded on a 10%–30% glycerol gradient. Sedimentation was performed in the SW41 rotor (Beckmann) for 20 h at 30,000 rpm. Fractions of 750 μL were collected from the top of the gradient.
Acknowledgments
We acknowledge with thanks the sequences of the Crithidia proteins prior to publication from R. Benne and a gift of the anti-gBP21 antiserum from U. Goringer. The TAP plasmid was obtained from CellZome. This work was supported in part by NIH Research Grant 09012 to L.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Ltp26/Ltp28 complex, 100-kD complex of Ltp26 and Ltp28
TUTase, terminal uridylyltransferase
AmS, ammonium sulfate
U, uridine
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2134303.
REFERENCES
- Allen, T.E., Heidmann, S., Reed, R., Myler, P.J., Goringer, H.U., and Stuart, K.D. 1998. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol. Cell. Biol. 18: 6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev, R. and Simpson, L. 2001. Isolation and characterization of a U-specific 3′-5′ exonuclease from mitochondria of Leishmania tarentolae. J. Biol. Chem. 276: 21280–21284. [DOI] [PubMed] [Google Scholar]
- Aphasizhev, R., Sbicego, S., Peris, M., Jang, S.H., Aphasizheva, I., Simpson, A.M., Rivlin, A., and Simpson, L. 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell 108: 637–648. [DOI] [PubMed] [Google Scholar]
- Blom, D., Burg, J., Breek, C.K., Speijer, D., Muijsers, A.O., and Benne, R. 2001. Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, a novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res. 29: 2950–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, B., Bakalara, N., and Simpson, L. 1990. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60: 189–198. [DOI] [PubMed] [Google Scholar]
- Braly, P., Simpson, L., and Kretzer, F. 1974. Isolation of kinetoplast-mitochondrial complexes from Leishmania tarentolae. J. Protozool. 21: 782–790. [DOI] [PubMed] [Google Scholar]
- Bringaud, F., Peris, M., Zen, K.H., and Simpson, L. 1995. Characterization of two nuclear-encoded protein components of mitochondrial ribonucleoprotein complexes from Leishmania tarentolae. Mol. Biochem. Parasitol. 71: 65–79. [DOI] [PubMed] [Google Scholar]
- Bringaud, F., Stripecke, R., Frech, G.C., Freedland, S., Turck, C., Byrne, E.M., and Simpson, L. 1997. Mitochondrial glutamate dehydrogenase from Leishmania tarentolae is a guide RNA-binding protein. Mol. Cell. Biol. 17: 3915–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, E.M., Connell, G.J., and Simpson, L. 1996. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 15: 6758–6765. [PMC free article] [PubMed] [Google Scholar]
- Chien, C.T., Bartel, P.L., Sternglanz, R., and Fields, S. 1991. The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. 88: 9578–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corell, R.A., Read, L.K., Riley, G.R., Nellissery, J.K., Allen, T.E., Kable, M.L., Wachal, M.D., Seiwert, S.D., Myler, P.J., and Stuart, K.D. 1996. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol. Cell. Biol. 16: 1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdz, M., Palazzo, S.S., Salavati, R., O’Rear, J., Clayton, C., and Stuart, K. 2002. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 21: 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez, A.M., Kierszenbaum, F., Wirtz, E., Bringaud, F., Grunstein, J., and Simpson, L. 1999. Knockout of the glutamate dehydrogenase gene in bloodstream Trypanosoma brucei in culture has no effect on editing of mitochondrial mRNAs. Mol. Biochem. Parasitol. 100: 5–17. [DOI] [PubMed] [Google Scholar]
- Favre, A., Moreno, G., Blondel, M.O., Kliber, J., Vinzens, F., and Salet, C. 1986. 4-Thiouridine photosensitized RNA-protein crosslinking in mammalian cells. Biochem. Biophys. Res. Commun. 141: 847–854. [DOI] [PubMed] [Google Scholar]
- Hayman, M.L. and Read, L.K. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 274: 12067–12074. [DOI] [PubMed] [Google Scholar]
- Huang, C.E., O’Hearn, S.F., and Sollner-Webb, B. 2002. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 22: 3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo, R.P., Palazzo, S.S., Burgess, M.L., Panigrahi, A.K., and Stuart, K. 2000. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell. Biol. 20: 8447–8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapushoc, S.T. and Simpson, L. 1999. In vitro uridine insertion RNA editing mediated by cis-acting guide RNAs. RNA 5: 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.H. and Abelson, J. 1996. Site-specific crosslinks of yeast U6 snRNA to the pre-mRNA near the 5′ splice site. RNA 2: 995–1010. [PMC free article] [PubMed] [Google Scholar]
- Koller, J., Muller, U.F., Schmid, B., Missel, A., Kruft, V., Stuart, K., and Goringer, H.U. 1997. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 272: 3749–3757. [DOI] [PubMed] [Google Scholar]
- Köller, J., Nörskau, G., Paul, A.S., Stuart, K., and Goringer, H.U. 1994. Different Trypanosoma brucei guide RNA molecules associate with an identical complement of mitochondrial proteins in vitro. Nucleic Acids Res. 22: 1988–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, L., Muller, U.F., Souza, A.E., and Goringer, H.U. 1999. The involvement of gRNA-binding protein gBP21 in RNA editing-an in vitro and in vivo analysis. Nucleic Acids Res. 27: 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBowitz, J.H., Coburn, C.M., McMahon-Pratt, D., and Beverley, S.M. 1990. Development of a stable Leishmania expression vector and application to the study of parasite surface antigen genes. Proc. Natl. Acad. Sci. 87: 9736–9740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegwater, P., Speijer, D., Benne, R. 1995. Identification by UV cross-linking of oligo(U)-binding proteins in mitochondria of the insect trypanosomatid Crithidia fasciculata. Eur. J. Biochem. 227: 780–786. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Sabatini, R.S., Pollard, V.W., and Hajduk, S.L. 1998. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): A novel editing complex protein with repetitive domains. EMBO J. 17: 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missel, A., Souza, A.E., Nörskau, G., and Göringer, H.U. 1997. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 17: 4895–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, U.F., Lambert, L., and Goringer, H.U. 2001. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 20: 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Gygi, S.P., Ernst, N.L., Igo, R.P., Palazzo, S.S., Schnaufer, A., Weston, D.S., Carmean, N., Salavati, R., Aebersold, R., et al. 2001. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21: 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, M., Miller, M.M., and Read, L.K. 2000. RNA-binding properties of the mitochondrial Y-box protein RBP16. Nucleic Acids Res. 28: 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris, M., Frech, G.C., Simpson, A.M., Bringaud, F., Byrne, E., Bakker, A., and Simpson, L. 1994. Characterization of two classes of ribonucleoprotein complexes possibly involved in RNA editing from Leishmania tarentolae mitochondria. EMBO J. 13: 1664–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris, M., Simpson, A.M., Grunstein, J., Liliental, J.E., Frech, G.C., and Simpson, L. 1997. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol. Biochem. Parasitol. 85: 9–24. [DOI] [PubMed] [Google Scholar]
- Pollard, V.W., Harris, M.E., Hajduk, S.L. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11: 4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman, D.S. and Dreyfuss, G. 1994. RNA annealing activities in HeLa nuclei. EMBO J. 13: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig, O., Caspary, F., Rigaut, G., Rutz, B., Bouveret, E., Bragado-Nilsson, E., Wilm, M., and Seraphin, B. 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods 24: 218–229. [DOI] [PubMed] [Google Scholar]
- Rusche, L.N., Cruz-Reyes, J., Piller, K.J., and Sollner-Webb, B. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16: 4069–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, R. and Hajduk, S.L. 1995. RNA ligase and its involvement in guide RNA/mRNA chimera formation. J. Biol. Chem. 270: 7233–7240. [DOI] [PubMed] [Google Scholar]
- Schnaufer, A., Panigrahi, A.K., Panicucci, B., Igo, R.P., Salavati, R., and Stuart, K. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291: 2159–2161. [DOI] [PubMed] [Google Scholar]
- Seiwert, S.D., Heidmann, S., and Stuart, K. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84: 831–841. [DOI] [PubMed] [Google Scholar]
- Simpson, L. and Kretzer, F. 1997. The mitochondrion in dividing Leishmania tarentolae cells is symmetric and becomes a single asymmetric tubule in non-dividing cells due to division of the kinetoplast portion. Mol. Biochem. Parasitol. 87: 71–78. [DOI] [PubMed] [Google Scholar]
- Vanhamme, L., Perez-Morga, D., Marchal, C., Speijer, D., Lambert, L., Geuskens, M., Alexandre, S., Ismaïli, N., Göringer, U., Benne, R., et al. 1998. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 273: 21825–21833. [DOI] [PubMed] [Google Scholar]