Abstract
The molecular basis for specific recognition of simple homopolymeric sequences like the polypyrimidine tract (Py tract) by multiple RNA recognition motifs (RRMs) is not well understood. The Drosophila splicing repressor Sex lethal (SXL), which has two RRMs, can directly compete with the essential splicing factor U2AF65, which has three RRMs, for binding to specific Py tracts. We have combined site-specific photocross-linking and chemical cleavage of the proteins to biochemically map cross-linking of each of the uracils within the Py tract to specific RRMs. For both proteins, RRM1 and RRM2 together constitute the minimal Py-tract recognition domain. The RRM3 of U2AF65 shows no cross-linking to the Py tract. Both RRM1 and RRM2 of U2AF65 and SXL can be cross-linked to certain residues, with RRM2 showing a surprisingly high number of residues cross-linked. The cross-linking data eliminate the possibility that shorter Py tracts are bound by fewer RRMs. We present a model to explain how the binding affinity can nonetheless change as a function of the length of the Py tract. The results indicate that multiple modes of binding result in an ensemble of RNA–protein complexes, which could allow tuning of the binding affinity without changing sequence specificity.
Keywords: Multiple registers and RNA looping, multiple RNA-binding domains, pre-mRNA splicing, single-stranded RNA, splicing regulation
INTRODUCTION
RNA-binding proteins recognize specific sequences to control various posttranscriptional RNA processing events. Uridine-rich sequences, present within introns adjacent to both 5′- and 3′-splice sites and within 5′- and 3′-untranslated regions (UTRs) of mRNAs, modulate various aspects of RNA processing, including splice-site selection, polyadenylation, RNA stability, and translation (for review, see Chen and Shyu 1995; Colgan and Manley 1997; Burge et al. 1999; Richter 1999; Moore 2000; Reed 2000; Vagner et al. 2000; Singh 2002). In higher eukaryotes, the polypyrimidine tract (Py tract) adjacent to the 3′-splice site is an essential splicing signal (Moore 2000; Reed 2000). It is specifically recognized by several proteins, including the U2 snRNP auxiliary factor (U2AF) and the Drosophila protein Sex lethal (SXL).
Human U2AF is composed of two subunits, the 65-kD large subunit (U2AF65) and the 35-kD small subunit (U2AF35). Early during spliceosome assembly (for reviews, see Hastings and Krainer 2001; Will and Luhrmann 2001), U2AF65 interacts with the Py tracts (Zamore et al. 1992) and facilitates the recruitment of U2 snRNP to the pre-mRNA branch site by promoting RNA–RNA base-pairing (Valcarcel et al. 1996). U2AF65 also collaborates with other splicing factors such as UAP56, p54, mBBP/SF1, and SAP155 for splicing (Zhang and Wu 1996; Fleckner et al. 1997; Berglund et al. 1998; Gozani et al. 1998). U2AF35 recognizes the invariant AG dinucleotide at the 3′-splice site, and plays an important role in the splicing of introns that have weak Py tracts (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999a). The fruit fly, nematode, and fission yeast orthologs of U2AF65 are essential for viability (Kanaar et al. 1993; Potashkin et al. 1993; Zorio and Blumenthal 1999b).
The Drosophila melanogaster master sex-switch protein SXL is another Py-tract-binding protein. Early during development, a small amount of SXL is synthesized in females from the transcripts initiated at the establishment promoter (Pe), which responds to the ratio of the number of X chromosomes to the number of autosomes (for reviews, see Cline and Meyer 1996; Schutt and Nothiger 2000). Subsequently, SXL controls the splicing or translation of Sxl, transformer (tra), and male-specific lethal-2 (msl2) premRNAs during sex determination and dosage compensation. Alternative splicing allows the synthesis of SXL and TRA proteins in females and MSL2 in males (for reviews, see Black 2000; Smith and Valcarcel 2000; Graveley 2001). SXL regulates tra through 3′-splice site switching by competing with the binding of U2AF65 to the non-sex-specific (NSS) Py tract of tra, thereby diverting U2AF65 to an otherwise weak, female-specific (FS), Py tract located further downstream, to which SXL does not bind (Sosnowski et al. 1989; Inoue et al. 1990; Valcarcel et al. 1993; Granadino et al. 1997). In the msl2 pre-mRNA, SXL competes for the binding of TIA-1 and U2AF65 proteins to the uridine-rich sequences near the 5′- and 3′-splice sites, respectively, causing retention of an intron in the 5′-UTR (Merendino et al. 1999; Forch et al. 2001). Moreover, the binding of SXL to uridine-rich sequences in both 5′- and 3′-UTRs blocks the translation of msl2 (Bashaw and Baker 1997; Kelley et al. 1997; Gebauer et al. 1998). SXL controls its own expression through exon skipping by cooperatively binding to distantly located uridine-rich sequences in flanking introns (Sakamoto et al. 1992; Horabin and Schedl 1993; Wang and Bell 1994; Penalva et al. 2001). Most interestingly, whereas splicing of tra and msl2 is regulated very early during spliceosome assembly, autoregulation of Sxl splicing involves its interaction with the second-step splicing factor SPF45 to block splicing at the second step (Lallena et al. 2002). In addition, SXL synthesis is tightly autoregulated by feedback inhibition of translation through uridine-rich sequences within its 3′-UTR (Yanowitz et al. 1999). Although there are differences in the precise mechanisms of regulation, and it is possible that different pre-mRNAs have fine tuned the SXL-binding sites to accommodate involvement of cofactors and to serve specific regulatory requirements, all known examples of SXL regulation involve its binding to uridine-rich sequences.
The SXL-binding site for the tra pre-mRNA has been defined in greater detail by iterative selection-amplification (Sakashita and Sakamoto 1994; Singh et al. 1995), mutagenesis (Inoue et al. 1990; Valcarcel et al. 1993; Sosnowski et al. 1994; Kanaar et al. 1995), phylogenetic comparison among five Drosophila species (O’Neil and Belote 1992), and chemical interference/protection analysis and phosphorothioate substitution (Singh et al. 2000). In addition, the binding of SXL to portions of the regulated Py tracts of tra and Sxl premRNAs have been characterized by X-ray crystallography (Handa et al. 1999) and NMR (Lee et al. 1994; Kanaar et al. 1995; Inoue et al. 1997; Kim et al. 2000). Although the structures of isolated RRM1 and RRM2 of U2AF65 are known (Ito et al. 1999), how they interact with each other when bound to a Py tract remains to be structurally characterized.
Both SXL and U2AF65 belong to the ribonucleoprotein-consensus motif or RNA-recognition motif (RRM) family of proteins. The RRM family is the largest family of RNA-binding proteins, and is found in all three domains of life (bacteria, archaea, and eukarya; Burd and Dreyfuss 1994; Varani and Nagai 1998; Antson 2000). The RRM motif is an 80–90 amino acid region that is characterized by a four-stranded antiparallel β-sheet and two α-helices. The two central β-strands correspond to the RNP-1 and RNP-2 motifs, which are highly conserved among members of this protein family. In general, RNA binds to the β-sheet platform by means of base intercalation, hydrophobic interactions, hydrogen bonding, and charge interactions. The majority of the RRM family members contain multiple RRM motifs (Varani and Nagai 1998); both RRMs of SXL and all three of U2AF65 are important for RNA binding (Zamore et al. 1992; Kanaar et al. 1995). Intriguingly, although SXL and U2AF65 have different number of RRMs, their preferred binding sites are uridine-rich consensus sequences of similar length, as determined by in vitro selection from a random pool of RNA (Singh et al. 1995). Given that a single RRM likely binds ∼4–7 nt (Shamoo et al. 1994; Varani and Nagai 1998), it is unclear how the two RRMs of SXL and the three RRMs of U2AF65 bind to the functional Py tracts, which widely differ in length. In general, RNA structure plays an important role in binding specificity (Frankel 1999; Williamson 2000). However, how multiple RRMs recognize simple, homopolymeric sequences such as poly(U) and poly(A), which lack a discrete RNA structure (Saenger 1994), is beginning to be understood (Antson 2000). Furthermore, it is well known that the binding affinity of these proteins correlates with the length of the Py tract. However, the basis for the relationship between the length of the Py tract and the number of RRMs bound or the binding affinity remains to be understood.
In the present study, we tested how multiple RRMs of U2AF65 and SXL contribute to Py-tract recognition in solution. We devised a biochemical strategy combining site-specific photochemical cross-linking and chemical cleavage to map the regions of interaction. Our detailed biochemical analysis provides important new information on Py-tract recognition.
RESULTS
Assignment of a site-specific photochemical cross-link to specific RRMs
To elucidate the role of multiple RRMs in RNA recognition in general, and to gain insight into the relationship between the length of the Py tract and the number of RRMs or the binding affinity for protein in particular, we developed an approach to biochemically map cross-linking of specific uridines to particular RRMs. First, we generated a series of RNA substrates in which a 5-iodouracil (5-IU) was introduced by chemical synthesis at various positions of three natural Py tracts: the tra NSS, the tra FS, and the adenoviral major late (AdML) Py tracts (Fig. 1A ▶). Because SXL and U2AF65 are efficiently cross-linked to Py tracts when exposed to short-wavelength UV light (Zamore et al. 1992; Valcarcel et al. 1993; Singh et al. 1995), it is necessary to ensure that the proteins cross-link at desired sites. A 5-IU can be cross-linked to certain amino acids (Tyr, Phe, His, Met) in the vicinity by using a 325-nm wavelength HeCd laser source, which eliminates photoagent-independent cross-links (Willis et al. 1993). The Py tracts differ in the length of their uridine tracts and binding affinity for U2AF65 and SXL. For example, SXL has ∼20- to 30-fold lower affinity for the AdML Py tract compared with that for the tra NSS Py tract, and no detectable binding for the tra FS Py tract (Valcarcel et al. 1993; H. Banerjee and R. Singh, unpubl. results). U2AF65 has about threefold lower affinity for the AdML Py tract and 100-fold lower affinity for the tra FS compared with that for the tra NSS Py tract (Valcarcel et al. 1993; H. Banerjee and R. Singh, unpubl. results). To account for the differences in binding affinities, protein concentrations were appropriately adjusted to obtain comparable binding (80%–90%) for all Py tracts, as determined experimentally by a gel mobility shift assay (data not shown). Second, we introduced, by site-directed mutagenesis, a single tryptophan residue within the linker region between RRMs at amino acid Y200 (SXL[W]) in SXL, and amino acids L235 (U2AF65[1W23]) or A339 (U2AF65[12W3]) in U2AF65 (Fig. 1B ▶). SXL and U2AF65 have no other tryptophan residues within the RNA-binding domain. Although no structural information was available for the two linker regions of U2AF65, the choice for the substitution of the Y200 position of SXL was guided by structure examination because it is located away from the RNA-binding face, and, based on the X-ray structure, its substitution with tryptophan would not be expected to perturb SXL structure (A. Rahn and R. Singh, unpubl. results). The RNA-binding properties of SXL and U2AF65 were not affected by the tryptophan substitutions, as determined by a filter-binding assay (A. Rahn and R. Singh, unpubl. results).
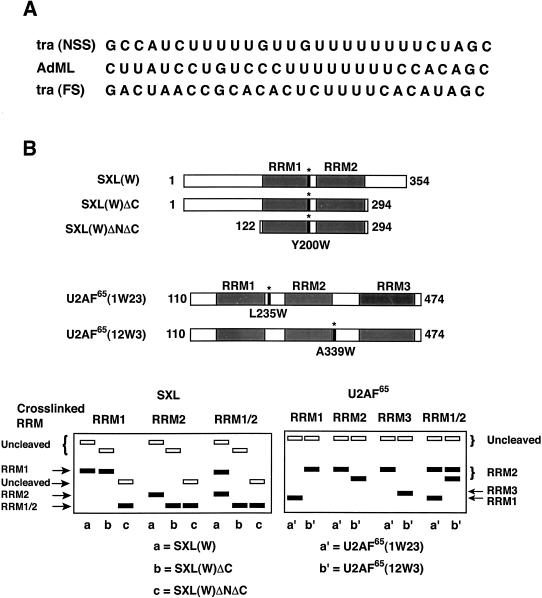
FIGURE 1.
Schematics of site-specific cross-linking assay. (A) Sequences of three natural Py tracts used for cross-linking. (B, top) Schematics of SXL(W), SXL(W)ΔC, SXL(W)ΔNΔC, U2AF65(1W23), and U2AF65(12W3) proteins. Shaded areas represent RRMs. Asterisks indicate the positions of tryptophan substitutions: tyrosine-200, SXL(W); leucine-235, U2AF65 (1W23); and alanine-339, U2AF65 (12W3). (Bottom) Diagrammatic representation of SDS–polyacrylamide gels showing the hypothetical positions of the radiolabeled protein fragments following the NCS cleavage of SXL(W), SXL(W)ΔC, SXL(W)ΔNΔC, U2AF65(1W23), and U2AF65(12W3). For SXL (left panel), if an RNA is cross-linked to RRM2, NCS cleavage of SXL(W)ΔC would generate a smaller labeled polypeptide than that observed with SXL(W). However, the size of the polypeptide would remain unaffected if it is cross-linked to RRM1. On the other hand, the NCS cleavage of SXL(W)ΔNΔC would generate a smaller labeled polypeptide fragment than that with SXL(W) regardless of whether RNA is cross-linked to RRM1 or RRM2. If an RNA is cross-linked to both RRMs (RRM1/RRM2), two cleavage products, of varying intensity, would appear in the same lane. For U2AF65 (right panel), if an RNA is cross-linked to RRM1, NCS cleavage would generate a smaller polypeptide for U2AF65(1W23) and a larger polypeptide for U2AF65(12W3). If an RNA is cross-linked to RRM3, NCS cleavage would generate a smaller polypeptide for U2AF65(12W3) and a larger polypeptide for U2AF65(1W23). If it is cross-linked to RRM2, NCS cleavage would generate a larger polypeptide for both proteins. If it is cross-linked to two RRMs (RRM1/RRM2), two cleavage products, of varying intensity, would appear in the same lane. For simplicity, other double combinations (RRM1/RRM3, RRM2/RRM3) are not shown. This strategy unambiguously identifies whether a given residue is cross-linked to RRM1, RRM2, or RRM3. Under these conditions, a portion of the input protein remains uncleaved (Mirfakhrai and Weiner 1993). Solid rectangles represent cleaved peptides corresponding to RRM1, RRM2, or RRM3, and empty rectangles represent uncleaved proteins.
SXL and U2AF65 proteins containing single tryptophan substitutions were used for site-specific cross-linking with each of the 5-IU RNAs that were radioactively labeled at the 5′-end. The cross-linked protein could be specifically cleaved at the tryptophan residue by using N-chlorosuccinimide (NCS; Mirfakhrai and Weiner 1993). It would generate different-size polypeptide fragments corresponding to RRM1 and RRM2 for SXL(W), RRM1 and RRM2–RRM3 for U2AF65(1W23), and RRM1–RRM2 and RRM3 for U2AF65(12W3). The identity of the cross-linked RRM was determined by electrophoretic separation of the NCS cleavage peptide fragments in a sodium dodecyl sulfate (SDS)–polyacrylamide gel and autoradiography. We also generated two small deletions in SXL(W), outside of the RRMs, in which either a portion of the C terminus (SXL[W]ΔC) or the N as well as the C termini (SXL[W]ΔNΔC) of SXL were deleted to alter their electrophoretic mobility, and thus allow for ready identification of the SXL RRMs; the SXL(W)ΔNΔC protein fragment corresponds to the SXL derivative that was used for the X-ray structure of SXL (Handa et al. 1999). The schematic of the NCS cleavage pattern shown in Figure 1B ▶ (bottom) was used for the assignment of cross-linked RRMs. Because the original protocol for NCS cleavage was technically cumbersome (Mirfakhrai and Weiner 1993), we modified this procedure such that extraction of the NCS cleavage reaction with chloroform circumvented several intermediate steps, allowing direct analysis in an SDS–polyacrylamide gel (see Materials and Methods for details).
SXL cross-linking
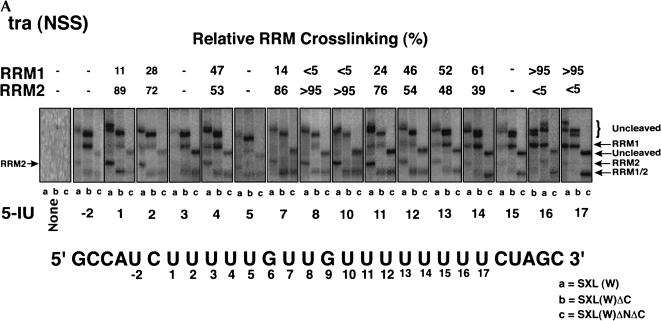
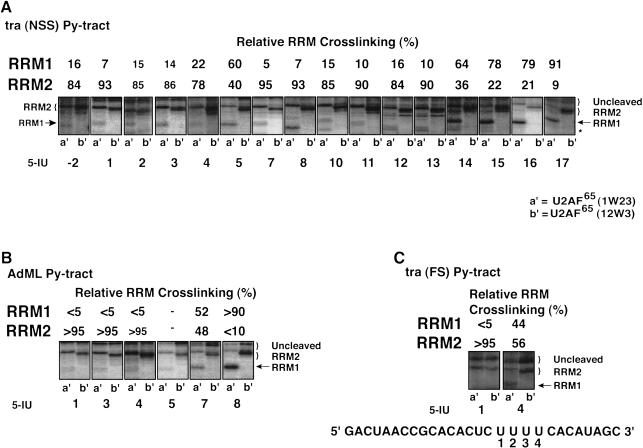
Sixteen RNAs that contained a single 5-IU at each of the positions within the NSS Py tract of tra were 5′-end-labeled, photocross-linked to SXL, and analyzed in an SDS–polyacrylamide gel after treatment with NCS (Fig. 2A ▶). As a test of specificity, there was no cross-linking in the absence of 5-IU (Fig.2A ▶, panel None). Positions U1, U7, U8, and U10 show particularly clear examples of the RRM2 pattern, in which the cleaved cross-linked species comigrated in lanes b,c. Positions 16 and 17 show exclusively the RRM1 pattern, in which the cleaved cross-linked species comigrated in lanes a,b, and had a faster mobility in lane c. Other positions gave a mixture of the RRM1 and RRM2 patterns, as indicated in Figure 2A ▶. For some positions such as U13, U14, and U15, the C-terminal deletion slightly increased cross-linking of RRM1 relative to RRM2 (Fig.2A ▶, cf. lanes a and b). It should be pointed out that these positions appear to lie at the junction of RRM1 and RRM2 binding (see below). We could not directly analyze the G6 and G9 positions because photocross-linking required a 5-IU.
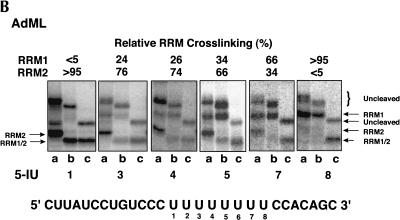
FIGURE 2.
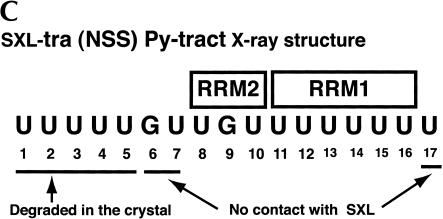
Site-specific cross-linking of SXL to 5-IU containing tra NSS (A) or AdML (B) Py tracts. Each of the 5′-end-labeled RNAs containing either no 5-IU (None) or a single 5-IU at various positions was cross-linked to SXL(W), SXL(W)ΔC, and SXL(W)ΔNΔC. The cross-linked protein was cleaved with NCS and resolved in an SDS–polyacrylamide gel. The positions of the 5-IU used for the cross-linking are shown below, and the percentage relative cross-linking to either RRM1 or RRM2 is shown above the autoradiograms; (–) weak cross-linking. For reference, the tra (NSS) and the AdML Py-tract sequences are shown. Lanes a, b, and c correspond to various SXL derivatives. The positions of the uncleaved protein (curly brackets) and of the cleaved fragments corresponding to RRM1 and RRM2 (arrows) are shown. (C) Summary of the X-ray structure of SXL and the NSS Py tract of tra (Handa et al. 1999). The boxes refer to RRM1 and RRM2, and lines below the sequence refer to the nucleotides that were either degraded or did not contact SXL in the X-ray structure.
We also analyzed the AdML Py tract, which has a uridine-octamer sequence similar to the SXL-binding sites found in Sxl and msl2 pre-mRNAs. Figure 2B ▶ shows that residues U1, U3, and U4 were primarily cross-linked to RRM2; residue U5 was cross-linked >60% to RRM2; residue U7 was cross-linked between 60% and 80% to RRM1; and residue U8 was cross-linked exclusively to RRM1. The tra FS Py tract could not be analyzed because SXL does not bind to this Py tract (Valcarcel et al. 1993).
Several conclusions can be drawn from the SXL photochemical cross-linking experiments. First, the orientation of SXL on both Py tracts is such that RRM1 is bound near the 3′-end of the Py tract and RRM2 near the 5′-end, consistent with the X-ray structure (Fig.2C ▶; Handa et al. 1999). Second, RRM1 is cross-linked to fewer residues than RRM2 on both Py tracts. Strong cross-linking of RRM1 is limited to the residues U16 and U17 of the tra NSS, and to the residues U7 and U8 of the AdML Py tract. Third, U11 and U12 of the tra NSS Py tract preferentially cross-link to RRM2, and U13 and U14 to both RRM1 and RRM2. Finally, U1–U5, U7, and U17 residues of the NSS Py tract, which are missing in the SXL X-ray structure (Fig.2C ▶; Handa et al. 1999), apparently interact with SXL.
U2AF65 cross-linking
Similar cross-linking experiments were performed with U2AF65 (see Fig.1B ▶ for schematic). Figure 3A ▶ shows that positions U-2, U1–U4, and U7–U13 of the tra NSS Py tract were preferentially cross-linked to RRM2. Residue U5 was weakly cross-linked and U14 was efficiently cross-linked to both RRM1 and RRM2, and residues U15–U17 were cross-linked to RRM1. For the AdML Py tract, residues U1, U3, and U4 were efficiently cross-linked to RRM2; residue U5 showed weak but preferential cross-linking to RRM2; residue U7 was cross-linked to both RRM1 and RRM2; and residue U8 was cross-linked only to RRM1 (Fig.3B ▶). For the tra FS Py tract, residue U1 was cross-linked to RRM2, and residue U4 was cross-linked to both RRMs (Fig.3C ▶).
FIGURE 3.
Site-specific cross-linking of U2AF65 to 5-IU containing tra NSS (A), AdML (B), and tra FS (C) Py tracts. The 5-IU RNAs were cross-linked to U2AF65(1W23) (lane a′) and U2AF65(12W3) (lane b′). Positions of various fragments are indicated; an asterisk represents positions of variant size band(s), which is prominent in lane 8a′, of unknown identity. For reference, the tra (FS) Py-tract sequence is shown in panel C. For further details, see legend to Figure 2 ▶.
The following conclusions can be drawn from the U2AF65 cross-linking experiments. First, on all three Py tracts, the orientation of the RRM1 and RRM2 of U2AF65 is similar to those of SXL. Second, RRM1 is cross-linked to fewer residues than RRM2, and strong cross-linking of RRM1 is limited to the last one–four residues, depending on the length of the Py tract. Third, both RRMs are cross-linked to certain residues, which are likely at the RRM junction. The basis for the increased cross-linking of the U4 and U5 residues of the NSS Py tract of tra to the RRM1 of SXL and U2AF65, respectively, remains unclear. Fourth, both RRM1 and RRM2 are cross-linked to all of the Py tracts, including the shortest Py tract (tra FS), which has only four contiguous uridines. Finally, surprisingly, RRM3 is not cross-linked to any of the 5-IU tested.
Importance of residues U1–U5 in SXL binding
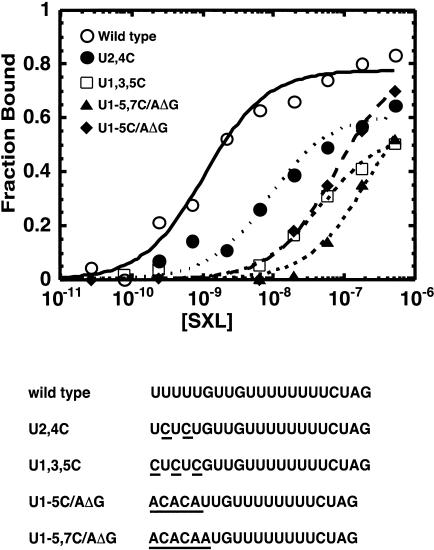
Figure 2A ▶ shows that residues U1–U5, U7, and U17 are cross-linked to SXL. These residues were either absent from or did not contact SXL in the X-ray structure (Fig.2C ▶; Handa et al. 1999). Because cross-linking does not necessarily correlate with binding affinity, residues U1–U5 and U17 were mutagenized, and the mutant RNAs were analyzed for SXL binding in vitro in a filter-binding assay. As shown in Figure 4 ▶, mutation of residues U1–U5 significantly reduced SXL binding. For example, mutation U2,4C reduced the binding affinity by ∼22-fold, and mutations U1,3,5C, U1-5C/AΔG, and U1-6C/AΔG reduced the binding affinity by 50-fold or more. The U17C mutation had a modest threefold effect on RNA binding (H. Banerjee and R. Singh, unpubl. data). These data show that residues U1–U5 are important for SXL binding in vitro, consistent with previous selection-amplification and chemical probing studies (Singh et al. 1995, 2000).
FIGURE 4.
RNA binding of SXL to wild type and various mutants of the tra NSS Py tract. Molar concentrations of recombinant GST–SXL are shown on the X-axis, and the fraction of RNA bound on the Y-axis. The sequences of various mutants are shown at the bottom, and the mutations are underlined. For simplicity, only the Py tracts are shown.
Importance of residues U1–U5 in splicing regulation by SXL
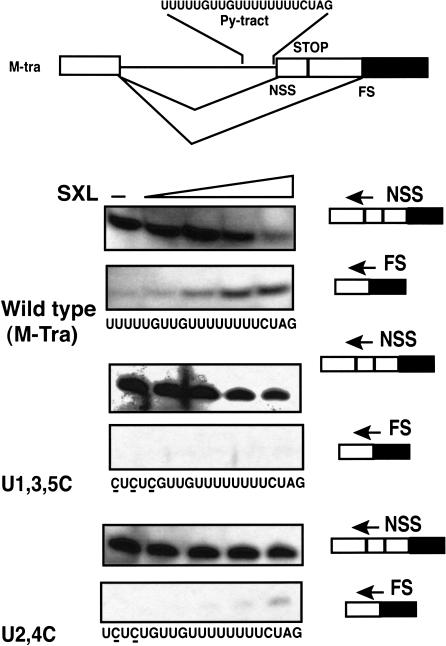
Next, we asked if residues U1–U5 are important for splicing regulation by SXL. We used an in vitro 3′-splice-site switching assay, which faithfully recapitulates SXL regulation in vivo (Valcarcel et al. 1993). As expected, the NSS 3′-splice site is used in the absence of SXL (Fig.5 ▶). For the wild-type pre-mRNA, addition of recombinant SXL mediated 3′-splice-site switching in a concentration-dependent manner, as seen by the decreased splicing to the proximal NSS 3′-splice site and increased splicing to the distal FS 3′-splice site. However, point mutations of U1–U5 residues significantly reduced splice-site switching by SXL. Therefore, we conclude that residues U1–U5 are also important for splicing regulation by SXL, consistent with the RNA-binding data (Fig.4 ▶).
FIGURE 5.
3′-splice-site switching by SXL. (Top) Schematics of the sex-specific alternative splicing of the wild-type pre-mRNA substrate. The boxes represent exons, and the lines represent introns. The alternative 3′-splice sites (NSS and FS), the NSS Py tract (SXL-binding site), and the translation stop codon (STOP) are shown. (Bottom) SXL-mediated splice-site switching for the wild-type (M-tra), and the U1,3,5C and U2,4C mutants (underlined) of the NSS Py tract of tra. Precursor RNAs were spliced in an HeLa nuclear extract in the absence (−) or presence of different concentrations of GST–SXL (0.08 μM, 0.25 μM, 0.76 μM, and 2.3 μM), and the spliced products were analyzed by primer extension assay using radiolabeled NSS or FS splice junction primers indicated by the arrows.
DISCUSSION
A systematic biochemical analysis with two proteins and three natural Py tracts revealed new information on Py-tract recognition by RRMs. Although it is possible that the nature of 5-IU cross-linking, which is efficient with only certain amino acids (Willis et al. 1993), may have influenced the result for a particular position, taken together, the large data set presented here supports a compelling trend (Figs.2,3 ▶ ▶). RRM1 is bound near the 3′-end of the Py tract, and RRM2 is bound near the 5′-end, which is consistent with all known X-ray structures of the proteins containing two RRMs (Varani and Nagai 1998; Antson 2000). Both RRMs of SXL and only RRM1 and RRM2 of U2AF65 together constitute the minimal Py-tract recognition domain; the RRM3 motif of U2AF65 is not cross-linked to any of the Py tracts. There are two unusual observations. First, certain 5-IU positions are cross-linked to both RRM1 and RRM2 for all of the Py tracts tested. Second, the size of the cross-linking site for RRM2 is variable and can greatly exceed the RNA site size for other RRMs (Varani and Nagai 1998; Antson 2000). The efficient cross-linking site of RRM1 is limited to two–four uridines at the 3′-end of the Py tract. Below, we present a model for Py-tract recognition that explains various observations, and discuss the biological significance of this mode of RNA recognition.
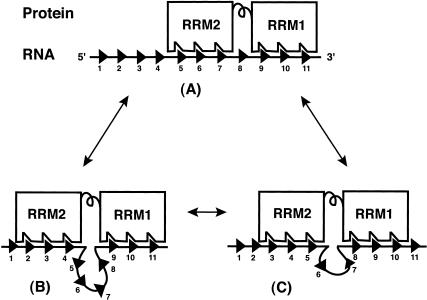
A model: Multiple registers and RNA looping
We postulate that RRM1 as well as RRM2 of both proteins can bind to the Py tract in multiple registers, and RNA at the junction of RRM1 and RRM2 can form a loop of variable length, resulting in an ensemble of complexes. Although the number of different possible RNA–protein complexes could exceed 40, assuming that each RRM contacts 4 residues in the 17-nt-long Py tract of tra, only three examples are shown in Figure 6 ▶. The actual number of residues contacted by each RRM could be different. In complex A, both RRMs are bound to two adjacent uridine stretches, which is similar to the sharp boundary observed at the RRM junction in the X-ray structure of SXL (Fig.2C ▶; Handa et al. 1999). In complex B, whereas the binding of RRM1 is unchanged, the binding of RRM2 is shifted by 3 nt upstream. As a consequence, residues 5–8 are looped out. In complex C, the binding of RRM1 is shifted by 1 nt upstream and of RRM2 by 2 nt, resulting in a loop of 2 nt. It should be emphasized that the size and location of the loop will vary depending on the interactions of each RRM. The complexes shown here as well as those not shown are likely in rapid equilibrium, shown by the arrows. It is possible that various RNA–protein complexes have different binding energies. This unusual situation of multiple modes of binding likely arises because it is hard for an RRM to discriminate between adjacent uridines. Several observations led to this proposal. First, the size of the cross-linking site is variable, which can be large for RRM2 on longer Py tracts. Second, both RRMs are cross-linked to certain residues on all of the Py tracts tested. Third, although efficient cross-linking of RRM1 is restricted to the 3′-end of the Py tract, it does not cross-link to a unique set of residues. The preference of RRM1 near the 3′-end of the Py tract could limit the number of possible complexes. Fourth, a lack of duplicated RRM2–RRM1 cross-linking pattern supports the possibility that in the majority of complexes a single protein molecule binds to the Py tract.
FIGURE 6.
Model for Py-tract recognition—multiple modes of binding. Both RRM1 and RRM2 contact adjacent uridine stretches (A). In comparison to complex A, only RRM2 binds in a different register in complex B, and both RRMs bind in different registers in complex C. Triangles represent nucleotides or uridines. Double arrows indicate that these complexes are in equilibrium. For simplicity, only three binding sites are shown for each RRM. The actual number of complexes will depend on the length of the Py tract and the number of nucleotides that interact with each RRM.
What does the model explain? First, it explains how certain residues can be cross-linked to both RRMs. The possibility of subpopulations of various complexes implies that a particular residue could contact either RRM1 or RRM2 in a given complex. However, the reason we observe cross-linking of the same residue to both RRMs is because the experiment reflects data from a mixture of complexes. Second, the model provides the basis for the extended site size of RRM2. Although the site size for each RRM is typically 4–7 residues for a given complex (Shamoo et al. 1994; Varani and Nagai 1998; Antson 2000), the extended site size for RRM2 can be explained by RNA looping for certain members of the ensemble. The malleable nature of uridine-rich sequences, which are known to be largely unstructured, makes them particularly suited for adopting flexible RNA loops (Saenger 1994). Third, this model could explain previous chemical interference/protection and saturation mutagenesis data for SXL, in which the binding site appeared larger than would have been expected for two RRMs (Singh et al. 2000). Fourth, in the absence of RRM3 cross-linking, we favor that only two of the three RRMs of U2AF65, and both RRMs of SXL, likely contributed to the selection of the consensus sequences (Singh et al. 1995). This suggestion is consistent with the interaction of RRM3 with other splicing factors such as mBBP/SF1 and SAP155 (Berglund et al. 1998; Gozani et al. 1998). However, we cannot exclude the possibility that RRM3, which was shown to be required for Py-tract binding (Zamore et al. 1992), lacks appropriate amino acids for 5-IU cross-linking. Fifth, the positioning of the RRM1 of U2AF65 at the 3′-end of the Py tract would allow interaction with the small subunit (U2AF35; Zhang et al. 1992; Rudner et al. 1998), and thus ready recognition of the 3′-splice-site AG dinucleotide by U2AF35 (Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999a). Sixth, the model explains how SXL could bind uridine tracts of variable length in the SXL-regulated pre-mRNAs, and how U2AF65 could bind to natural Py tracts that differ widely in length. Finally, although a comparison of the cross-linking pattern (Fig.2A ▶) and the SXL X-ray structure (Fig.2C ▶) indicates differences in binding, we favor the idea that the SXL structure represents only one member of the ensemble, perhaps chosen because of the crystal contacts that favored crystallization (Handa et al. 1999).
The cross-linking pattern observed here is inconsistent with an alternative model(s) in which RRM1 and RRM2 would contact a fixed site in a single register with a sharp boundary at the junction of two RRMs. In this scenario, somehow RRM2 would contact a much larger site at the same time, which is inconsistent with the known interactions for RRMs (Varani and Nagai 1998; Antson 2000), including the SXL structure in which RRM2 contacts only 3 residues (Handa et al. 1999). Alternatively, if RRM1 and RRM2 are constrained with respect to each other upon RNA binding, the entire protein could bind at different locations. This would result in an increased site size for RRM1 and an increased number of residues cross-linked to both RRMs. The observed cross-linking pattern—restricted cross-linking of RRM1 to the 3′-end of the Py tract and cross-linking of only 2–4 residues to both RRMs—is incompatible with the alternative model.
Although recognition of a Py tract in multiple modes explains several observations, it begs the question of whether or how one member of the ensemble might convert to another. Either RRM2 could slide on the RNA with respect to RRM1 or the protein may undergo dissociation/reassociation. We also do not understand the molecular basis for the preferential cross-linking of RRM1 to the 3′-end of the Py tract; perhaps there is a signal at the 3′ boundary. The exact site size for an RRM or the amount of each complex cannot be accurately determined because the observed cross-linking depends on the product of occupancy and the intrinsic cross-linking efficiency of a given binding site. We cannot distinguish whether RRM1 is flexible or constrained with respect to RRM2 when bound to RNA in solution. The X-ray structure shows that although two RRMs of SXL are tethered by a flexible linker in the absence of RNA (Crowder et al. 1999), the linker region forms a short 310-helix upon RNA binding. In addition, the RRM2 of SXL when bound to RNA interacts with RRM1 as well as the linker region (Handa et al. 1999). Similar interactions are also seen for HuD (Wang and Tanaka Hall 2001). However, the energetics of these interactions for SXL as well as the structure of the first linker region of U2AF65 when bound to RNA remain to be determined.
Biological significance
Our model has important biological consequences. In general, the strength of 3′-splice sites correlates well with the length of the adjacent Py tracts, and the binding affinity for U2AF65. Two possibilities for this correlation have been envisioned. All three RRMs of U2AF65 could contact longer Py tracts, whereas only a subset of the RRMs contact shorter Py tracts. Alternatively, all three RRMs of U2AF65 could contact Py tracts, regardless of the length of the Py tract (Zamore et al. 1992), but the number of interactions differ depending on the length of the Py tract. We find that both RRM1 and RRM2 of U2AF65 are cross-linked to all three Py tracts, including the shortest FS Py tract of tra, and that RRM3 is not cross-linked to any of the Py tracts tested, including the longest, NSS Py tract of tra. Therefore, we propose that changes in the number of interactions with only RRM1 and RRM2, the number of possible complexes or both, rather than interactions with a subset of RRMs (one, two, or three RRMs), provide the most likely basis for different affinities for various-length Py tracts, and thus 3′-splice-site strength. In this scenario, longer Py tracts would provide additional registers or binding sites, thereby resulting in increased apparent binding affinity. For example, if an RNA offers a single register for binding, only one of the possible encounters with the protein will lead to productive binding; others would require continued sampling until the correct register is found. In contrast, if there are multiple correct registers, encounters with any of them will be productive, thereby increasing the chances of finding the binding site. A homopolymeric sequence like poly(U) provides a much larger set of binding sites because different registers, rather than being contiguous, extensively overlap, thereby offering a significantly large advantage in increasing the apparent binding affinity (for further discussion, see Kelly et al. 1976; Draper and von Hippel 1978).
Our proposal that an ensemble of complexes recognizes the Py tracts is also relevant to RNA recognition by other proteins that recognize less complex, repeating sequences, such as the mRNA poly(A) tails (Deo et al. 1999), the AU-rich sequences that control mRNA stability (Wang and Tanaka Hall 2001), the GU-rich sequences that modulate polyadenylation efficiency (Colgan and Manley 1997; Richter 1999), the U-rich sequences that control translation (Millard et al. 2000), and the CUG repeats that have been implicated in human diseases (Philips et al. 1998). To our knowledge, recognition of single-stranded RNAs that lack intramolecular base-pairing is very different from RNA recognition by other well-characterized RRM proteins that specifically recognize unique sequences and/or RNA structures such as hairpins and bulges (Nagai 1996; Frankel 1999; Williamson 2000), and from DNA recognition by most of the sequence-specific DNA-binding proteins that bind in a single register (Tan and Richmond 1998). However, individual features of this model share resemblance with other well-studied systems. For example, the nucleosomes and single-stranded DNA-binding proteins (ssb), which lack sequence specificity, can bind in multiple registers (Lohman and Ferrari 1994; Flaus et al. 1996). In addition, the σ factor of the bacterial RNA polymerase binds to two sequence motifs that can be separated by a spacer that shows limited (2–3 nt) variability in length (Reznikoff et al. 1985), and many transcription regulators bind, usually as homo- or heterodimers, to two sequence motifs that can be separated by a spacer (loop) that shows considerable variability in length (Pashne and Gann 1997; Schleif 2000). However, each motif has a single register. Finally, it is tempting to speculate that this situation of binding in multiple modes may have evolved because it could allow fine tuning of the binding affinity without changing sequence specificity, and that certain sequence defects could modify some registers without affecting others. This situation is particularly important in higher organisms, where multiple factors or subunits bind to overlapping sites to regulate gene expression.
In conclusion, these studies provide insight into Py-tract recognition. These findings should be applicable to the entire family of proteins that recognize uridine-rich sequences, contain multiple RRMs, and show sequence and structural similarities with SXL (Chen and Shyu 1995; Keene 1999). The modified NCS cleavage protocol and the tryptophan-based domain mapping strategy described here provide a useful means for defining recognition of RNA, DNA, or protein sequences by any protein that has multiple recognition domains. Our detailed biochemical analysis underscores the importance of independent evaluation of conclusions from structural studies.
MATERIALS AND METHODS
Plasmid constructs
U2AF65Δ1–63 (Zamore et al. 1992) contained two tryptophans at positions 92 and 475. To eliminate the tryptophans, the sequence corresponding to amino acids 110–474 was PCR-amplified using primers F (GCGGGATCCATGCAAGCTGCGGGTCAGA) and R (CGGAATTCAAGATCCACGCGGAACCAGGTCGACGA AGTCCCGGCGGTGATAA). The PCR product was digested with BamHI/EcoRI and cloned into pGEX-2T to obtain GST–U2AF65–110–474. For oligonucleotide-directed mutagenesis, inverse PCR was carried out using GST–SXL, M-Tra (Valcarcel et al. 1993), or GST–U2AF65–110–474 as templates and the following sets of primers. The PCR product was blunt-ended by T4 DNA polymerase, ligated, and transformed into Escherichia coli.
SXL(W) (in pGEX-2T): F (5′-TGGGCACGTCCCGGCGGAG A-3′) and R (5′-GGAAACCTTAAGCCGCTTG-3′).
U2AF65(1W23) (in pGEX-2T): F (TGGCTGGTAGTCGTGA GG) and R (TGGCCTGGCATGTCAGAGAAC).
U2AF65(12W3) (in pGEX-2T): F (TCCCACACTCGCCCTC TG) and R (TGGAAGAATGCCACGCTGGTG).
U1,3,5C: F (5′-GTTGTTTTTTTTCTAGTGTCATATTG-3′) and R (5′-GAGAGGATGGCACTGGATCAGAATCTG-3′).
U2,4C: F (5′-GTTGTTTTTTTTCTAGTGTCATATTG-3′) and R (5′-AGAGAGATGGCACTGGATCAGAATCTG-3′).
U1-5 A/CΔG: F (5′-GTTGTTTTTTTTCTAGTGTCATATTG-3′) and R (5′-TGTGTGATGGCACTGGATCAGAATCTG-3′).
U1-5,7 A/CΔG: F (5′-CATGTTTTTTTTCTAGTGTCATATGT G-3′) and R (5′-TGTGTGATGGCACTGGATCAGAATCTG-3′).
U17C: F (5′-CCTAGTGTCATATTGTGTGAA-3′) and R (5′-AAAAAAACAACAAAAAGATGGCA-3′).
Sequences corresponding to amino acids 1–294 (SXL[W]ΔC) and 122–294 (SXL[W]ΔNΔC) were PCR-amplified from SXL(W) in pGEX–2T using the following sets of primers:
Sxl(W)ΔC: F (5′-ATCCTCCAAAATCGGATCTG-3′) and R (5′-GGAATTCTCACTTGCCATGCTCCTCAGCCAAC-3′).
Sxl(W)ΔNΔC: F (5′-CGGGATCCGCAAGCAACACCAACGA TTGT-3′) and R (5′-GGAATTCTCACTTGCCATGCTCCTC AGCCAAC-3′).
The BamH1/EcoR1 fragment of SXL(W), U2AF65(1W23), and U2AF65(12W3), in a pGEX–2T plasmid, and the PCR products for SXL(W)ΔC and SXL(W)ΔNΔC were cloned into a pGEX–6P vector. This vector provided the cleavage site for PreScission protease for the synthesis of recombinant proteins lacking GST, which has four undesirable tryptophans. The transcripts in Figure 4 ▶ contained an additional 26 nt upstream and 45 nt downstream of the tra NSS Py tract. All of the clones were confirmed by restriction digestion and direct sequencing.
Expression of recombinant SXL and U2AF65 proteins
Expression of GST fusion proteins and their purification was essentially as described (Valcarcel et al. 1993). For SXL or U2AF65 derivatives lacking GST, the recombinant proteins (SXL[W], SXL[W]ΔC, SXL[W]ΔNΔC, U2AF65[1W23], and U2AF65[12W3]) were expressed in bacteria. Following the manufacturer’s protocol, the recombinant proteins were cleaved from the glutathione–Sepharose matrix by using PreScission protease (Amersham Pharmacia Biotech.), and dialyzed overnight against Buffer D (20 mM HEPES at pH 8.0, 0.2 mM EDTA, 20% glycerol, 0.05% NP-40, and 1 mM dithiothreitol).
RNA–protein cross-linking assay
RNAs containing a single 5-IU at various positions were chemically synthesized on an Applied Biosystems DNA synthesizer. The RNA was 5′-end-labeled and incubated with protein in a 100-μL reaction containing 10 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 50 mM KCl, 0.5 U/μL RNasin, 0.09 μg/μL acetylated bovine serum albumin, 0.15 μg/μL tRNA, and appropriate dilutions of recombinant proteins in Buffer D. Protein concentrations were estimated by the staining of an SDS–polyacrylamide gel with Coomassie Brilliant Blue R-250, with bovine serum albumin as a standard. The RNA–protein mix was aliquoted into a semi-UV cuvette (Fisher Scientific) and irradiated with a 325-nm HeCd laser source (300 mW/cm2) at room temperature for 10 min (Willis et al. 1993). A 10-μL aliquot of the cross-linked reaction was mixed with 40 μL of 50% acetic acid/urea and 10 mg/mL N-chlorosuccinimide. This mixture was vortexed and incubated at room temperature for 2.5 min, and extracted twice with chloroform:isoamyl alcohol (24:1). The aqueous phase was vacuum dried at 65°C for 20 min. The lyophilized samples were resuspended in 100 μL of 2× SDS loading buffer, and 10 μL was separated by electrophoresis in a 15% SDS–polyacrylamide gel. The SDS–polyacrylamide gel was dried, exposed to a phosphorimager screen, and quantified using ImageQuant (Molecular Dynamics).
RNA–protein filter binding assay
Binding, under conditions described in the RNA–protein cross-linking section above, was done in triplicates and quantified using ImageQuant (Molecular Dynamics) for the bound fraction (nitrocellulose membrane) and the unbound fraction (DEAE membrane; Wong and Lohman 1993). The curves were generated using a standard two-state binding model in Kaleidagraph.
Transcription and RNA splicing assay
RNA transcription, splicing, and primer extension were carried out as described (Valcarcel et al. 1993; Singh et al. 1995).
Acknowledgments
We thank Tom Cech, Mike Yarus, Olke Uhlenbeck, Tom Blumenthal, Art Pardi, Debra Wuttke, Bob Boswell, Jens Lykke-Anderson, and members of the Singh laboratory for critical reading of the manuscript; and Olke Uhlenbeck for discussions on the model. We are also grateful to Anne Gooding for generously synthesizing a large number of RNAs, and Tad Koch for the laser source. A.R. and W.D. were supported in part by training grant GM07135 from the National Institutes of Health. This work was supported by grant GM58576 from the National Institutes of Health to R.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2131603.
REFERENCES
- Antson, A.A. 2000. Single-stranded-RNA binding proteins. Curr. Opin. Struct. Biol. 10: 87–94. [DOI] [PubMed] [Google Scholar]
- Bashaw, G.J. and Baker, B.S. 1997. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89: 789–798. [DOI] [PubMed] [Google Scholar]
- Berglund, J.A., Abovich, N., and Rosbash, M. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes & Dev. 12: 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103: 367–370. [DOI] [PubMed] [Google Scholar]
- Burd, C.G. and Dreyfuss, G. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621. [DOI] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA world (eds. R.F. Gesteland et al.), pp. 525–560. Cold Spring Harbor Laboratory Press, New York.
- Chen, C.Y. and Shyu, A.B. 1995. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 20: 465–470. [DOI] [PubMed] [Google Scholar]
- Cline, T.W. and Meyer, B.J. 1996. Vive la difference: Males vs females in flies vs worms. Annu. Rev. Genet. 30: 637–702. [DOI] [PubMed] [Google Scholar]
- Colgan, D.F. and Manley, J.L. 1997. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 11: 2755–2766. [DOI] [PubMed] [Google Scholar]
- Crowder, S.M., Kanaar, R., Rio, D.C., and Alber, T. 1999. Absence of interdomain contacts in the crystal structure of the RNA recognition motifs of Sex-lethal. Proc. Natl. Acad. Sci. 96: 4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo, R.C., Bonanno, J.B., Sonenberg, N., and Burley, S.K. 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98: 835–845. [DOI] [PubMed] [Google Scholar]
- Draper, D.E. and von Hippel, P.H. 1978. Nucleic acid binding properties of Escherichia coli ribosomal protein S1. I. Structure and interactions of binding site I. J. Mol. Biol. 122: 321–338. [DOI] [PubMed] [Google Scholar]
- Flaus, A., Luger, K., Tan, S., and Richmond, T.J. 1996. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. 93: 1370–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner, J., Zhang, M., Valcarcel, J., and Green, M.R. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP–branchpoint interaction. Genes & Dev. 11: 1864–1872. [DOI] [PubMed] [Google Scholar]
- Forch, P., Merendino, L., Martinez, C., and Valcarcel, J. 2001. Modulation of msl-2 5′ splice site recognition by Sex-lethal. RNA 7: 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel, A.D. 1999. If the loop fits. Nat. Struct. Biol. 6: 1081–1083. [DOI] [PubMed] [Google Scholar]
- Gebauer, F., Merendino, L., Hentze, M.W., and Valcarcel, J. 1998. The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA 4: 142–150. [PMC free article] [PubMed] [Google Scholar]
- Gozani, O., Potashkin, J., and Reed, R. 1998. A potential role for U2AF–SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18: 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granadino, B., Penalva, L.O.F., Green, M.R., Valcarcel, J., and Sanchez, L. 1997. Distinct mechanisms of splicing regulation in vivo by the Drosophila protein Sex-lethal. Proc. Natl. Acad. Sci. 94: 7343–7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17: 100–107. [DOI] [PubMed] [Google Scholar]
- Handa, N., Nureki, O., Kurimoto, K., Kim, I., Sakamoto, H., Shimura, Y., Muto, Y., and Yokoyama, S. 1999. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 398: 579–585. [DOI] [PubMed] [Google Scholar]
- Hastings, M.L. and Krainer, A.R. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13: 302–309. [DOI] [PubMed] [Google Scholar]
- Horabin, J.I. and Schedl, P. 1993. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol. Cell. Biol. 13: 7734–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., Hoshijima, K., Sakamoto, H., and Shimura, Y. 1990. Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature 344: 461–463. [DOI] [PubMed] [Google Scholar]
- Inoue, M., Muto, Y., Sakamoto, H., Kigawa, T., Takio, K., Shimura, Y., and Yokoyama, S. 1997. A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal. J. Mol. Biol. 272: 82–94. [DOI] [PubMed] [Google Scholar]
- Ito, T., Muto, Y., Green, M.R., and Yokoyama, S. 1999. Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor. EMBO J. 18: 4523–4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaar, R., Roche, S.E., Beall, E.L., Green, M.R., and Rio, D.C. 1993. The conserved pre-mRNA splicing factor U2AF from Drosophila: Requirement for viability. Science 262: 569–573. [DOI] [PubMed] [Google Scholar]
- Kanaar, R., Lee, A.L., Rudner, D.Z., Wemmer, D.E., and Rio, D.C. 1995. Interaction of the sex-lethal RNA binding domains with RNA. EMBO J. 14: 4530–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, J.D. 1999. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc. Natl. Acad. Sci. 96: 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, R.L., Wang, J., Bell, L., and Kuroda, M.I. 1997. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387: 195–199. [DOI] [PubMed] [Google Scholar]
- Kelly, R.C., Jensen, D.E., and von Hippel, P.H. 1976. DNA “melting” proteins. IV. Fluorescence measurements of binding parameters for bacteriophage T4 gene 32-protein to mono-, oligo-, and polynucleotides. J. Biol. Chem. 251: 7240–7250. [PubMed] [Google Scholar]
- Kim, I., Muto, Y., Watanabe, S., Kitamura, A., Futamura, Y., Yokoyama, S., Hosono, K., Kawai, G., Takaku, H., Dohmae, N., et al. 2000. Interactions of a didomain fragment of the Drosophila sex-lethal protein with single-stranded uridine-rich oligoribonucleotides derived from the transformer and Sex-lethal messenger RNA precursors: NMR with residue-selective [5–2H]uridine substitutions. J. Biomol. NMR 17: 153–165. [DOI] [PubMed] [Google Scholar]
- Lallena, M.J., Chalmers, K.J., Llamazares, S., Lamond, A.I., and Valcarcel, J. 2002. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 109: 285–296. [DOI] [PubMed] [Google Scholar]
- Lee, A.L., Kanaar, R., Rio, D.C., and Wemmer, D.E. 1994. Resonance assignments and solution structure of the second RNA-binding domain of sex-lethal determined by multidimensional heteronuclear magnetic resonance. Biochemistry 33: 13775–13786. [DOI] [PubMed] [Google Scholar]
- Lohman, T.M. and Ferrari, M.E. 1994. Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63: 527–570. [DOI] [PubMed] [Google Scholar]
- Merendino, L., Guth, S., Bilbao, D., Martinez, C., and Valcarcel, J. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Millard, S.S., Vidal, A., Markus, M., and Koff, A. 2000. A U-rich element in the 5′ untranslated region is necessary for the translation of p27 mRNA. Mol. Cell. Biol. 20: 5947–5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirfakhrai, M. and Weiner, A.M. 1993. Chemical Cleveland mapping: A rapid technique for characterization of crosslinked nucleic acid–protein complexes. Nucleic Acids Res. 21: 3591–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J. 2000. Intron recognition comes of AGe. Nat. Struct. Biol. 7: 14–16. [DOI] [PubMed] [Google Scholar]
- Nagai, K. 1996. RNA–protein complexes. Curr. Opin. Struct. Biol. 6: 53–61. [DOI] [PubMed] [Google Scholar]
- O’Neil, M.T. and Belote, J.M. 1992. Interspecific comparison of the transformer gene of Drosophila reveals an unusually high degree of evolutionary divergence. Genetics 131: 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashne, M. and Gann, A. 1997. Transcriptional activation by recruitment. Nature 386: 569–577. [DOI] [PubMed] [Google Scholar]
- Penalva, L.O., Lallena, M.J., and Valcarcel, J. 2001. Switch in 3′ splice site recognition between exon definition and splicing catalysis is important for sex-lethal autoregulation. Mol. Cell. Biol. 21: 1986–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips, A.V., Timchenko, L.T., and Cooper, T.A. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280: 737–741. [DOI] [PubMed] [Google Scholar]
- Potashkin, J., Naik, K., and Wentz-Hunter, K. 1993. U2AF homolog required for splicing in vivo. Science 262: 573–575. [DOI] [PubMed] [Google Scholar]
- Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12: 340–345. [DOI] [PubMed] [Google Scholar]
- Reznikoff, W.S., Siegele, D.A., Cowing, D.W., and Gross, C.A. 1985. The regulation of transcription initiation in bacteria. Annu. Rev. Genet. 19: 355–387. [DOI] [PubMed] [Google Scholar]
- Richter, J.D. 1999. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 63: 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner, D.Z., Kanaar, R., Breger, K.S., and Rio, D.C. 1998. Interaction between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol. Cell. Biol. 18: 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger, W. 1994. Defining terms for nucleic acids. In Principles of nucleic acid structure (ed. C.R. Cantor), pp. 21–23. Springer-Verlag, New York.
- Sakamoto, H., Inoue, K., Higuchi, I., Ono, Y., and Shimura, Y. 1992. Control of Drosophila Sex-lethal pre-mRNA splicing by its own female-specific product. Nucleic Acids Res. 20: 5533–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita, E. and Sakamoto, H. 1994. Characterization of RNA binding specificity of the Drosophila sex-lethal protein by in vitro ligand selection. Nucleic Acids Res. 22: 4082–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif, R. 2000. Regulation of the L-arabinose operon of Escherichia coli. Trends Genet. 16: 559–565. [DOI] [PubMed] [Google Scholar]
- Schutt, C. and Nothiger, R. 2000. Structure, function and evolution of sex-determining systems in Dipteran insects. Development 127: 667–677. [DOI] [PubMed] [Google Scholar]
- Shamoo, Y., Abdul-Manan, N., Patten, A.M., Crawford, J.K., Pellegrini, M.C., and Williams, K.R. 1994. Both RNA-binding domains in heterogeneous nuclear ribonucleoprotein A1 contribute toward single-stranded-RNA binding. Biochemistry 33: 8272–8281. [DOI] [PubMed] [Google Scholar]
- Singh, R. 2002. RNA–protein interactions that regulate pre-mRNA splicing. Gene Expression 10: 79–92. [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J., and Green, M.R. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176. [DOI] [PubMed] [Google Scholar]
- Singh, R., Banerjee, H., and Green, M.R. 2000. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor sex-lethal. RNA 6: 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, C.W. and Valcarcel, J. 2000. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 25: 381–388. [DOI] [PubMed] [Google Scholar]
- Sosnowski, B.A., Belote, J.M., and McKeown, M. 1989. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 58: 449–459. [DOI] [PubMed] [Google Scholar]
- Sosnowski, B.A., Davis, D.D., Boggs, R.T., Madigan, S.J., and McKeown, M. 1994. Multiple portions of a small region of the Drosophila transformer gene are required for efficient in vivo sex-specific regulated RNA splicing and in vitro sex-lethal binding. Dev. Biol. 161: 302–312. [DOI] [PubMed] [Google Scholar]
- Tan, S. and Richmond, T.J. 1998. Eukaryotic transcription factors. Curr. Opin. Struct. Biol. 8: 41–48. [DOI] [PubMed] [Google Scholar]
- Vagner, S., Vagner, C., and Mattaj, I.W. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF65 to couple 3′-end processing and splicing. Genes & Dev. 14: 403–413. [PMC free article] [PubMed] [Google Scholar]
- Valcarcel, J., Singh, R., Zamore, P.D., and Green, M.R. 1993. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362: 171–175. [DOI] [PubMed] [Google Scholar]
- Valcarcel, J., Gaur, R.K., Singh, R., and Green, M.R. 1996. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA. Science 273: 1706–1709. [DOI] [PubMed] [Google Scholar]
- Varani, G. and Nagai, K. 1998. RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct. 27: 407–445. [DOI] [PubMed] [Google Scholar]
- Wang, J. and Bell, L.R. 1994. The Sex-lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes & Dev. 8: 2072–2085. [DOI] [PubMed] [Google Scholar]
- Wang, X. and Tanaka Hall, T.M. 2001. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 8: 141–145. [DOI] [PubMed] [Google Scholar]
- Will, C.L. and Luhrmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Williamson, J.R. 2000. Induced fit in RNA–protein recognition. Nat. Struct. Biol. 7: 834–837. [DOI] [PubMed] [Google Scholar]
- Willis, M.C., Hicke, B.J., Uhlenbeck, O.C., Cech, T.R., and Koch, T.H. 1993. Photocrosslinking of 5-iodouracil-substituted RNA and DNA to proteins. Science 262: 1255–1257. [DOI] [PubMed] [Google Scholar]
- Wong, I. and Lohman, T.M. 1993. A double-filter method for nitrocellulose-filter binding: Application to protein–nucleic acid interactions. Proc. Natl. Acad. Sci. 90: 5428–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S., Romfo, C.M., Nilsen, T.W., and Green, M.R. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. [DOI] [PubMed] [Google Scholar]
- Yanowitz, J.L., Deshpande, G., Calhoun, G., and Schedl, P.D. 1999. An N-terminal truncation uncouples the sex-transforming and dosage compensation functions of sex-lethal. Mol. Cell. Biol. 19: 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Patton, J.G., and Green, M.R. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614. [DOI] [PubMed] [Google Scholar]
- Zhang, M., Zamore, P.D., Carmo-Fonseca, M., Lamond, A.I., and Green, M.R. 1992. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc. Natl. Acad. Sci. 89: 8769–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W.J. and Wu, J.Y. 1996. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol. Cell. Biol. 16: 5400–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio, D.A. and Blumenthal, T. 1999a. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838. [DOI] [PubMed] [Google Scholar]
- ———. 1999b. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5: 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]