Abstract
The poliovirus cis-acting replication element (CRE) templates the uridylylation of VPg, the protein primer for genome replication. The CRE is a highly conserved structural RNA element in the enteroviruses and located within the polyprotein-coding region of the genome. We have determined the native structure of the CRE, defined the regions of the structure critical for activity, and investigated the influence of genomic location on function. Our results demonstrate that a 14-nucleotide unpaired terminal loop, presented on a suitably stable stem, is all that is required for function. These conclusions complement the recent analysis of the 14-nucleotide terminal loop in the CRE of human rhinovirus type 14. The CRE can be translocated to the 5′ noncoding region of the genome, at least 3.7-kb distant from the native location, without adversely influencing activity, and CRE duplications do not adversely influence replication. We do not have evidence for a specific interaction between the CRE and the RNA-binding 3CDpro complex, an essential component of the uridylylation reaction, and the mechanism by which the CRE is coordinated and orientated during the reaction remains unclear. These studies provide a detailed overview of the structural determinants required for CRE function, and will facilitate a better understanding of the requirements for picornavirus replication.
Keywords: Picornavirus, enterovirus, polymerase, replication, protease, uridylylation, VPg
INTRODUCTION
Poliovirus is the prototype picornavirus, and serves as a model for our understanding of the structure and replication of a range of related human and animal pathogens. The single-stranded 7.4-kb positive (messenger)-sense RNA genome contains all the necessary signals for virus replication, and is infectious as a cDNA, enabling the application of reverse genetic approaches to dissect structure and function (Racaniello and Baltimore 1981; Evans 1999). The poliovirus genome encodes a single polyprotein of 220 kD, which is cotranslationally processed by the virus-encoded 2Apro and 3Cpro proteases, and protease-containing precursors. The primary proteolytic cleavage events divide the genome into three precursors: P1, P2, and P3. P1 is subsequently processed to yield the four structural proteins, multiple copies of which assemble to form the ∼30-nm diameter icosahedral virus particle. The P1 region is dispensable for replication, and can be replaced with a reporter gene to facilitate the direct quantification of genome replication by enzymatic or colorimetric assay (Percy et al. 1992). The P2 and P3 regions contain the cis-acting nonstructural proteins required for virus replication, including the 2Apro and 3Cpro proteases, the 3Dpol RNA-dependent RNA polymerase, and VPg (3B), a 22 amino acid peptide that serves as a primer for the initiation of positive and negative strand genome replication and that is covalently attached to the virus genome (Flanegan et al. 1977; Nomoto et al. 1977; Kitamura et al. 1980).
The 5′ and 3′ noncoding regions (NCR) are extensively structured domains implicated in genome replication and translation, the latter via direct recruitment of ribosomes to the internal ribosome entry site (IRES) in the 5′ NCR (Pelletier and Sonenberg 1988; Skinner et al. 1989; Andino et al. 1999). The IRES is preceded by an 88-nt ‘‘cloverleaf’’ (CL) structure that forms a ribonucleoprotein complex with a cellular protein (PCBP2) and the virus-encoded 3CDpro precursor (Gamarnik and Andino 1997; Parsley et al. 1997). The precise role of the 3′ NCR in virus replication remains unclear; there is a high degree of sequence and structural conservation in the region, and defined mutations that disrupt the structure have a negative influence on replication (Rohll et al. 1995; Pilipenko et al. 1996; Melchers et al. 1997; Wang et al. 1999). However, the 3′ NCR is not absolutely required for replication—picornavirus 3′ NCR sequences can be functionally interchanged, and deletion of the 3′ NCR does not prevent virus replication (Todd et al. 1997; Meredith et al. 1999).
A third class of cis-acting replication elements (CRE) have recently been identified in picornavirus genomes. These consist of short stem-loop structures predicted to contain an internal or terminal loop with three unpaired A nucleotides. There is little other sequence conservation, and the CRE structures are located in diverse regions of the genome—in the 2C-encoding region of poliovirus (and by homology and covariance analysis, other human enteroviruses) (Goodfellow et al. 2000), in the capsid-encoding region of human rhinovirus type 14 (McKnight and Lemon 1998) and the cardioviruses (Lobert et al. 1999), the region encoding the 2Apro of human rhinovirus type 2 (Gerber et al. 2001a) and the 5′ noncoding region (NCR) of foot and mouth disease virus (Mason et al. 2002). To prime the initiation of replication, VPg must be uridylylated, a process involving the covalent addition of two uridine nucleotides to a tyrosine residue conserved in all picornaviruses (Crawford and Baltimore 1983; Kuhn et al. 1988). This reaction can be reproduced in vitro using synthetic VPg, purified 3Dpol, 3CDpro, 32P-UTP, and a poly(A) template (Paul et al. 1998). More recently, it has been demonstrated that the CRE is a specific and efficient template for VPg uridylylation, and that sequences in the conserved terminal loop are critical for this function (Paul et al. 2000; Rieder et al. 2000; Gerber et al. 2001a).
In this study we have extended our original observations on the structure and function of the poliovirus CRE. We have confirmed the computationally predicted CRE structure by enzymatic mapping, performed a rational analysis of the structural requirements for CRE function, including generating truncated elements that exhibit wild-type activity, and demonstrated that the conserved location of the CRE within the polyprotein-encoding region of the poliovirus genome is not a prerequisite for function.
RESULTS
Biochemical mapping of poliovirus CRE structure
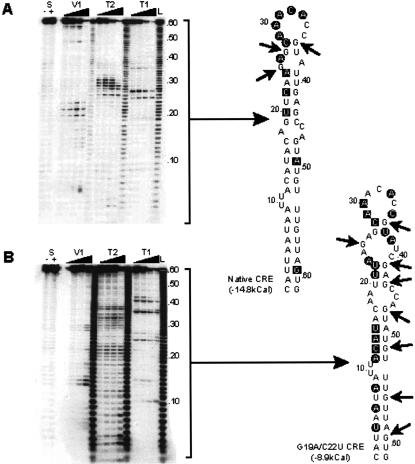
The poliovirus CRE (Fig. 1A ▶) was originally defined as a 61-nt structure consisting of an incompletely base-paired stem and terminal loop. The structure of the stem-loop was predicted by computational and covariance analysis, supported by limited site-directed mutagenesis (Goodfellow et al. 2000). We mapped the native structure enzymatically using single- and double-strand specific ribonucleases to verify the modeled structure. End-labeled CRE transcripts were refolded and subjected to partial digestion with RNAse T1, T2, or V1, which preferentially cleave at unpaired G nucleotides, any unpaired nucleotide and base-paired regions, respectively. Two CRE variants (G19A/U40C and G19A/C22U/U40C1) previously shown to not support replication (Goodfellow et al. 2000) were also analyzed to determine whether the mutations exerted their effect by grossly altering the CRE structure, or by disrupting key contact nucleotides required for function.
FIGURE 1.
Poliovirus CRE predicted structure and modification. (A) Predicted structure of the native poliovirus type 3 CRE. The designation and location of the stem regions (based on Goodfellow et al. 2000) are indicated together with modifications made to these regions. Nucleotides in bold italics in Stem 3 represent defined mutations, the substitution of two or more of which render the CRE inactive (Goodfellow et al. 2000). The G nucleotide in bold italics in Inv1 (inversion of the Stem 1 region) represents a substitution made to avoid introducing a termination codon to the sequence. The conserved functional A1A2A3CA motif in the terminal loop of the CRE is highlighted in bold. (B) Schematic diagram of virus genomes and subgenomic replicons used in this study. The noncoding regions are indicated as thin solid horizontal lines. Coding regions are indicated as wide open (unmodified poliovirus sequence) or filled (modified by replacement or mutagenesis) boxes. The region encoding chloramphenicol acetyl transferase expressed by subgenomic replicons is indicated as a solid black filled box labeled CAT. The disrupted native CRE sequence (located within the region encoding the virus 2C protein) is shown with an X. Subgenomic replicon-based cassette vectors (pT7/Rep3/SL3c and pT7/Rep3c) were constructed by the introduction of unique restriction sites flanking a ‘‘stuffer region’’ (indicated by the character ∼), which introduces a frameshift to the encoded polyprotein (see D). Relevant naturally present restriction sites used to introduce sequences into the 5′ NCR are indicated. The panel on the right-hand side defines the role the particular construct was used for in these studies, and whether there is a functional CRE in the native location within the region of the polyprotein encoding 2C. (C) The cassette region of pT7/Rep3c and pT7/Rep3/SL3c. The sequence through the engineered cassette region is shown, together with relevant translation products (indicated using single-letter amino acid codes). Without modification the cassette region is translated in the open reading frame (ORF) highlighted ‘‘0’’, which terminates 19 residues after the carboxyl terminus of the CAT protein. Digestion with Bss HII and Sma I allows replacement of the removed cassette stuffer region (shown in bold text) with complementary oligonucleotides that include the CRE sequence of interest. The oligonucleotides were designed to restore the polyprotein open reading frame to the +1 frame as shown. The YG dipeptide cleaved by the virus 2Apro protease is shown in double height type, below the filled blocks indicating the short spacer region that flanks the inserted sequence and the start of the 2Apro protease open reading frame.
The enzymatic mapping of the native CRE structure strongly supported the computer-generated model, with the exception that the short stem (Stem 4, Fig. 1A ▶) at the distal end of the structure was not base-paired under the conditions used in the assay (Fig. 2A ▶). The two or three substitutions within Stem 3 of the CRE in G19A/U40C and G19A/C22U/U40C resulted in many additional single- and double-strand cleavages (Fig. 2B,C ▶). The additional T1 cleavages (unpaired G nucleotides) throughout the CRE suggest considerable instability of the structure. In certain positions, for example C13 and G51 or C28 and G36 in G19A/U40C, the different RNAse cleavages generated contradictory results, which probably indicates that the RNA adopts multiple conformations due to its inherent instability. The proposed structure of the enzymatically mapped native poliovirus type 3 CRE is shown in Figure 2D ▶.
FIGURE 2.
Structural mapping of the poliovirus CRE. (A) The autoradiograph of a representative mapping acrylamide gel is shown alongside a diagram of the ribonuclease cleavages indicated on the predicted structure of the CRE. T1, V1, and T2 cleavages are indicated with an arrow, black square, or black circle, respectively. The few positions cleaved with both V1 and T2 are indicated with an open square. Lane L is a hydroxidegenerated 1bp ladder produced from the same probe, lanes S are the probe alone incubated in the absence (−) or presence (+) of 100 mM sodium chloride (which is required for V1 ribonuclease activity). (A) The native poliovirus type 3 CRE. (B) The poliovirus type 3 CRE containing two substitutions in the Stem 3 region (see Fig. 1A ▶) of G19A and C22U. (C) The poliovirus type 3 CRE containing three substitutions in the Stem 3 region of G19A, C22U, and U40C. (D) The proposed structure of the native poliovirus type 3 CRE derived from ribonuclease mapping studies.
Functional analysis of the poliovirus CRE
Construction and use of a cassette system for CRE mutagenesis and duplication
pT7/Rep3 is a plasmid containing a cDNA for a poliovirus subgenomic replicon2 previously constructed by replacing the majority of the P1-encoding region (the virus capsid proteins) with a chloramphenicol acetyltransferase (CAT) reporter gene, engineered in-frame with the truncated polyprotein (Percy et al. 1992; Barclay et al. 1998).
RNA generated in vitro from pT7/Rep3 replicates following transfection of suitable mammalian cells; the level of resulting CAT activity can be readily quantified as a measure of genome replication (Percy et al. 1992). We have previously shown that the CRE functions when inserted between the CAT and P2-encoding regions of pT7/Rep3 (in the cDNAs pT7/Rep3/SL3/#1 and pT7/Rep3/SL3/#3, see Goodfellow et al. 2000). To facilitate the analysis of CRE function in this location (1.1-kb 5′ to the native position) a cassette system was constructed by introducing unique Bss HII and Sma I sites flanking a stuffer sequence that disrupted the reading frame of the encoded polyprotein (Fig. 1B,C ▶). The stuffer fragment could then be replaced with complementary annealed oligonucleotides forming the CRE sequence of interest that restore the correct reading frame to the polyprotein.
The cassette modification was made to pT7/Rep3 and a derivative replicon cDNA, designated pT7/Rep3/SL3, which contains the SL3 mutation within the native CRE sequence in the region of the cDNA encoding the 2C protein. The SL3 mutation consists of eight substitutions within the CRE, designed to disrupt the structure but not the encoded protein, which we have previously demonstrated destroys the ability of the RNA (either genomic length, derived from the cDNA pT7/FLC/SL3, or subgenomic from pT7/Rep3/SL3) to replicate after transfection of suitable mammalian cells (Goodfellow et al. 2000).
The cassette vectors were distinguished by the addition of a ‘‘c’’ suffix to the replicon name (pT7/Rep3c and pT7/Rep3/SL3c; Fig. 1B,C ▶), digested with Bss HII and Sma I, and the stuffer region replaced with complementary annealed oligonucleotides (Table 1 ▶) forming native (WT) or nonfunctional (SL3) CRE sequences. The identity of the inserted sequence and restoration of the open reading frame (Fig. 1C ▶) was confirmed by DNA sequencing, and the resulting cDNA name amended by appending a superscripted suffix to indicate the insert. RNA generated in vitro was transfected into HeLa cells, and the level of CAT activity was quantified following transfection of HeLa cells (Fig. 3B ▶). Due to the presence of the frameshift, RNA generated from the cassette vectors (pT7/Rep3c or pT7/Rep3/SL3c) could not replicate, as indicated by the failure to produce any CAT activity (Fig. 3B ▶). The control cDNAs, pT7/Rep3/SL3/#1 and pT7/Rep3/SL3/#3, which contain native CRE sequences in the noncassette backbone pT7/Rep3/SL3 (Goodfellow et al. 2000), generated CAT levels sufficient to convert 50 or 80%, respectively, of the substrate under the conditions used for this assay. This was optimized to exhibit maximal CAT activity in the linear range, to emphasize potential differences between replicons carrying one or two functional CRE sequences, and is equivalent to ∼20%–30% conversion when the control RNA from pT7/Rep3 does not exhaust the substrate (data not shown). RNA generated from the cassette-derived cDNA pT7/Rep3/SL3cWT replicated at a level equivalent to RNA from pT7/Rep3/SL3/#3 (from which it essentially differs only in the presence of the restriction sites used to insert the CRE sequence, see Goodfellow et al. 2000; Fig. 1C ▶). As expected, the nonfunctional CRE sequence SL3 did not restore replication competence to RNA generated from pT7/Rep3/SL3cSL3.
TABLE 1.
Oligonucleotides used during this study
| Name | Oligonucleotide sequence | Notes |
| Cassette Vector oligonucleotides | ||
| WT | CGCGCCATTAATAATTACATACAGTTCAAGAGCAACACCGTATTGAGCCAGTATGTTTGTTAGTG | a |
| SL3 | CGCGCCATTAATAATTATATCCAATTTAAATCCAAACACCGTATCGAGCCAGTATGTTTGTTAGTG | |
| Clal | CGCGCCATTAATAATTACATACAATCGATGAGCAAACACCGTAATCGATCAGTATGTTTGTTAGTG | |
| Synth2 | CGCGCCCGGGTAAGAGCAAACACCGTATTACCCGGG | |
| Synth2mut7 | CGCGCCCGGGTAAGAGCAACACCGTATTACCCGGG | b |
| Synth3 | CGCGTCCGGGTAAGAGCAAACACCGTATTACCCGGG | |
| Synth4 | CGCGTTCGGGTAAGAGCAAACACCGTATTACCCGGG | |
| Synth5 | CGCGTTTGGGTAAGAGCAAACACCGTATTACCCGGG | |
| Inv1 | CGCGCGTGTTGTTTTACATACAGTTCAAGAGCAAACACCGTATTGAGCCAGTATGTAATAATTAC | c |
| Inv3 | CGCGCCATTAATAATTACATACACGAGTTGAGCAAACACCGTAAACTTGCAGTATGTTTGTTAGTG | |
| Oligonucleotides for insertion into the 5′ NCR | ||
| WTAUG | GATCATTAATAATTACAACAGTTCAAGAGCAAACACCGTATTGAGCCAGTATGTTGTTAGTGATGGGAGCT | d |
| Synth2 | GATCCCGGGTAAGAGCAAACACCGTATTACCCGGGATGCGAGCT | |
| Synth6 | GATCCCGGTATCAAACACCGATACCGGGATGGGAGCT | |
| Oligonucleotides used to generate T7 transcripts | e | |
| Synth2 | CCCGGGTAAGAGCAAACACCGTATTACCCGGG | |
| Synth2mut7 | CCCGGGTAAGAGCAACACCGTATTACCCGGG | |
| Synth3 | TCCGGGTAAGAGCAAACACCGTATTACCCGGG | |
| Synth4 | TTCGGGTAAGAGCAAACACCGTATTACCCGGG | |
| Synth5 | TTTGGGTAAGAGCAAACACCGTATTACCCGGG | |
| C28U | CATTAATAATTACATACAGTTCAAGAGAAACACCGTATTGAGCCAGTATGTTTGTTAGTG | |
| Mut7 (A29C) | CATTAATAATTACATACAGTTCAAGAGCAACACCGTATTGAGCCAGTATGTTTGTTAGTG | |
| Mut6 (A30C) | CATTAATAATTACATACAGTTCAAGAGCAACACCGTATTGAGCCAGTATGTTTGTTAGTG | |
| Mut2 (A31G) | CATTAATAATTACATACAGTTCAAGAGCAACACCGTATTGAGCCAGTATGTTTGTTAGTG | |
| G19A/C22U | CATTAATAATTACATACATTAAGAGCAAACACCGTATTGAGCCAGTATGTTTGTTAGTG | |
| G19A/C22U/U40C | CATTAATAATTACATACATTAAGAGCAAACACCGTATGAGCCAGTATGTTTGTTAGTG | |
| Miscellaneous oligonucleotides | ||
| IG73 | TAATACGACTCACTATAGGG (A) | f |
aThe poliovirus genome-sense strand only is shown. In the case of oligonucleotide pairs, designed for insertion into either the replicon-based cassette region or the 5′ NCR hypervariable region, the complementary oligonucleotide sequence is indicated for clarity.
bThe double underlined C nucleotide is identical to the mut7 mutation defined in (Rieder et al. 2000).
cThe double underline G nucleotide replaces a T, which would have introduced a termination codon to the sequence. The complementary base pairing is retained.
dInsertions into the hypervariable region include the underlined initiation codon for the polyprotein. The double underlined nucleotides were exchanged from the native sequence to avoid introducing an initiation condon (see Fig. 1A ▶).
eT7 transcripts were generated from antisense templates containing a 3′ extension complementary to the T7 promoter oligonucleotide IG73. The sequence complementary to the template oligonucleotide is shown, excluding the T7 promoter region. See text and Figure 1 ▶ for a description of variations from the native sequence that are indicated with double underlining.
fT7 promoter oligonucleotide. In certain cases an additional A nucleotide (complementary to a T included in the antisense partner oligonucleotide) was included at the 3′ end to increase T7 promoter activity and yield.
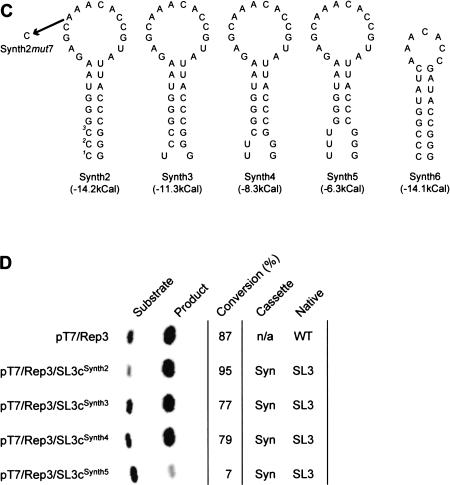
FIGURE 3.
CAT activity of CRE cassette vector-derived replicons. (A) Reference diagram indicating the cassette and native locations for CRE sequences. Refer to Figure 1B ▶ for further details. (B) Replication activity of CAT-expressing subgenomic replicons containing duplications of the CRE. The name of the replicon cDNA is indicated in the left-hand column next to a phosphorimager-generated image of the 14C-labeled substrate and monoacetylated CAT products separated by thin-layer chromatography. The proportion of substrate converted to product is indicated (in percent). The columns labeled ‘‘Cassette’’ and ‘‘Native’’ indicate the CRE sequences present in the two positions of the genome; see (A). WT and SL3, respectively, refer to the native CRE sequence or a CRE containing eight nucleotide substitutions designed to disrupt the CRE structure (Goodfellow et al. 2000). WT* indicates native CRE sequences as previously published (Goodfellow et al. 2000). N/A indicates not applicable, where no CRE sequences are present at a particular location. (C) Predicted structures of synthetic CRE sequences used in replication studies. Synth3 to Synth5 are derived from Synth2 by sequential replacement of one to three CG base pairs (highlighted with numerical superscripts in Synth2) with UG pairs. These are shown separated to emphasize the differences, but are predicted to adopt standard Watson-Crick base pairing. The single-point mutation of A1 to C in the 1A2A3CA motif, introduced during the construction of Synth2mut7 is also indicated. (D) Replication of CAT-expressing subgenomic replicons containing synthetic CRE sequences. Layout and labeling as in (B), Syn indicates the presence of a synthetic CRE sequence. Each assay shown in (B) or (D) used normalized levels of total cell protein (5–50 μg). Comparison of replication activity between (B) and (D) cannot be made as the control (pT7/Rep3) reaction in (B) was allowed to progress to completion to emphasize differences in the replication of dual CRE containing replicons.
Duplication of a functional CRE in a replicon
The cassette vector (pT7/Rep3c) constructed in a replication-competent backbone allowed the influence of two functional CRE elements on genome replication to be investigated. These studies were conducted to determine if there was competition between CRE elements. Complementary oligonucleotides for the native and nonfunctional CREs were inserted into pT7/Rep3c, producing pT7/Rep3cWT and pT7/Rep3cSL3, respectively, and the replication competence of resulting RNA determined as before. Monoacetylated CAT substrate conversion of ∼70%–80% was similar to the levels observed with replicons containing a single functional CRE, such as RNA generated from pT7/Rep3/SL3/#3 or pT7/Rep3/SL3cWT (Fig. 3B ▶). The demonstration that RNA from pT7/Rep3/SL3cWT and pT7/Rep3cWT replicate to equivalent levels suggests that there is no significant competition between duplicated CRE sequences in a poliovirus subgenomic replicon. We further investigated the reduced level of replication by the cassette-derived replicons (and the related pT7/Rep3/SL3/#1 or pT7/Rep3/SL3/#3), as we suspected that the modifications near the junction of the 2Apro cleavage site (between P1 and P2, see Fig. 1C ▶) may influence polyprotein processing. Quantification of the products of in vitro coupled transcription/translation assays demonstrated significantly reduced processing at the P1/P2 junction of proteins encoded by CRE sequences identical to or derived from the wild-type sequence (data not shown).
Structural requirements for CRE function in replicon-based cassette systems
The cassette replicons were used to analyze the structural requirements for CRE function by the replacement of the cassette region with CRE-derived sequences containing inversions, deletions, and substitutions (Fig. 1A ▶; Tables 1, 2 ▶ ▶), followed by in vitro transcription, RNA transfection, and CAT assay. CRE sequences from which Stem 1 or Stem 1 and Stem 2 were precisely deleted were incapable of supporting replication (pT7/Rep3/SL3cΔStem1, pT7/Rep3/SL3cΔStem1+2; Table 2 ▶). In contrast, inversion of Stem 1 or Stem 3 (in pT7/Rep3/SL3cInv1, pT7/Rep3/SL3cInv3; Fig. 1A ▶; Table 2 ▶), or replacement of Stem 3 with a synthetic complementary sequence exhibiting similar stability (pT7/Rep3/SL3cClaI), restored replication to levels comparable to RNA produced from the positive control pT7/Rep3/SL3cWT (Table 2 ▶). These results implied that the majority of the ‘‘stalk’’ region of the CRE, and certainly Stems 1 and 3, provide a structural framework upon which the functional terminal loop is presented.
TABLE 2.
CAT activity of CRE-containing replicons
| Replicon cDNA template | Activity |
| pT7/Rep3 | +++ |
| pT7/Rep3/SL3 | − |
| pT7/Rep3/SL3/#1 | + |
| pT7/Rep3/SL3/#3 | + |
| pT7/Rep3c | − |
| pT7/Rep3cWT | + |
| pT7/Rep3cSL3 | + |
| pT7/Rep3/SL3c | − |
| pT7/Rep3/SL3cWT | + |
| pT7/Rep3/SL3cSL3 | − |
| pT7/Rep3/SL3cΔStem1 | − |
| pT7/Rep3/SL3cΔStem1+2 | − |
| pT7/Rep3/SL3cInv1 | + |
| pT7/Rep3/SL3cInv3 | + |
| pT7/Rep3/SL3cClal | + |
| pT7/Rep3/SL3cSynth2 | +++ |
| pT7/Rep3/SL3cSynth3 | +++ |
| pT7/Rep3/SL3cSynth4 | +++ |
| pT7/Rep3/SL3cSynth5 | +/− |
| pT7/Rep3/SL3cSynth6 | − |
Notes: CAT activity indicated in the range +++ (100%; native activity of Rep3 replicon) to − (no detectable activity). + indicates ∼30% wild-type activity relative to the pT7/Rep3 replicon. See also Figure 3B ▶ and Figure 3D ▶. We have previously quantified both the RNA levels and the resulting CAT activity of replicating RNA generated from pT7/Rep3/SL3/#1 and pT7/Rep3/SL3/#1 (Goodfellow et al. 2000).
To test this conclusion further a ‘‘synthetic’’ CRE, designated Synth2, consisting of the terminal loop sequences (nucleotides 23–40 in Fig. 2D ▶) on an artificial Stem of comparable overall stability but reduced length was generated (Fig. 3C ▶). Transfection of HeLa cells with RNA generated in vitro from pT7/Rep3/SL3cSynth2 yielded similar levels of CAT activity to the parental replicon pT7/Rep3 (Fig. 3D ▶). To confirm that the CRE in pT7/Rep3/SL3cSynth2 functioned in essentially the same manner as the native structure a single point mutation of an A to a C within the terminal loop (A29C—analogous to the mut7 mutation of Rieder et al. 2000, which destroys CRE-templated uridylylation activity) was introduced to generate pT7/Rep3/SL3cSynth2mut7. HeLa cells transfected with RNA from pT7/Rep3/SL3cSynth2mut7 did not generate a CAT signal (data not shown). This result implies that the Synth2 CRE, containing only 18 nucleotides from the native CRE, is sufficient to confer wild-type CRE function if presented on a suitably stable stem structure. To investigate the influence of the predicted stem stability, we generated variants of pT7/Rep3/SL3cSynth2 lacking one, two, or three hydrogen bonds from the base of the stem (Fig. 3C ▶). Substitution of C1U, C1U/C2U, or C1U/C2U/C3U, generated pT7/Rep3/SL3cSynth3, pT7/Rep3/SL3cSynth4, and pT7/Rep3/SL3cSynth5, respectively, which were assayed for replication as before. With the exception of RNA generated from pT7/Rep3/SL3cSynth5, which exhibited 5% wild-type activity, all could replicate to the same level as RNA generated from pT7/Rep3 (Fig. 3D ▶).
Biochemical analysis of VPg uridylylation in poliovirus type 3
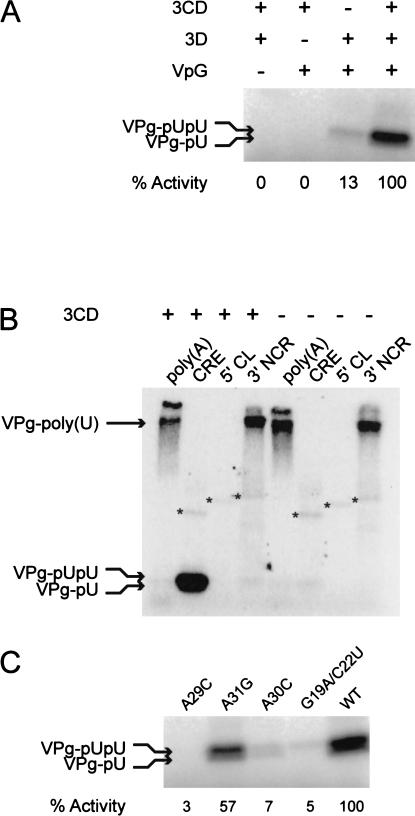
The CRE functions as a template for the uridylylation of VPg. We have reproduced the poliovirus type 1 (Mahoney) uridylylation assay reported by Wimmer and colleagues (Paul et al. 1998,2000; Rieder et al. 2000), using poliovirus type 3-derived components. The assay is essentially the same other than the use of 3.5 mM manganese rather than magnesium for optimal activity; formation of VPg-pU and VPg-pUpU necessitates the presence of the CRE and 3CDpro (Fig. 4A ▶), whereas VPg-poly(U) is the predominant product in reactions in which the CRE is replaced with poly(A) RNA, or polyadenylated 3′ NCR templates, and the 5′ CL cannot serve as a template (Fig. 4B ▶). 3Dpol-catalyzed terminal-transferase activity is apparent with all the templates tested other than poly(A) RNA (Fig. 4B ▶).
FIGURE 4.
The poliovirus type 3-based in vitro uridylylation assay. (A) CRE-mediated uridylylation of VPg requires the presence of 3CDpro and 3Dpol. The assay was constituted as described in Materials and Methods, with the exclusion of individual components as indicated in the table. Activity was quantified by measuring 32P-UMP incorporation onto VPg using a Bio-Rad phosphorimager and is shown normalized to the activity observed in the presence of all three proteins. (B) Formation of VPg-pU(pU) requires the CRE template and 3CDpro. The production of uridylylated VPg is dependent upon the presence of a template containing an unpaired A triplet (CRE, poly[A] or the 3′NCR) and CDpro, whereas polyadenylated RNA templates (poly[A] and the 3′NCR) yield VPg-poly(U) in the presence or absence of 3CDpro (indicated above the column labels). The reaction products of the 3Dpol terminal transferase activity are indicated with an asterisk. (C) CRE-templated uridylylation of VPg requires the A triplet in the A1A2A3CA motif and a base-paired stem. VPg-pU(pU) was quantified by Bio-Rad phosphorimager and normalized to the levels observed with an unmodified CRE template (WT). The mutations present in the CRE RNA template are indicated above the autoradiogram, and relate to the numbering scheme shown in Figure 1A ▶. The G19A/C22U mutations disrupt Stem 3 (Figs. 1A, 2B ▶ ▶; Goodfellow et al. 2000).
The requirement for a specific CRE structure and for sequences within the conserved loop was demonstrated using short RNA transcripts encompassing variant CREs in the uridylylation reaction (Table 1 ▶). The double mutant A19C/C22U, which renders the CRE nonfunctional in the replicon system (Goodfellow et al. 2000), exhibited only 5% activity of the WT CRE for the formation of VPg-pU and VPg-pUpU (VPg-pU[pU]). The substitution of A29C or A30C reduced formation of VPg-pU(pU) to less than 10% of WT CRE activity, whereas the A31G replacement reduced VPg-pU(pU) formation by over 40% (Fig. 4C ▶); a full-length cDNA bearing this mutation is known to yield quasi-infectious virus (Rieder et al. 2000). These results confirm the authenticity of a poliovirus type 3-based uridylylation assay for analysing CRE function in vitro.
The ability of the truncated Synth2 CRE elements to function in the in vitro uridylylation assay was determined. Derivatives of Synth2 with reduced stability had already been tested in the replicon system (Fig. 3D ▶; Table 2 ▶), and were assayed for uridylylation activity at temperatures ranging from 26–42°C to further investigate the influence of CRE stability upon function (Fig. 5 ▶). Of the temperatures used, maximal activity of the native CRE (WT) was observed at 29°C, to which all other activities were normalized. The Synth2 CRE maximally reached 88% of the uridylylation activity of the native CRE, but appeared to exhibit a slightly higher optimum temperature. In contrast, Synth3–5 showed significantly reduced activities at 29°C, and the temperature at which activity dropped below 10% of the native CRE structure was reduced in line with the stability of the stem-loop (Fig. 5 ▶). These results clearly demonstrate that the structure and stability of the CRE is critical for function. The apparent discrepancy between the CRE activity of Synth2–4 in subgenomic replicons and in the in vitro uridylylation assay (cf. Fig. 3D and 5A ▶ ▶, respectively) is probably due to the stabilizing influence of sequences flanking CRE in the replicon. Priming the in vitro uridylylation assay with full-length replicon-derived RNA transcripts containing Synth2, Synth3 (data not shown), or Synth4 generated wild-type levels of VPg-pU(pU), with only Synth5 being nonfunctional (Fig. 5B ▶).
FIGURE 5.
Uridylylation of VPg does not require the stem region of the CRE, but is influenced by CRE stability. In both panels the reaction temperature is indicated in the row labeled °C, and in (A) the amount of product is shown normalized to the maximal level observed with the unmodified native CRE (WT). The in vitro uridylylation assay was constituted with RNA templates containing a synthetic stem region and the native CRE terminal loop (Synth2–Synth5). Synth3 to Synth5 differ only in the number of hydrogen bonds stabilizing the base of the stem (see Fig. 3C ▶). (B) In vitro uridylylation of VPg primed with full-length RNA (∼5.5 kb) from subgenomic replicon cDNAs pT7/Rep3, pT7/Rep3/SL3, pT7/Rep3/SL3cSynth4, and pT7/Rep3/SL3cSynth5, labeled WT, SL3, Synth4, and Synth5, respectively.
RNA–protein interactions of the poliovirus CRE
The presence of 3CDpro is a requirement for the in vitro uridylylation of VPg (see above and Paul et al. 1998). The 3Cpro component of this complex is known to be an RNA binding protein (Andino et al. 1990), and this activity has been shown to be required for uridylylation (Paul et al. 2000). Because this suggests that 3CDpro may directly and specifically bind the CRE, we investigated this interaction in an electrophoretic mobility shift assay (EMSA). The positive control (5′ CL) readily formed a complex with recombinant 3CDpro (Fig. 6A ▶), the specificity of which was demonstrated by competition with unlabeled 5′ CL (data not shown). Under identical conditions 3CDpro only poorly formed a complex with the CRE (Fig. 6A ▶), and then only when present at high concentrations. To investigate the specificity of the 3CDpro–CRE ribonucleoprotein complex, unlabeled competitor RNA (in addition to the 5 μg tRNA already present) was added to each reaction at 10× or 100× molar excess. Two categories of competitor were used, consisting of predominantly structured (5′ CL, 3′ NCR, and CRE) or linear (CRE variants SL3, G19A/C22U, and G19A/C22U/U40C) RNA, all of which are characterized in this study, or previous work. All competitors readily disrupted the 3CDpro–CRE complex, with the unstructured CRE variants SL3, G19A/C22U, or G19A/C22U/U40C being particularly effective at even low molar excess (Fig. 6B ▶).
FIGURE 6.
Protein–RNA interactions of the poliovirus CRE. (A) Electrophoretic mobility shift assay (EMSA) of the interaction of the 5′CL or CRE with poliovirus CDpro. The identity of the input probe is indicated below the autoradiograph, with the position of the free probe indicated with the label ‘‘Probe’’. The lanes labeled (−) contain no added 3CDpro, the relative concentration of which is indicated with a triangle. The 3CDpro-containing complex is indicated with an asterisk (*). (B) Specificity of the 3CDpro interaction with the CRE. Formation of the 3CDpro/CRE ribonucleoprotein complex (indicated *) was assayed in the presence of no competitor (lane labeled 0) or competing (10× and 100×) molar excess of the native CRE, the 5′CL, 3′ NCR, or the nonfunctional CRE variants, SL3, G19A/C22U, or G19A/C22U/U40C. All lanes contain 3CDpro, as indicated by the bar below the figure, with the exception of the lane containing probe alone, which is labeled (−).
Positional dependence of the CRE
Our previous results suggest that there is no absolute positional dependence for CRE function, as the element can be moved at least 1.1 kb from the native position within the genome (Goodfellow et al. 2000). However, because in vivo studies of CRE function would be considerably facilitated by relocating the CRE to a noncoding region (where substitutions could be made without regard to the translation product) we investigated whether the CRE would function when engineered into the 5′ NCR. Taking advantage of two naturally occurring restriction sites in the poliovirus type 3 cDNA (Bam HI at 674 and Sac I at 752), the ∼100 nt hypervariable region of pT7/FLC/SL3 (Fig. 1B ▶) immediately preceding the translational initiation codon was replaced with complementary oligonucleotides forming a CRE-derived sequence. The oligonucleotides were designed to restore the sequence encoding the amino terminus of the polyprotein, but differed from the native CRE sequence by modification of the AUG triplet (at position 49–51) by the double substitution U15A/A49U (Fig. 1A ▶). This was necessary, as the presence of initiation codons within this region of the genome are known to be deleterious (Pelletier et al. 1988). This cDNA was designated pT7/FLC/SL3/HVCRE-AUG. Similarly, the truncated Synth2 CRE sequence (Fig. 3C ▶), previously shown to be functional in uridylylation assays (Fig. 5A ▶) and in replicon-based assays of virus replication (Fig. 3D ▶), and a Synth2-derivative lacking one of the three critical A residues A1A2A3CA in the unpaired terminal loop, were introduced into the hypervariable region to generate cDNAs designated pT7/FLC/SL3/HVSynth2 and pT7/FLC/SL3/HVSynth2ΔA, respectively.
Transfection of HeLa cells with normalized levels of RNA generated from pT7/FLC, pT7/FLC/SL3/HVCRE-AUG, or pT7/FLC/SL3/HVSynth2 yielded plaques at similar dilutions, indicating that the CRE was functional when positioned in the 5′ NCR (Fig. 7B,D,E ▶). The comparable specific infectivity and plaque size suggests that the CRE can contribute equally well to poliovirus replication when located over 3.7 kb away from the native location. As previously shown, the mutations in the CRE of pT7/FLC/SL3 rendered the cDNA noninfectious (Goodfellow et al. 2000; Fig. 7C ▶). In contrast, RNA generated from pT7/FLC/SL3/HVSynth2ΔA was poorly infectious, generating plaques about 1000-fold less frequently than pT7/FLC, pT7/FLC/SL3/HVCRE-AUG, or pT7/FLC/SL3/HVSynth2 (Fig. 7F ▶). This frequency suggests that a single compensating mutation was required to generate viable virus from RNA derived from pT7/FLC/SL3/HVSynth2ΔA. Six plaques were picked, the region between the Bam HI and Sac I sites (Fig. 7A ▶) was amplified by PCR, and the sequence determined—all viruses had acquired an additional A nucleotide, reforming the A1A2A3CA motif in the terminal loop of the Synth2 CRE. There were no other insertions or substitutions within the Synth2 CRE sequence (data not shown). A final cDNA containing a truncated CRE (Synth6; Fig. 3C ▶) inserted into the hypervariable region of pT7/FLC/SL3 did not yield plaques and could not be recovered by serial blind passage (data not shown). This suggests that the A1A2A3CA motif and the presence of a stable stem-loop (equivalent in stability to Synth2) are not the only features required for CRE function.
FIGURE 7.
The poliovirus CRE functions when located in the 5′ NCR of the genome. Complementary oligonucleotides for the CRE were directionally cloned between the Bam HI and Sac I sites located, respectively, in the 5′ NCR and region encoding the amino-terminus of VP4. The CRE sequence was modified by substitution of U15A/A49U (Fig. 1A ▶) to replace the AUG49–51 to generate pT7/SL3HVCRE-AUG. The same region of pT7/SL3 was also replaced with oligonucleotides encoding the Synth2 CRE, and a Synth2-derived CRE in which one of the conserved A triplet was deleted (generating pT7/SL3-HVSynth2 and pT7/SL3-HVSynth2ΔA, respectively). Normalized levels of RNA generated in vitro, and serial 10-fold dilutions, was transfected into HeLa cells and overlaid with agar. Plaques were visualized after 3 d by staining with crystal violet.
DISCUSSION
Cis-acting replication elements have been identified in the polyprotein-coding regions of enterovirus (Goodfellow et al. 2000), rhinovirus (McKnight and Lemon 1998; Gerber et al. 2001a), cardiovirus (Lobert et al. 1999), and aphthovirus (Mason et al. 2002) genomes. Despite the differences in absolute location and the relatively limited sequence similarity between the different CREs, it seems probable that the Picornaviridae replicate using an evolutionarily conserved mechanism that predates the divergence of the different genera. The CRE functions to template the addition of two uridines to VPg, a reaction that can be replicated in vitro using VPg peptide, 3CDpro, 3Dpol, and 32P-UTP supplemented with either poly(A) or CRE RNA. This assay has been used to identify novel CRE elements (Gerber et al. 2001a) and to define features of the CRE (Paul et al. 2000) necessary for the formation of VPg-pU(pU). These include the requirement for an A1A2A3CA motif in the terminal loop of the CRE, and a proposed mechanism by which A1 and A2 respectively function to template the addition of UMP to VPg (and VPg-pU) and to provide specificity to the reaction, likened to the ‘‘slide-back’’ mechanism described for nucleotidylylation reactions of bacteriophage and viral DNA polymerases (Salas et al. 1996; Gerber et al. 2001a). More recently, Yang et al. (2002) have used this assay to map functional residues of the rhinovirus type 14 CRE by demonstrating a direct relationship between the ability of the CRE to function in vitro with the ability to support replication.
We have used a combination of biochemical and virological approaches to better define the structural determinants of poliovirus CRE function, including the enzymatic mapping of the CRE and deletion/modification analysis of the base-paired CRE stem predicted by covariance analysis.
The ribonuclease mapping of the native CRE strongly supported our previous computer-predicted model of the structure (Fig. 2A ▶; Goodfellow et al. 2000), with the exception of the size of the loop at the top of the base-paired stem. Notably, single-strand specific T2 nuclease cleavages were observed at each nucleotide in the conserved A1A2A3CA motif. This clearly demonstrates that this critical functional domain of the CRE, which must intimately interact with VPg and 3Dpol during the uridylylation reaction, is not base paired. The only significant difference between the modeled CRE structure and that demonstrated by ribonuclease mapping is the status of the nucleotides that form Stem 4 (Fig. 1A ▶). Ribonuclease T1 (single-strand G nucleotides) and T2 cleavages at G27, G36, and C28, respectively, indicate that this region is not base paired. This would suggest that, taking account of the single-strand specific cleavages at G25 and A26, and the predicted unpaired A38, the poliovirus CRE is terminated by a 14-nucleotide unpaired loop. This conclusion is in line with the sequence conservation, covariance analysis of Stem 4, and the predicted structure of the CREs from all four lineages of human enteroviruses (Goodfellow et al. 2000, and data not shown), and is in agreement with the predictions made recently by Yang et al. (2002) following analysis of the rhinovirus type 14 CRE. Our studies show that 10 of the 14 nucleotides that form the terminal loop of the poliovirus CRE are unpaired (underlined in the enterovirus/rhinovirus consensus RNNNAARNNNNNNR with residues that influence function identified by Yang et al. 2002, highlighted in bold), including the critical functional residues in the first, fifth, and sixth positions, respectively, the G at the 5′ end of the loop (G25; Fig. 2A,D ▶) and the two A residues that template uridylylation (A29 and A30; Fig. 2A,D ▶). We do not have direct evidence that residues G25 and A38, which close the terminal loop (Fig. 2D ▶), are critical for poliovirus CRE function as they are in rhinovirus type 14 (Yang et al. 2002). We have only modified G25 in the context of the nonfunctional SL3 CRE (Goodfellow et al. 2000), which contains seven additional substitutions. However, a truncated synthetic CRE, designated Synth6 (Fig. 3C ▶), containing a six nucleotide unpaired loop, but lacking both the G and A residues in the correct positions relative to the A triplet, was unable to restore replication when inserted into the hypervariable region of the 5′ NCR (data not shown).
In contrast to the native CRE, the data from the mapping of the two mutant forms (G19A/C22U and G19A/C22U/U40C; Fig. 2B,C ▶) did not allow rational structural models to be developed. The significant increase in T1 cleavages throughout the sequence, generated as a consequence of the two or three introduced substitutions in Stem 3, suggests that the RNA is predominantly single stranded. We infer that Stem 3 plays a critical role in the overall stability of the CRE.
Although the structural stability contributed by Stem 3 is critical for poliovirus CRE function, the sequence per se is not. This was demonstrated using a subgenomic replicon-based cassette system that enabled the effects of deletions and modifications of the CRE to be monitored, and enabled us to investigate competition/interference between duplicated functional CREs (Fig. 1A ▶; Table 2 ▶). We propose that Stems 1, 2, and 3 (Fig. 1A ▶) of the poliovirus CRE provide the structural framework upon which the functional terminal loop sequences (nucleotides 25 to 38; Fig. 1A ▶) are presented, with neither the base-paired stem regions, nor the bulge loops between Stem 1/2 and Stem 2/3 contributing directly to CRE function.
This conclusion is further supported by the demonstration that the entire stem region of the CRE, with the exception of the 14-nt unpaired loop, could be replaced in a subgenomic replicon with an artificial stem of a similar stability without compromising function (Synth2; Fig. 3C,D ▶). Replicons that contain native CRE sequences in the cassette region replicate less well than those that contain synthetic CRE sequences; cf. pT7/Rep3/SL3cWT or pT7/Rep3/SL3/#3 with pT7/Rep3 in Figure 3B ▶ and pT7/Rep3/SL3cSynth2 with pT7/Rep3 in Figure 3D ▶. We have determined that this is because the amino acid sequence encoded by native CRE sequences at this position inhibit the 2Apro-mediated polyprotein processing at the P1/P2 junction (data not shown). Taking this into account, we see no evidence for significant competition for cellular or viral factors between CREs present in the same replicon. The replicons derived from pT7/Rep3cWT, pT7/Rep3cSL3, and pT7/Rep3/SL3cWT, which respectively contain WT/WT, SL3/WT, or WT/SL3 CREs in the cassette and native locations, generate similar levels of CAT activity (Fig. 3A,B ▶). However, although our data demonstrates that the CRE is functional in both locations, we cannot draw any conclusions as to whether one or both are being used to template VPg uridylylation in dual CRE-containing replicons.
The poliovirus CRE—either native or synthetic forms—also functions when translocated to the 5′ NCR (pT7/FLC/SL3c-HVCRE-AUG and pT7/FLC/SL3c-HVSynth2; Fig. 7 ▶). The efficiency with which virus is recovered and the characterisation of revertants recovered from pT7/FLC/SL3c-HVSynth2ΔA suggests that the CRE in the 5′ NCR functions with similar activity and in a similar manner when located 3.7 kb away from the native position. Because the genomic location does not significantly influence CRE function and some picornavirus CREs are naturally located in noncoding regions (Mason et al. 2002), it is notable that the enterovirus, cardiovirus, and rhinovirus CREs are all located in coding regions of the genome. This suggests that the additional constraints imposed on sequence variation by the coding requirements could be important—a scenario supported by the restricted synonymous codon variation possible in the poliovirus CRE due to the base pairing of the stem. Alternatively, the presence of a CRE within the capsid-coding region may restrict the formation of defective interfering particles (Nomoto et al. 1979; Kuge et al. 1986) or (if located within the P2 region) the generation of virus recombinants (Kirkegaard and Baltimore 1986; Cammack et al. 1988), or, as suggested recently, may influence the temporal control of translation and replication (Barton et al. 2001).
We have established the in vitro assay for VPg uridylylation using poliovirus type 3 components. There are no significant differences between this assay and the poliovirus type 1-based assay originally developed by Wimmer and colleagues (Paul et al. 1998), although we observe optimal activity using Mn++ rather than Mg++ as the divalent cation. In this regard the poliovirus type 3 assay more closely resembles that recently developed for rhinovirus type 2 (Gerber et al. 2001b). Formation of VPg-pU(pU) requires three proteins—3Dpol, 3CDpro, and VPg—together with a suitable RNA to template the uridylylation. Polyadenylated templates such as poly(A) RNA or a 3′ NCR sequence (which has nine As on it) will template the formation of VPg-poly(U), but VPg-pU(pU) is only formed in the presence of a CRE-derived template. CRE sequences known to be nonfunctional in a replicon-based assay (G19A/C22U; Fig. 5C ▶), or replacement of A1 or A2 within the A1A2A3CA motif (corresponding to A29C, A30C; Fig. 5C ▶) also do not work in vitro, whereas substitution of A3 with a G reduces activity by approximately 50%. This substitution is present in bovine enterovirus (Earle et al. 1988) and the poliovirus type 2 Sabin vaccine strain, but appears not to have a role in the attenuated phenotype of the latter (Moss et al. 1989; Pollard et al. 1989). Despite the presence of an AAACA motif, the 5′ CL (nucleotides 1 to 88) cannot serve as a template for uridylylation (Fig. 5B ▶). It is probable that this is due to the flanking nucleotides and the local structure adopted by the AAACA sequence in the 5′ CL, which is predicted to be wholly base paired (Andino et al. 1990,1993).
The base-paired stem of the CRE presumably functions to present the terminal loop to the proteins involved in the uridylylation reaction. Although the sequence of the stem is unimportant, for example, demonstrated by the function of the synthetic CRE Synth2 (Fig. 3D ▶), our studies demonstrate that the stability of the stem significantly influences CRE function. This was first suggested by the phenotype of genomes containing double or treble mutations in Stem 3, which disrupt CRE structure (see Goodfellow et al. 2000; Fig. 2B,C ▶), and was further investigated using derivatives of Synth2 that differed by one to three hydrogen bonds (Figs. 3C, 5 ▶ ▶). Sequential replacement of one, two, or three CG base pairs with UG pairs reduced in vitro uridylylation activity at the optimal temperature by approximately 50% for each hydrogen bond removed, and activity became increasingly sensitive to elevated temperatures (Fig. 5A ▶). The optimum in vitro uridylylation temperature of ∼30°C is similar to that shown for the HRV 2 3Dpol catalyzed reaction with the HRV 2 CRE, but activity is more stable at elevated temperatures (Gerber et al. 2001a). In this respect the poliovirus type 3 uridylylation reaction resembles that previously reported for poliovirus type 1 (Mahoney) in an earlier analysis of the temperature sensitivity of uridylylation. It may be possible to use the in vitro uridylylation assay to determine the lower (and possibly upper) limits of CRE stability compatible with function. However, within a replicon, the sequences flanking the CRE may provide an additional stabilizing influence. Only the least stable of the four synthetic CREs (Synth2–Synth5; Fig. 3C,D ▶) did not support reasonable levels of genome replication. We ascribe this apparent discrepancy with the in vitro uridylylation data to the influence of the GC-rich cassette region, a conclusion supported by the wild-type uridylylation activities of RNA generated by full-length Synth2–4 replicons (Fig. 5B ▶).
How is the CRE orientated and presented during the uridylylation reaction? If the in vitro assay precisely mimics the interactions that occur within a membrane-associated virus replication complex in an infected cell, it should be possible to demonstrate specific interactions between the various components using a suitable assay. We have so far been unable to demonstrate such specificity. Complexes that form weakly between the RNA-binding 3CDpro protein and the CRE are efficiently competed by a 5′ CL RNA (which EMSA studies suggest has a significantly higher affinity for 3CD; Fig. 7A ▶) and the G19A/C22U mutant form of the CRE, which is structurally disrupted (Fig. 2B ▶) and nonfunctional (Goodfellow et al. 2000; Rieder et al. 2000). Alternatively, these low specificity interactions may be sufficient in the in vitro assay where protein and RNA components are abundant, a situation clearly different from the early stages of cell infection. In the cellular environment, it is likely that the confines of the replication complex or the presence of additional virus or cellular factors (proteins or RNA), may provide the specificity necessary to allow the precise interactions of polymerase, CRE, and VPg required for uridylylation. Further studies are needed to identify such components, but we have not been able to demonstrate a specific interaction between proteins from uninfected cells and the CRE by EMSA, UV cross linking, Northwestern blotting or using the yeast-3-hybrid system (data not shown).
Unlike the other cis-acting replication elements located in the 5′ and 3′ noncoding regions of picornavirus genomes, the location of the CRE exhibits considerable natural diversity, and—from studies presented here and recent work on FMDV (Mason et al. 2002)—apparent plasticity. It will be intriguing to correlate the latter observations with the probable noncovalent association of the noncoding extremes of the genome (Barton et al. 2001; Herold and Andino 2001). Does the CRE function independently of this association, or are the three elements coordinated within the replication complex ? The uridylylation of picornavirus VPgs is a necessary early stage of virus replication, and a unique reaction catalyzed by a virus enzyme. A better understanding of CRE structure and function may identify targets for therapeutic intervention against the picornaviruses, and will provide important insights into the replication of the positive strand RNA viruses.
MATERIALS AND METHODS
Computing
RNA structural predictions used the Zuker energy minimization algorithm (Zuker 1989) as implemented in the MFOLD program of the GCG suite (Wisconsin Package Version 10.0, Genetics Computer Group) or the RNAdraw implementation of FoldRNA from the Vienna RNA package (Hofacker et al. 1994).
Plasmid construction and analysis
Construction of CAT-encoding replicon-based cassette vectors
PCR mutagenesis was used to introduce a unique Sma I site adjacent to the Bss HII site near the in-frame fusion of the chloramphenicol acetyl transferase (CAT) gene and the region encoding the carboxyl-terminus of VP1 in the poliovirus type 3 (Leon)-derived replicon, pT7/Rep3 (described previously by Barclay et al. 1998; Goodfellow et al. 2000). Similar modifications were made to pT7/Rep3/SL3, a replication-incompetent version of pT7/Rep3 containing eight noncoding substitutions within the region of poliovirus 2C that forms the CRE (Goodfellow et al. 2000). The cassette vectors were designated pT7/Rep3c and pT7/Rep3/SL3c, respectively.
Insertion of CRE sequences in cassette vectors
Complementary oligonucleotides (Table 1 ▶) were designed to replace the stuffer region in the cassette vectors pT7/Rep3c or pT7/Rep3/SL3c and restore the open reading frame to the replicon cDNA. Oligonucleotides were annealed and directionally cloned between the Bss HII and Sma I sites of pT7/Rep3c or pT7/Rep3/SL3c and recombinants were sequenced prior to further analysis.
Insertion of CRE sequences in the 5′ noncoding region
pT7/Rep3 or pT7/Rep3/SL3 were digested with Bam HI and Sac I and ligated with annealed oligonucleotides encoding the CRE sequences of interest (Table 1 ▶). Native CRE sequences were designed with two substitutions U15A/A49U to destroy the translation initiation codon that would otherwise be included. The modified 5′ NCR was engineered into a full length infectious clone (pT7/FLC or pT7/FLC/SL3) on a Not I–Sac I fragment, which respectively cut within the plasmid vector sequences and the beginning of the polyprotein-coding region.
RNA transfection, CAT assays, and recovery of recombinant viruses
Transfection of HeLa cells with RNA generated in vitro was carried out essentially as described previously (Evans and Almond 1991; Goodfellow et al. 2000) with RNA quantified by 14C-rATP incorporation and normalization to 2 to 4 μg of RNA per dish as necessary. CAT assays were performed as described previously (Percy et al. 1992; Goodfellow et al. 2000), and quantified by phosphorimager. Virus plaques were visualized following transfection of HeLa cells with 10-fold serial dilutions of RNA, overlaying with agar and staining with crystal violet after 3 d, as described previously (Evans and Almond 1991; Meredith et al. 1999).
Biochemical analysis of CRE structure
Ribonuclease (RNAse) V1, T1, and T2 were obtained from US Biochemicals, Roche, and Sigma, respectively. RNA transcripts from annealed oligonucleotide templates were generated using the MegaShortScript kit (Ambion), purified on 10% denaturing acrylamide gels, dephoshorylated with calf intestinal phosphatase (Roche) and 5′ end-labeled using T4 polynucleotide kinase (Roche) and γP32-ATP (Amersham-Pharmacia), prior to final purification on a 10% denaturing acrylamide gel. Before cleavage with RNAse, 50,000 c.p.m of labeled RNA was annealed in buffer A (10 mM Tris at pH 7.5, 20 mM MgCl2) followed by the addition of 1 μg of tRNA (brewers yeast; Sigma). RNAse digestion was performed at 28°C for 10 min using predetermined concentrations of enzyme that generated approximately one cleavage per transcript (three doubling dilutions of a 1:20 dilution from the manufacturer’s stock; maximal levels corresponding to 5 units of T1, 0.5 units of T2 and 0.036 units of V1). Buffer A was supplemented with 100 mM NaCl for RNAse V1 digestions. Following RNAse treatment, reactions were extracted with acid-phenol, precipitated with ice-cold ethanol with a glycogen carrier (Roche) and resuspended in 3 μL of denaturing loading dye. RNAse-generated fragments were separated by 12% denaturing acrylamide gel electrophoresis, fixed, and visualized by exposure to imaging screens (Bio-Rad).
Protein expression and purification
The Escherichia coli expression and purification of native poliovirus type 3 3Dpol involved the IPTG-induced expression of untagged 3Dpol in E. coli BL21 DE3 pLysS (Novagen) for 5 h at 37°C, pelleting cells by centrifugation and washing in buffer A (50 mM Tris at pH 7.5, 1 mM DTT, 10% glycerol) before resuspension at 20 mL/L in buffer I (50 mM Tris at pH 8.0, 0.5 mM EDTA, 1 mM DTT, 0.1% NP40) and storage at −20°C for 16 h. Cells were subsequently thawed, lysed by sonication, centrifuged, and the supernatant retained as soluble fraction 1. The pellet was resuspended in buffer I supplemented with 1 M NaCl, solublized by sonication, and recentrifuged to remove debris. The supernatant was combined with soluble fraction1, diluted to a final salt concentration of 50 mM, and passed through a 1.5-mL SP-sepharose column (Pharmacia) by gravity. After four column volume washes with buffer I and four column volume washes with buffer A, bound protein was eluted using a step gradient of buffer A containing 50, 100, 200, or 500 mM NaCl. Over 60% of 3Dpol eluted in 200 mM NaCl and was at least 95% pure as determined by gel electrophoresis. The resulting protein was dialyzed against storage buffer (50 mM Tris at pH 8.0, 2 mM DTT 45 mM KCl, 20% glycerol), aliquoted and stored at −70°C. Typical yield was 0.5 mg/L.
An amino-terminal poly-histidine-tagged fusion of poliovirus type 3 (Leon) 3CDpro was generated following 3Cpro inactivation by mutagenesis of the histidine in the catalytic triad to alanine (H40A; based upon previous studies; Andino et al. 1993), cloning in pQE30 (Qiagen) and expression and purification from E. coli according to the manufacturer’s instructions. All proteins were visualized following denaturing polyacrylamide electrophoresis by Coomassie blue staining and estimated to be at least 95% pure.
Electrophoretic mobility shift assays
RNA–protein interactions were analyszd by electrophoretic mobility shift assays (EMSA) (described previously in Mellits et al. 1998). γP32-ATP-labeled transcripts were generated and purified as described above. HeLa S10 extracts (prepared according to Barton et al. 1995) were titrated into reactions to leave ∼60% free probe. All reactions contained 5 or 10 μg of tRNA (Brewers yeast, Sigma) and were visualized following electrophoresis in 10% native polyacrylamide gels using a Bio-Rad phosphorimager.
Uridylylation reactions
Reactions were carried out essentially as described (Paul et al. 1998) with the replacement of magnesium acetate with 3.5 mM manganese chloride as this was shown to increase the activity of poliovirus type 3 3Dpol (I.G. Goodfellow and D.J. Evans, unpubl.). Short RNA templates for the uridylylation reaction were synthesized using the Ambion MegaShortScript kit as described above and purified by denaturing polyacrylamide gel electrophoresis. 32P-UTP-radiolabeled uridylylation reaction products were separated by 10% tricine gel electrophoresis, fixed, and visualized and quantified by phosphorimaging.
Acknowledgments
This work was supported by awards to D.J.E. from the Biotechnology and Biological Sciences Research Council (Project Grant #17/G11681) and the Medical Research Council (Programme Grant #G9901250). The authors thank Peter Simmonds for interesting discussions on CRE evolution.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2950603.
Footnotes
The nomenclature used to define bases within the CRE is in the form of native nucleotide, position, substituted nucleotide, with all numbering starting at the base of the CRE, which corresponds to nucleotide 4435 of the genomic sequence of poliovirus type 3 (P3/Leon/37), GenBank accession no. K01392.
Standard naming conventions: A pT7/prefix indicates a plasmid with a T7 promoter containing the virus or subgenomic replicon cDNA, pT7/Rep3 was derived from pT7/FLC (Barclay et al. 1998), a ‘‘full-length (infectious) clone’’ of poliovirus type 3 Leon. The resulting replicon or virus generated following transfection are designated Rep3 or FLC, respectively.
REFERENCES
- Andino, R., Böddeker, N., Silvera, D., and Gamarnik, A.V. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 7: 76–82. [DOI] [PubMed] [Google Scholar]
- Andino, R., Rieckhof, G.E., and Baltimore, D. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63: 369–380. [DOI] [PubMed] [Google Scholar]
- Andino, R., Rieckhof, G.E., Achacoso, P.L., and Baltimore, D. 1993. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 12: 3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay, W., Li, Q., Hutchinson, G., Moon, D., Richardson, A., Percy, N., Almond, J., and Evans, D. 1998. Encapsidation studies of poliovirus subgenomic replicons. J. Gen. Virol. 79: 1725–1734. [DOI] [PubMed] [Google Scholar]
- Barton, D.J., Black, E.P., and Flanegan, J.B. 1995. Complete replication of poliovirus in vitro: Preinitiation RNA replication complexes require soluble cellular factors for the synthesis of VPg-linked RNA. J. Virol. 69: 5516–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, D.J., O’Donnell, B.J., and Flanegan, J.B. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20: 1439–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammack, N., Phillips, A., Dunn, G., Patel, V., and Minor, P.D. 1988. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology 167: 507–514. [PubMed] [Google Scholar]
- Crawford, N.M. and Baltimore, D. 1983. Genome-linked protein VPg of poliovirus is present as free VPg and VPg-pUpU in poliovirus-infected cells. Proc. Natl. Acad. Sci. 80: 7452–7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle, J.A., Skuce, R.A., Fleming, C.S., Hoey, E.M., and Martin, S.J. 1988. The complete nucleotide sequence of a bovine enterovirus. J. Gen. Virol. 69: 253–263. [DOI] [PubMed] [Google Scholar]
- Evans, D.J. 1999. Reverse genetics of picornaviruses. Adv. Virus Res. 53: 209–228. [DOI] [PubMed] [Google Scholar]
- Evans, D.J. and Almond, J.W. 1991. Design, construction and characterization of poliovirus antigen chimeras. Methods Enzymol. 203: 387–400. [DOI] [PubMed] [Google Scholar]
- Flanegan, J.B., Petterson, R.F., Ambros, V., Hewlett, M.J., and Baltimore, D. 1977. Covalent linkage of a protein to a defined nucleotide sequence at the 5′-terminus of virion and replicative intermediate RNAs of poliovirus. Proc. Natl. Acad. Sci. 74: 961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik, A.V. and Andino, R. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3: 882–892. [PMC free article] [PubMed] [Google Scholar]
- Gerber, K., Wimmer, E., and Paul, A.V. 2001a. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: Identification of a cis-replicating element in the coding sequence of 2A(pro). J. Virol. 75: 10979–10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001b. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: Purification and enzymatic analysis of the RNA-dependent RNA polymerase 3D(pol). J. Virol. 75: 10969–10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow, I.G., Chaudhry, Y., Richardson, A., Meredith, J.M., Almond, J.W., Barclay, W.S., and Evans, D.J. 2000. Identification of a cis-acting replication element (CRE) within the poliovirus coding region. J. Virol. 74: 4590–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold, J. and Andino, R. 2001. Poliovirus RNA replication requires genome circularization through a protein–protein bridge. Mol. Cell 7: 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker, I.L., Fontana, W., Stadler, P.F., Bonhoeffer, L.S., Tacker, M., and Schuster, P. 1994. Fast folding and comparison of RNA secondary structures. Monatshefte Chem. 125: 167–188. [Google Scholar]
- Kirkegaard, K. and Baltimore, D. 1986. The mechanism of RNA recombination in poliovirus. Cell 47: 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura, N., Alder, C., Martinko, J., Nathenson, S.G., and Wimmer, E. 1980. The genome-linked protein of picornaviruses. VII. Genetic mapping of poliovirus VPg by protein and RNA sequence studies. Cell 21: 295–302. [DOI] [PubMed] [Google Scholar]
- Kuge, S., Saito, I.. and Nomoto, A. 1986. Primary structure of poliovirus defective-interfering particle genomes and possible generation mechanisms of the particles. J. Mol. Biol. 192: 473–487. [DOI] [PubMed] [Google Scholar]
- Kuhn, R.J., Tada, H., Ypma Wong, M.F., Semler, B.L., and Wimmer, E. 1988. Mutational analysis of the genome-linked protein VPg of poliovirus. J. Virol. 62: 4207–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobert, P.E., Escriou, N., Ruelle, J., and Michiels, T. 1999. A coding RNA sequence acts as a replication signal in cardioviruses. Proc. Natl. Acad. Sci. 96: 11560–11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, P.W., Bezborodova, S.V., and Henry, T.M. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76: 9686–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight, K.L. and Lemon, S.M. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4: 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers, W.J.G., Hoenderop, J.G.J., Slot, H.J.B., Pleij, C.W.A., Pilipenko, E.V., Agol, V.I., and Galama, J.M.D. 1997. Kissing of the 2 predominant hairpin loops in the coxsackie-b virus 3′-untranslated region is the essential structural feature of the origin of replication required for negative-strand rna-synthesis. J. Virol. 71: 686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellits, K.H., Meredith, J.M., Rohll, J.B., Evans, D.J., and Almond, J.W. 1998. Binding of a cellular factor to the 3′ untranslated region of the RNA genomes of entero- and rhinoviruses plays a role in virus replication. J. Gen. Virol. 79: 1715–1723. [DOI] [PubMed] [Google Scholar]
- Meredith, J.M., Rohll, J.B., Almond, J.W., and Evans, D.J. 1999. Similar interactions of the poliovirus and rhinovirus 3D polymerase with the 3′ untranslated region of rhinovirus 14. J. Virol. 73: 9952–9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, E.G., O’Neill, R.E., and Racaniello, V.R. 1989. Mapping of attenuating sequences of an avirulent poliovirus type 2 strain. J. Virol. 63: 1884–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto, A., Kitamura, N., Golini, F., and Wimmer, E. 1977. The 5′ terminal structures of poliovirus RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc. Natl. Acad. Sci. 77: 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto, A., Jacobson, A., Lee, Y.F., Dunn, J., and Wimmer, E. 1979. Defective interfering particles of poliovirus: Mapping of the deletion and evidence that the deletions in the genomes of DI(1), (2) and (3) are located in the same region. J. Mol. Biol. 128: 179–196. [DOI] [PubMed] [Google Scholar]
- Parsley, T.B., Towner, J.S., Blyn, L.B., Ehrenfeld, E., and Semler, B.L. 1997. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3: 1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Paul, A.V., vanBoom, J.H., Filippov, D., and Wimmer, E. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393: 280–284. [DOI] [PubMed] [Google Scholar]
- Paul, A.V., Rieder, E., Kim, D.W., van Boom, J.H., and Wimmer, E. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74: 10359–10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier, J. and Sonenberg, N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- Pelletier, J., Flynn, M.E., Kaplan, G., Racaniello, V., and Sonenberg, N. 1988. Mutational analysis of upstream AUG codons of poliovirus RNA. J. Virol. 62: 4486–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percy, N., Barclay, W.S., Sullivan, M., and Almond, J.W. 1992. A poliovirus replicon containing the chloramphenicol Acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J. Virol. 66: 5040–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Poperechny, K.V., Maslova, S.V., Melchers, W.J., Slot, H.J., and Agol, V.I. 1996. cis-Element, oriR, involved in the initiation of (−) strand poliovirus RNA: A quasi-globular multi-domain RNA structure maintained by tertiary (‘‘kissing’’) interactions. EMBO J. 15: 5428–5436. [PMC free article] [PubMed] [Google Scholar]
- Pollard, S.R., Dunn, G., Cammack, N., Minor, P.D., and Almond, J.W. 1989. Nucleotide sequence of a neurovirulent variant of the type 2 oral poliovirus vaccine. J. Virol. 63: 4949–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello, V.R. and Baltimore, D. 1981. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214: 916–919. [DOI] [PubMed] [Google Scholar]
- Rieder, E., Paul, A.V., Kim, D.W., van Boom, J.H., and Wimmer, E. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74: 10371–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohll, J.B., Moon, D.H., Evans, D.J., and Almond, J.W. 1995. The 3′-untranslated region of picornavirus RNA—Features required for efficient genome replication. J. Virol. 69: 7835–7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas, M., Miller, J.T., Leis, J., and DePamphilis, M. 1996. Mechanisms of priming DNA synthesis. In DNA replication in eukaryotic cells (ed. M.L. DePamphilis), pp. 131–176. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Skinner, M.A., Racaniello, V.R., Dunn, G., Cooper, J., Minor, P.D., and Almond, J.W. 1989. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 207: 379–392. [DOI] [PubMed] [Google Scholar]
- Todd, S., Towner, J.S., Brown, D.M., and Semler, B.L. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71: 8868–8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Bakkers, J.M., Galama, J.M., Bruins Slot, H.J., Pilipenko, E.V., Agol, V.I., and Melchers, W.J. 1999. Structural requirements of the higher order RNA kissing element in the enteroviral 3′UTR. Nucleic Acids Res. 27: 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y., Rijnbrand, R., McKnight, K.L., Wimmer, E., Paul, A., Martin, A., and Lemon, S.M. 2002. Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J. Virol. 76: 7485–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244: 48–52. [DOI] [PubMed] [Google Scholar]