Abstract
Through exhaustive two-hybrid screens using a budding yeast genomic library, and starting with the splicing factor and DEAH-box RNA helicase Prp22p as bait, we identified yeast Prp45p and Prp46p. We show that as well as interacting in two-hybrid screens, Prp45p and Prp46p interact with each other in vitro. We demonstrate that Prp45p and Prp46p are spliceosome associated throughout the splicing process and both are essential for pre-mRNA splicing. Under nonsplicing conditions they also associate in coprecipitation assays with low levels of the U2, U5, and U6 snRNAs that may indicate their presence in endogenous activated spliceosomes or in a postsplicing snRNP complex.
Keywords: Protein interaction, snRNP, spliceosome, two-hybrid, yeast
INTRODUCTION
The excision of introns from precursor messenger RNAs (pre-mRNAs) occurs by two consecutive trans-esterification reactions in the spliceosome, a large and highly dynamic, ribonucleoprotein complex. In many in vitro studies, spliceosomes have been observed to assemble de novo on each substrate pre-mRNA in an ordered stepwise manner. In a series of ATP-dependent steps, five small RNAs, the U1, U2, U4, U5, and U6 snRNAs, in the form of small ribonucleoprotein particles (snRNPs), plus a variety of non-snRNP proteins associate with the pre-mRNA (for review, see Burge et al. 1999; Will and Lührmann 2001). Prior to activation of the spliceosome and during the splicing process, a number of conformational rearrangements take place, including critical changes to RNA–RNA interactions (for review, see Nilsen 1998). These rearrangements include dissociation of an extensively base-paired U4/U6 snRNA heterodimer, displacement of a U1 snRNA:5′ splice site interaction, and a consequent loosening of the associations of the U1 and U4 snRNPs with the spliceosome immediately prior to the initiation of the splicing process. Following the completion of the two splicing reactions, the spliceosome appears to dissociate, at least in vitro.
The splicing factors have been identified through genetic approaches in the budding yeast Saccharomyces cerevisiae (for review, see Beggs 1995), fractionation of HeLa cell nuclear extracts and reconstitution of activities (Caceres and Krainer 1997), two-hybrid screens (e.g., Fromont-Racine et al. 1997) or by affinity purification of complexes under mild, nondenaturing conditions and identification of the protein components by mass spectrometry (Neubauer et al. 1997, 1998; Stevens and Abelson 1999). Using the latter approach, Stevens et al. (2002) isolated a large penta-snRNP complex that contains all five spliceosomal snRNAs and over 60 protein splicing factors. This complex supports pre-mRNA splicing when complemented with soluble factors, and it was proposed that the spliceosomal snRNPs may associate to form a penta-snRNP complex independently of the substrate pre-mRNA (for review, see Nilsen 2002).
In addition to aiding the identification of novel splicing factors, two-hybrid screens provide information about interactions between known splicing factors, and can also define regions of interaction between two binding partners (e.g., Fromont-Racine et al. 1997; Ben-Yehuda et al. 2000). Through exhaustive two-hybrid screens using a budding yeast genomic library and starting with the DEAH-box RNA helicase Prp22p as bait, we identified yeast Prp45p and Prp46p. We show that Prp45p and Prp46p interact with each other in vitro. We demonstrate that Prp45p and Prp46p are spliceosome associated throughout the splicing process and both are essential for pre-mRNA splicing. Under nonsplicing conditions they associate in coprecipitation assays with a low level of U2, U5, and U6 snRNAs that may indicate their presence in endogenous activated spliceosomes or, more likely, in a postsplicing snRNP complex. These data are compatible with the recent detection of these proteins in the penta-snRNP complex (Stevens et al. 2002) and in another high molecular weight complex that contains a subset of spliceosomal proteins (Ohi et al. 2002).
RESULTS
Two-hybrid analysis of Prp45p
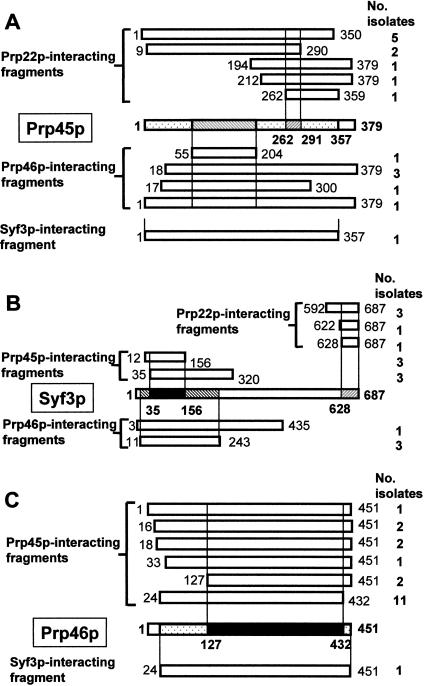
With the aim of identifying novel splicing factors and to shed light into the myriad of protein–protein interactions within the spliceosome, a number of two-hybrid screens of a yeast genomic library (FRYL; Fromont-Racine et al. 1997) were performed. Using the full-length Prp22 protein of S. cerevisiae as bait, 47 million potential interactions were tested, and a total of 20 clones were identified as positive for activation of both of the reporter genes used (Table 1 ▶). Ten isolates encoded five independent fragments of Prp45p (ORF YAL032c). All the Prp45p prey fragments share a small common region between amino acids 262 and 291 (Fig. 1A ▶), which therefore corresponds to a minimum region of interaction although it is not clear that this is sufficient for the interaction.
TABLE 1.
Interactions identified in the yeast two-hybrid screensa
| Protein | ORF | Protein length (amino acids) | MRIb | Isolatesc | Categoryd |
| Prp22p two-hybrid screen isolates | |||||
| Prp45p | YAL032c | 379 | 262–291 | 10 | A1 |
| Syf3p | YLR117c | 687 | 628–687 | 5 | A1 |
| YNL274c | 350 | 33–350 | 1 | A3 | |
| Pmi40p | YER003c | 460 | 342–460 | 1 | A4 |
| Dbp7p | YKR024c | 742 | 327–621 | 2 | A4 |
| Prp45p two-hybrid screen isolates | |||||
| YDR141c | 1698 | 580–* | 2 | A1 | |
| Fir1p | YER032w | 925 | 709–831 | 4 | A1 |
| YGR198w | 817 | 748–817 | 3 | A1 | |
| Yta7p | YGR270w | 1379 | 835–1198 | 4 | A1 |
| Ubr2p | YLR024c | 1872 | 834–* | 4 | A1 |
| Syf3p | YLR117c | 687 | 35–156 | 6 | A1 |
| Vac14p | YLR386w | 880 | 331–* | 5 | A1 |
| Sgs1p | YMR190c | 1447 | 503–739 | 2 | A1 |
| Prp46p | YPL151c | 451 | 127–432 | 19 | A1 |
| Cdc27p | YBL084c | 758 | 30–412 | 2 | A2 |
| YLR424w | 708 | 31–* | 1 | A2/A3 | |
| YMR031c | 843 | 1–* | 1 | A2 | |
| Vps75p | YNL246w | 264 | 26–* | 1 | A2 |
| Ahc1p | YOR023c | 566 | 19–* | 1 | A2 |
| Pkc1p | YBL105c | 1151 | 301–* | 1 | A3 |
| Syf1p | YDR416w | 859 | 442–827 | 2 | A3 |
| Ade6p | YGR061c | 1358 | 792–* | 1 | A3 |
| YGR071c | 860 | 301–* | 1 | A3 | |
| Sec6p | YIL068c | 806 | 100–* | 1 | A3 |
| Apl1p | YJR005w | 700 | 159–* | 1 | A3 |
| YJR061w | 935 | 395–* | 1 | A3 | |
| Dyn1p | YKR054c | 4092 | 690–* | 2 | A3 |
| Mms22p | YLR320w | 1454 | 611–* | 2 | A3 |
| Nup188p | YML103c | 1655 | 1291–* | 1 | A3 |
| Ecm5p | YMR176w | 1411 | 790–* | 1 | A3 |
| Sla2p | YNL243w | 968 | 114–* | 1 | A3 |
| Ski3p | YPR189w | 1432 | 463–* | 1 | A3 |
| Rad16p | YBR114w | 790 | 59–* | 1 | A4 |
| YDL001w | 430 | 362–* | 1 | A4 | |
| Nup84p | YDL116w | 726 | 170–* | 1 | A4 |
| Ngg1p | YDR176w | 702 | 508–* | 1 | A4 |
| Trp5p | YGL026c | 707 | 617–* | 1 | A4 |
| Yap1801 | YHR161c | 637 | 112–* | 1 | A4 |
| YJL103c | 618 | 219–* | 1 | A4 | |
| YJL149w | 663 | 508–* | 1 | A4 | |
| YLR187w | 1026 | 167–* | 3 | A4 | |
| Nis1p | YNL078w | 407 | 270–* | 1 | A4 |
| Bni1p | YNL271c | 1953 | 1481–* | 1 | A4 |
| Mnt4p | YNR059w | 580 | 195–* | 1 | A4 |
| Nif1p | YOR156c | 726 | 361–* | 1 | A4 |
| Eaf3p | YPR023c | 401 | 228–* | 1 | A4 |
| Prp46p two-hybrid screen isolates | |||||
| Prp45 | YAL032c | 379 | 54–204 | 5 | A1 |
| Syf3 | YLR117c | 687 | 10–243 | 2 | A1 |
| HFM1 | YGL251c | 1046 | 699–880 | 2 | A1 |
| Pkc1 | YBL105c | 1151 | 301–801 | 1 | A3 |
| YLR426w | 202 | 202 | 1 | A2 | |
| VAP | YBR069c | 619 | 290–574 | 1 | A4 |
| PEP12 | YOR036w | 288 | 288 | 1 | A4 |
aThe open reading frames that were identified as prey in two-hybrid screens with full-length Prp22p, Prp45p, and Prp46p bait fusions. Indicated are systematic protein and ORF names as given in the Saccharomyces Genome Database.
bMRI: minimal region of interaction—amino acid boundaries of the region shared by all isolates of a given protein (* C termini of these inserts have not been determined).
cIsolates: total number of prey isolates.
dCategory: Prey fusions were classified as defined by Fromont-Racine et al. (1997). A1 (statistically the most significant): multiple coding fusions that share a common region; A2: fusion starts close to the translation initiation codon; A3: contains a large coding insert; A4: single coding fusion not represented in A2 or A3 category, and possibly the least significant, although this may also indicate that only a precisely defined peptide can mediate an interaction.
FIGURE 1.
Protein–protein interactions involving Prp45p, Syf3p, and Prp46p. (A) Prp45p; (B) Syf3p; (C) Prp46p. Numbers represent amino acid positions. Vertical lines delineate minimal regions found in common in overlapping prey fusions.
A second candidate interactor, Syf3 protein, was isolated in the Prp22p two-hybrid screen a total of five times as three independent fusions. The C-terminal region, consisting of amino acids 628 to 687, corresponds to the minimum region of Prp22p interaction in Syf3p (Fig. 1B ▶). Syf3p (also called Clf1p) was shown earlier to be involved in spliceosome assembly as well as in cell cycle progression from G2 to M phase (Chung et al. 1999; Russell et al. 2000).
Full-length Prp45p was used in turn as bait to screen the FRYL genomic library, and 34 million potential interactors were tested. By far the most statistically significant interactor was the protein Prp46 encoded by ORF YPL151c (Table 1 ▶); 19 plasmids encoding six independent protein fragments were isolated (Fig. 1C ▶). The Prp45p interacting region appears to be within the amino acid boundaries 127 and 432 of Prp46p, as deduced from the overlapping region of the isolated fragments. This region encodes seven copies of a conserved WD repeat motif that is thought to mediate protein–protein interactions (Smith et al. 1999). None of the other prey isolated in this two-hybrid screen corresponds to a WD repeat protein. Prp46p is the yeast homolog of the recently identified human splicing factor PLRG1 and the Schizosaccharomyces pombe protein prp5, for both of which association with subspliceosomal complexes and their requirement for pre-mRNA splicing has been demonstrated (Potashkin et al. 1998; McDonald et al. 1999; Ajuh et al. 2000, 2001).
Intriguingly, the Prp45p bait also found a total of six isolates of Syf3p. These represented two independent fragments of Syf3p that are situated close to the N terminus, sharing a common region between amino acids 35 and 156 (Fig. 1B ▶). Notably, this region is different from the Prp22p interacting region, which is located at the extreme C terminus of Syf3p.
In addition, another known splicing factor with a cell cycle link, namely the Syf1 protein (Russell et al. 2000), was found to interact with the Prp45p bait. This interaction is statistically less significant, possibly indicating that this interaction is weaker or indirect (Table 1 ▶).
In conclusion, these results suggest an association of Prp45p with four known or potential splicing factors, although this two-hybrid assay may not distinguish between direct and indirect interactions.
In this iterative approach to identify interacting factors, full-length Prp46p was used next as bait in a two-hybrid screen, testing 21 million potential interactors. Reassuringly, among the 13 identified positive clones (Table 1 ▶), Prp45p was the most statistically significant, occurring as five distinct fusions. Interestingly, Prp46p interacts with a region of Prp45p that is not required for the Prp22p interaction (Fig. 1A ▶), indicating that the Prp46p and Prp22p interactions with Prp45p are distinct. Furthermore, two distinct fusions of Syf3p were found as prey with Prp46p (Fig. 1B ▶). The Prp46p-interacting region in Syf3p overlapped the region for interaction with Prp45p. Thus Syf3p may interact indirectly with Prp45p and/or Prp46p (or cooperatively with both), but the interaction with Prp22p is distinct. The fact that in a previous screen with Syf3p as bait (Ben-Yehuda et al. 2000; Fig. 1A,C ▶) the prey fusions of Prp45p and of Prp46p that were isolated contained almost the full-length proteins may also reflect indirect or cooperative interactions. The other A1 category (and therefore statistically significant) prey with Prp46p was Hfm1p/Mer3p, a DEAH-box protein and putative helicase that is implicated in meiotic recombination (Nakagawa and Ogawa 1999).
Depletion of Prp45p causes a splicing defect in vivo
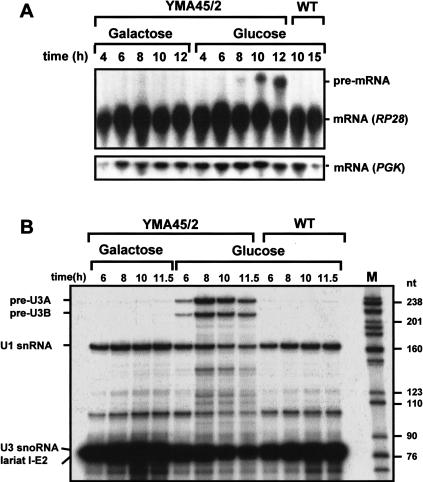
The two-hybrid associations of Prp45p with a set of splicing proteins led us to ask whether Prp45p is also required for pre-mRNA splicing. Because it was reported earlier (Harris et al. 1992) that PRP45 (FUN20) is a gene essential for cell viability, we constructed strain YMA45/2 with a galactose-inducible and protein A-tagged chromosomal PRP45 allele. Cultures of YMA45/2 and of the parental wild-type strain BMA38a were transfered to either permissive (galactose) or nonpermissive (glucose) conditions, and growth and pre-mRNA splicing were monitored. About 8 h after the transfer of the cultures to the nonpermissive conditions, the growth rate of YMA45/2 cells started to decline, and growth stopped at around 12 h (data not shown), unlike wild-type cells and YMA45/2 cells grown under permissive conditions, which continued to grow logarithmically. Northern analysis (Fig. 2A ▶) showed that unspliced RP28 RNA accumulated in YMA45/2 cells at 8 h, just as the growth rate started to decline, whereas under permissive growth conditions, strain YMA45/2 behaved like wild type. Thus cells depleted of Prp45p display a clear splicing defect, indicating that Prp45p is indeed required for pre-mRNA splicing.
FIGURE 2.
Effect of Prp45p depletion on pre-mRNA splicing in vivo. (A) Northern analysis of splicing. RNA was extracted from aliquots of the cultures grown for the indicated times under permissive or nonpermissive conditions. RNA (10 μg) was fractionated on a 1% (w/v) formaldehyde gel, blotted to Hybond-N membrane (Amersham), probed with a radiolabeled DNA fragment complementary to exon 1 of the RP28 gene, and analyzed by autoradiography. The positions of the RP28 pre-mRNA and mRNA are indicated. The blot was stripped and reprobed for the intronless PGK gene as a control for loading. (B) Primer extension analysis of splicing of U3 precursor RNA. RNA (10 μg) was used in a primer extension reaction with a radiolabeled oligonucleotide primer complementary to the extreme 5′ end of exon 2 of the U3 snoRNA. As a control, a primer for the intronless U1snRNA was used in the same reaction. The products were resolved on a 6% (w/v) polyacrylamide gel and visualized by autoradiography. The positions of the extension products are indicated.
As Prp45p was identified as a Prp22p-interacting protein, the question arises whether it is a general splicing factor, required for splicing of all introns, or whether its requirement might be dictated by the distance between the branchpoint and the 3′ splice site, as reported for the second-step splicing factors Prp22p, Slu7p, and Prp18p. Prp22p, for instance, is only required for splicing of introns in which the branchpoint–3′ splice site distance is larger than or equal to 21 nt (Schwer and Gross 1998). If Prp45p interacted with Prp22p during 3′ splice site recognition and cleavage, it would not be surprising to see a similar pattern of selective Prp45p requirement.
The RP28 pre-mRNA intron, which was assayed by Northern analysis, has a branchpoint–3′ splice site distance of 39 nt. To test whether Prp45p might be dispensable for splicing of introns with very short branchpoint–3′ splice site distances, splicing of U3 pre-snoRNA was investigated, in which the distance between the branchpoint sequence and the 3′ terminal CAG is only 7 nt. Figure 2B ▶ shows the results of primer extension analysis of U3 RNA and, as a control, of U1 snRNA. Already after 6 h growth under nonpermissive conditions, there was a strong accumulation of the two U3 precursor RNAs in strain YMA45/2, but not in the control cultures. Thus Prp45p may be required for the splicing of all spliceosomal introns.
Spliceosomal association of Prp45p
To determine whether and at what particular stage of the splicing process Prp45p interacts with spliceosomes, the association of protein A-tagged Prp45p (protA:Prp45p) with pre-mRNA, intermediates, or products of splicing was tested by coprecipitation. Whole-cell extract produced from strain YMA45/2 grown under permissive conditions (i.e., producing protA:Prp45p) was incubated under splicing conditions with radiolabeled actin pre-mRNA. An aliquot (10%) of each sample was analyzed for splicing activity (‘‘splicing’’), while the remainder was incubated with IgG-agarose (‘‘I’’) or, as controls, agarose beads (‘‘B’’) or anti-Prp8p antibodies (‘‘C’’). Prp8p is a U5 snRNP protein that is associated with the spliceosomes throughout the splicing reactions and should therefore precipitate the pre-mRNA, intermediates, and products of the splicing reaction as shown previously by Teigelkamp et al. (1995b). To assess background precipitation by the protein A epitope, extract was prepared from strain BMA38a (pNOPPATAIL) in which the protein A epitope alone (‘‘protA’’) was produced. To aid the determination of the particular stage of the splicing process at which Prp45p associates with the spliceosome, the precipitations were also performed with extracts to which a purified recombinant form of dominant negative (dn) Prp2p (‘‘+Prp2LATp’’) had been added, which stalls splicing prior to the first step (Plumpton et al. 1994).
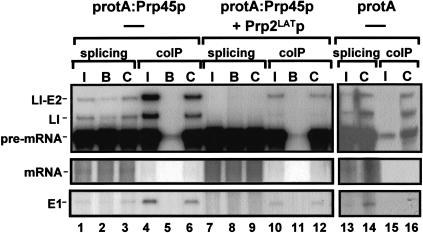
Compared with the Prp8p control (Fig. 3 ▶, lane 6), protA:Prp45p efficiently coprecipitated (‘‘coIP’’; Fig. 3 ▶, lane 4) the pre-mRNA, the lariat intron-exon 2 (LI-E2), and exon 1 (E1) intermediates as well as the excised intron (LI; lane 4). As also observed for Prp8p, spliced mRNA was not enriched in the protA:Prp45p precipitate, although some coprecipitation of mRNA cannot be ruled out, as detection of the mRNA was poor. In both negative controls, using agarose beads without antibody (Fig. 3 ▶, lane 5) and providing the protein A epitope but no Prp45p (Fig. 3 ▶, lane 21), only very low levels of pre-mRNA were found in the precipitate, demonstrating specific precipitation by Prp45p. These findings showed that Prp45p is associated with the spliceosome throughout the splicing reactions and that it stays associated with the spliceosome until after the second catalytic step, when it is found associated with the excised intron.
FIGURE 3.
Coprecipitation of spliceosomes by Prp45p. Whole yeast cell extract (splicing extract) was prepared from cells of strain YMA45/2 grown in galactose-based medium, producing protA:Prp45p. As a control, extract was prepared from the parental wild-type strain carrying vector pNOPPATAIL, producing a double protein A epitope (protA). Splicing (50 μL total volume) was performed using 32P-labeled actin pre-mRNA. The reactions were stopped and 5 μL were removed as splicing controls (input). The remaining 45-μL samples were mixed with an equal volume of precipitation buffer containing either IgG-agarose beads (I), agarose beads without antibody (B) or protein A-Sepharose beads with prebound anti-Prp8p antibodies (C) and incubated at 4°C for 2 h. Beads were washed in buffer containing 150 mM NaCl, deproteinized, and the RNAs precipitated. The samples from the immunoprecipitations (coIP) as well as from the input samples (5 μL; splicing) were then resuspended in formamide loading buffer, resolved on a 6% (w/v) polyacrylamide gel, and labeled RNAs were visualized by autoradiography. In additional samples, recombinant dominant negative Prp2p was added to the extract prior to splicing (+ Prp2LATp) and the samples were treated as above. The positions of the RNA species are indicated. (lariat I-E2) Lariat intron-exon2; (LI) Lariat-intron; (E1) exon1.
When splicing was stalled due to prior addition of Prp2LATp to the extracts (Fig. 3 ▶, lanes 7–9), the Prp45 protein still precipitated almost equivalent quantities of pre-mRNA compared to untreated extracts (Fig. 3 ▶, lane 10 compared to 4). Therefore, Prp45p is already in the spliceosome before Prp2p acts to promote the first transesterification reaction. Small amounts of precipitated splicing intermediates and excised intron from these stalled extracts must be the result of a low residual splicing activity in the extracts due to the presence of wild-type Prp2p.
These data demonstrate that Prp45p associates with the splicing machinery throughout the splicing process, and upon dissociation of the spliced mRNA, Prp45p may segregate with the excised intron.
Coimmunoprecipitation of spliceosomal snRNAs with Prp45p
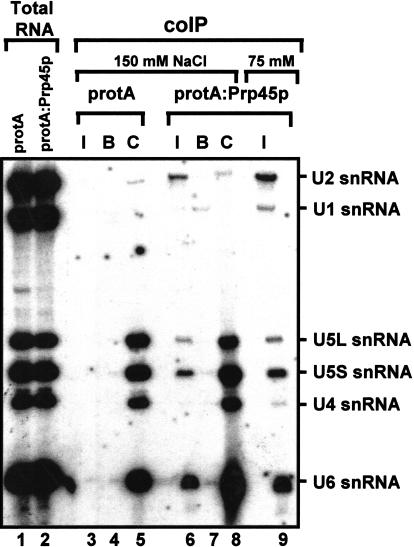
To investigate whether Prp45p is snRNA associated, coimmunoprecipitation of the spliceosomal U1, U2, U4, U5, and U6 snRNAs with protA:Prp45p was tested. Splicing extracts of strain YMA45/2 producing protA:Prp45p and, as a negative control, strain BMA38a-pNOPPATAIL were either incubated with IgG-agarose (I), or, as controls, with agarose beads (B) or anti-Prp8p antibodies, prebound to protein A-Sepharose beads (C). After the immunoprecipitation with IgG agarose, the coprecipitated snRNAs were detected by Northern analysis.
As shown in Figure 4 ▶, lanes 5 and 8, under normal precipitation conditions, washing with 150 mM salt, Prp8p strongly coprecipitated U4, U5(L and S), and U6 snRNAs, as expected for this U5 snRNP protein that is also present in U4/U6.U5 tri-snRNPs. Significantly, under these conditions, protA:Prp45p pulled down approximately equal amounts of the U2, U5, and U6 snRNAs, but no U1 or U4 snRNAs (Fig. 4 ▶, lane 6). Low amounts of U1 and U4 snRNAs were only detected when the precipitates were washed at lower stringency (75 mM salt; Fig. 4 ▶, lane 9). This suggests that Prp45p associates with U2, U5, and U6 snRNAs under nonsplicing conditions (no ATP was added to the extracts), and has a lower level of salt-sensitive interaction with U1 and U4 snRNPs.
FIGURE 4.
Coprecipitation of snRNAs by Prp45p. Whole yeast cell extract (splicing extract) was prepared from cells of strain YMA45/2 grown in galactose-based medium, producing protA:Prp45p. As a control, extract was prepared from the parental wild-type strain carrying vector pNOPPATAIL, producing a double protein A epitope (protA). Extracts (50 μL) were mixed with an equal volume of precipitation buffer containing either IgG-agarose beads (I; lanes 3, 6, 9), agarose beads without antibody (B; lanes 4, 7), or protein A-Sepharose beads with prebound anti-Prp8p antibodies (C; lanes 5, 8) and incubated at 4°C for 2 h. The beads were washed in buffer containing 150 mM NaCl (or 75 mM where indicated), deproteinized, and the RNAs precipitated. The samples were resuspended in formamide loading buffer, resolved on a 6% (w/v) polyacrylamide gel, electroblotted to a Hybond-N nylon membrane (Amersham), and probed for U1, U2, U4, U5, and U6snRNAs. Total RNA extracted from 20 μL of extract is shown in lanes 1 and 2.
Analysis of the Prp46p knockout
The finding that Prp46p interacted with Prp45p as well as with Syf3p in the two-hybrid screens and considering the link with splicing for the putative human and fission yeast homologs, PRLG1 and prp5, respectively (Potashkin et al. 1998; McDonald et al. 1999; Ajuh et al. 2000, 2001), an involvement of Prp46p in splicing seemed likely.
To approach a functional analysis of Prp46p, a gene deletion experiment was performed. A diploid strain, YMA151KO1, was constructed in which the ORF of one allele of PRP46 was entirely replaced by the HIS3 marker gene and the PRP46/prp46Δ::HIS3 strain was sporulated. Among 14 spore tetrads that were dissected, none produced more than two viable spores, all of which were His− (data not shown), demonstrating that PRP46 encodes an essential gene in S. cerevisiae.
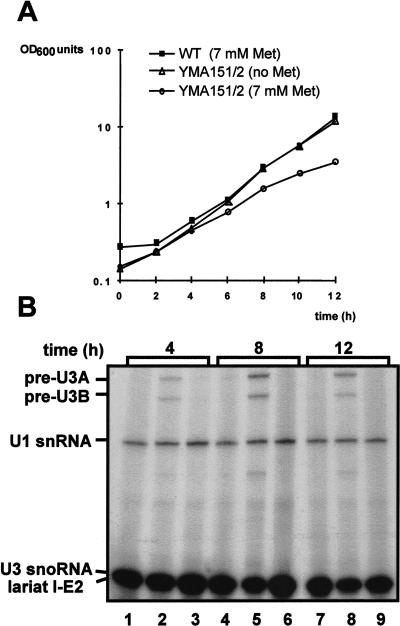
To facilitate further functional characterization of the essential Prp46 protein, strain YMA151/2 was constructed in which the chromosomal PRP46 gene was transcribed under control of the regulatable MET3 promotor, which is repressed by methionine. Cultures of YMA151/2 and parental strain BMA64α were grown either in the absence (permissive conditions) or in the presence (nonpermissive conditions) of 7 mM methionine. At various time points, RNA was extracted and assayed by primer extension analysis for splicing of precursor U3 snoRNA (Fig. 5 ▶).
FIGURE 5.
Effect of Prp46p depletion on cell growth and pre-mRNA splicing. (A) Effect of Prp46p depletion on cell growth. Strains YMA151/2 and BMA64α (WT) were grown at 30°C in minimal medium lacking methionine (inducing conditions for Prp46p production in YMA151/2) or with 7 mM methionine (repressing condition) as indicated. The O.D.600 was monitored and cultures were diluted at intervals to maintain logarithmic growth. The indicated O.D.600 is therefore the calculated theoretical O.D. (B) Effect of Prp46p depletion on in vivo splicing. RNA (10 μg) extracted from aliquots of the cultures shown in A was used as template in a primer extension reaction using radiolabeled oligonucleotide primers complementary to the extreme 5′ end of exon 2 of the U3 snoRNA and to the (intronless) U1 snRNA as control. The products were resolved on a 6% (w/v) polyacrylamide gel and visualized by autoradiography. The positions of the extension products are indicated. (time [h]) Time point of sample. (Lanes 1,4,7) YMA151/2 minus methionine; (lanes 2,5,8) YMA151/2 plus 7 mM methionine; (lanes 3,6,9) BMA64α plus 7 mM methionine. (Lariat I-E2) lariat intron-exon2.
Already after 4 h growth of strain YMA45/2 under nonpermissive conditions, just as the growth rate began to decline compared to the permissive state (Fig. 5A ▶), accumulation of the U3A and U3B precursor RNAs was detectable (Fig. 5B ▶, lane 2). Thus, repressing PRP46 expression, thereby depleting the protein from the cells, leads to a splicing defect that causes the accumulation of unspliced precursor RNAs. Therefore, we conclude that Prp46p is required for pre-mRNA splicing in S. cerevisiae.
Prp45p and Prp46p interact in vitro
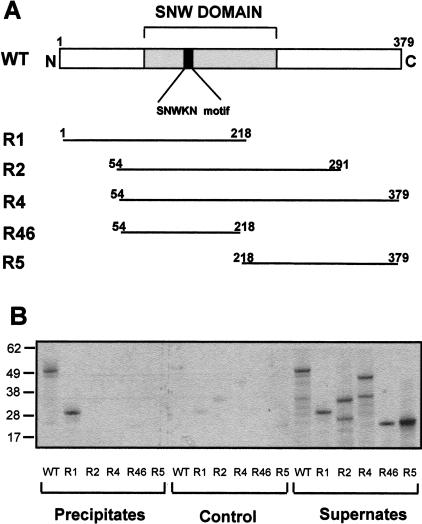
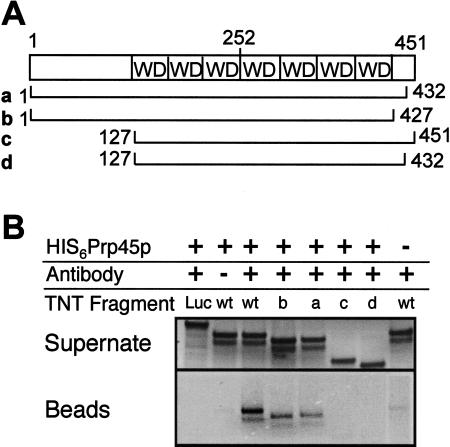
To test whether the two-hybrid interactions between Prp45p and Prp46p could be reproduced in vitro, a coupled in vitro transcription–translation system was used to produce 35S-labeled Prp45p or fragments thereof and these were incubated with yeast extract containing HA2-tagged Prp46p. As shown in Figure 6 ▶, HA2:Prp46p coprecipitated full-length Prp45p as well as an N-terminal fragment, R1, that contains amino acids 1 to 218, but not fragments R2, R4, or R46, all of which include the minimum region of interaction (amino acids 54–204) determined in the two-hybrid screen. To investigate the region of Prp46p required for this interaction and to determine whether these proteins interact directly, 35S-labeled fragments of Prp46p were produced in vitro and incubated with His6-tagged Prp45p that had been produced in Escherichia coli and affinity purified. As shown in Figure 7 ▶, His6:Prp45p coprecipitated full-length Prp46p as well as fragments ‘‘a’’ and ‘‘b’’ that lack the C-terminal 19 and 24 amino acids, respectively. Neither fragments ‘‘c’’ nor ‘‘d’’ was coprecipitated with His6-Prp45p, although they contain all seven WD repeats, and ‘‘d’’ corresponds to the minimum region of interaction determined in the two-hybrid screens. Other fragments ‘‘e,’’ ‘‘f,’’ ‘‘g,’’ and ‘‘h’’ that start at the N terminus, are truncated at the C end, and contain 6, 5, 3, or no WD repeats also failed to coprecipitate with Prp45p (data not shown).
FIGURE 6.
Prp45p coprecipitates with Prp46p in vitro. (A) 35S-methionine-labeled full-length Prp45p or fragments of the regions indicated were produced in vitro. (B) The in vitro -produced polypeptides were incubated with ATP-depleted yeast splicing extract containing HA2-tagged Prp46p and with anti-HA antibodies on ice for 1 h before adding protein A-Sepharose beads. The pellets and supernates (1.6%) were analyzed by SDS-PAGE and the 35S-labeled Prp45 polypeptides were visualized by autoradiography.
FIGURE 7.
Prp46p coprecipitates with Prp45p in vitro. (A) 35S-methionine-labeled full-length Prp46p or fragments of the regions indicated were produced in vitro. (B) His6-Prp45p affinity purified from E. coli extract was incubated with the in vitro produced Prp46 polypeptides on ice for 1 h before adding antipentahistine antibodies bound to protein A-Sepharose beads, or beads alone as a control. The pellets and supernates (1.6%) were analyzed by SDS-PAGE and the 35S-labeled Prp45 polypeptides were visualized by autoradiography.
DISCUSSION
Prp45-like proteins have been identified and characterized in other organisms, including SKIP/SNW1/NCoA-62 in humans (Dahl et al. 1998), Bx42 in Drosophila melanogaster (Wieland et al. 1992), SnwA in Dictyostelium discoideum (Folk et al. 1996), and Cwf13p/spSNW1 in fission yeast (Ambrozkova et al. 2001; Ohi et al. 2002). All these proteins share a conserved SNW domain (Baudino et al. 1998) that contains a 100% conserved SNWKN motif (Fig. 6A ▶). Experimental data have shown the involvement of SKIP as either an activator or a repressor for a number of different transcription pathways (Zhou et al. 2000; Leong et al. 2001; Zhang et al. 2001). On the other hand, SKIP has been demonstrated to be a component of human spliceosomes (Neubauer et al. 1998) although, until now, a role for SKIP/Prp45p in splicing had not been demonstrated.
Prp46p is a member of the ancient family of WD repeat proteins that have diverse functions in a variety of cellular processes (Smith et al. 1999). Prp46p shares greatest similarity with a subset of these proteins that are likely to be functional homologs. This includes the human protein PLRG1, which was demonstrated to be a component of human spliceosomes (Neubauer et al. 1998) and essential for splicing in HeLa nuclear extract (Ajuh et al. 2001), and the S. pombe prp5 protein. Mutation of the prp5 gene results in pre-mRNA splicing defects as well as a block to cell cycle progression at the G2/M phase (Potashkin et al. 1998; McDonald et al. 1999). The related Arabidopsis thaliana PRL1 protein regulates glucose and hormone responses (Nemeth et al. 1998). All these proteins contain a C-terminally located cluster of seven WD repeat units that are thought to mediate protein–protein interactions.
The interactions described here of Prp22p with Prp45p, of Prp45p with Prp46p, and of both Prp45p and Prp46p with Syf3p in two-hybrid screens also suggested possible roles for budding yeast Prp45p and Prp46p in the splicing process, and we demonstrate that metabolic depletion of either Prp45p or Prp46p results in a splicing defect (Figs. 2 and 5 ▶ ▶). Compatible with a role as a splicing factor, protA:Prp45p and HA-tagged Prp46p coprecipitated low levels of U2, U5, and U6 snRNAs (Fig. 4 ▶; data not shown); however, the amounts of these coprecipitated snRNAs were significantly lower than the levels of U4, U5, and U6 snRNAs coprecipitated with Prp8p, a component of U5 snRNPs and U4/U6.U5 tri-snRNPs. Because protA:Prp45p could be efficiently precipitated by IgG-agarose (confirmed by Western analysis; data not shown), and strongly coprecipitated spliceosomes, these data suggest that only a portion of the total pool of Prp45p or Prp46p was associated with the snRNAs under these conditions.
A number of proteins including Cef1p, Isy1p, Syf1p, Syf2p, and Syf3p have now been shown to interact with each other and with Prp45p and Prp46p in two-hybrid screens and coprecipitation assays (Dix et al. 1999; Ben-Yehuda et al. 2000; this work) and have been detected together with Prp45p and Prp46p in large protein complexes, including the Cef1p-associated ‘‘Cwc’’ complex (Ohi et al. 2002) and a penta-snRNP complex that may represent a form of recycled spliceosome (Stevens et al. 2002). In a high-throughput mass-spectrometric analysis (Gavin et al. 2002), Prp45p and Prp46p were detected in multiple complexes that contained overlapping subsets of these and other proteins; however, both proteins were rarely present in the same complex, and the functional significance of these different complexes is unclear.
It was previously reported that Isy1p, Syf1p, Syf2p, and Syf3p associate with low levels of U5 and U6 snRNAs, and under some conditions also with U2 snRNA (Dix et al. 1999; Russell et al. 2000), and it was proposed that this might indicate an association with a postsplicing complex (Dix et al. 1999). The salt-sensitive association of Prp45p with lower levels of U1 and U4 snRNAs supports this hypothesis, as following activation of the spliceosome, these two snRNAs are less strongly associated with this complex. Like Prp8p, Prp45p coprecipitated the excised intron lariat (Fig. 3 ▶). Thus, these data may indicate the presence of Prp45p and Prp46p in a postsplicing complex that contains the excised intron and the U2, U5, and U6 snRNPs, although the coexistence of all these factors in a single complex is not demonstrated here. Interestingly, the Cef1p-associated ‘‘Cwc’’ complex that includes Prp45p and Prp46p also contains U2, U5, and U6 snRNAs, which was reported to indicate spliceosome association (Ohi et al. 2002).
Significantly, Prp45p and Prp46p found each other as the predominant binding partners in two-hybrid screens, and both found Syf3p as a highly significant interactor (A1 category; Fromont-Racine et al. 1997). We have shown that in addition to their interaction in two-hybrid screens, Prp45p and Prp46p interact in vitro, most likely through direct protein–protein contact. In addition, the use of truncated forms of these proteins produced in vitro confirmed the approximate region of the proteins required for the interaction. In each case, however, the interaction required more of the in vitro produced proteins towards the N termini compared to the minimum regions found in the two-hybrid screens. As the two-hybrid prey are fusion proteins, fused at their N-terminal ends, this might simply reflect a requirement for additional sequence at the N termini of these proteins to promote correct folding and/or stability.
Ajuh et al. (2001) demonstrated that PLRG1 interacts with CDC5L, the human homolog of S. cerevisiae Cef1p (Xu et al. 1996). The interaction of PLRG1 with CDC5L was localized to the WD40 domain of PLRG1, the same region that in the yeast Prp46p interacts with Prp45p. Similarly, Cef1p has been shown to interact with the WD repeat region of Prp46p in budding yeast (Ohi and Gould 2002). Thus, Prp46p might act as a platform for protein–protein interactions at different stages in the splicing reaction, explaining why it has been found in several complexes.
Cef1p was not found as a binding partner in our two-hybrid screens with Prp45p or Prp46p, although it was identified in two-hybrid screens with Isy1p, Syf1p, Syf2p, Syf3p, and Ntc20p, which form a network of two-hybrid interactions that is compatible with the existence of a complex (Ben-Yehuda et al. 2000). The results presented here extend this network of interactions to include Prp45p, Prp46p, and Prp22p. Indeed, these correspond to a subset of the Cwc complex of proteins (McDonald et al. 1999; Ohi and Gould 2002; Ohi et al. 2002) and some of these proteins were identified as components of a Prp19p-associated (Ntc) complex (Chen et al. 2002).
The presence of Prp45p and Prp46p in a salt-sensitive penta-snRNP complex (Stevens et al. 2002) could explain the salt-sensitive snRNA associations observed here. Curiously, Prp22p was not present in the penta-snRNP complex, although it was detected by mass spectrometry as a potential Cwc component (Ohi et al. 2002). Thus the two-hybrid interactions that we detected between Prp22p as bait and Prp45p and Syf3p may represent transient interactions of Prp22p with these proteins in the spliceosome.
Analysis of the splicing of two different introns in cells that had been metabolically depleted of Prp45p indicated that, unlike Prp22p, Prp45p may be required for splicing irrespective of the different branchpoint–3′ splice site distances of the introns. This result argues against a possible role for Prp45p in directly modulating Prp22p function in 3′ splice site selection and cleavage, as in that case, one would expect to see a similar selective requirement for Prp45p only for splicing of introns with short branchpoint–3′ splice site distances. However, recent experiments using an intron-specific microarray to obtain an overview of the splicing of all pre-mRNAs in S. cerevisiae indicated that analyzing the effects of a mutant splicing factor on a few selected pre-mRNAs can be misleading (Clark et al. 2002). Prp18p, previously reported to be dispensable for splicing introns with branchpoint–3′ splice site distances of 17 nt or more in vitro, showed no such correlation when the whole genome was analyzed in vivo. It remains a possibility that Prp45p could act as a general second-step splicing factor in close proximity or even in contact with Prp22p at a stage late in the splicing process. Indeed it is conceivable that Prp45p might activate Prp22p in its reported function of releasing the spliced mRNA from the spliceosome (Company et al. 1991), leaving Prp45p with the residual intron-containing postsplicing complex.
Although the A1 category prey (those that are found as multiple fusions) in a two-hybrid screen are considered to be statistically more significant than others (Fromont-Racine et al. 1997), prey that occur as single fusions may also be functionally significant and may not arise as multiple fusions if the N terminus of the protein is essential for the interaction (A2 category), if most of the open reading frame is required (A3 category), or if the interaction only works with a specific fragment of the protein, no more and no less (A4 category). When multiple two-hybrid screens are performed, such single prey fusions become more statistically significant if they are found in several screens with functionally related proteins as bait (but not as nonspecific prey with unrelated bait proteins). This is the case with the budding yeast protein kinase Pkc1p, which was found as an interactor with Prp45p (A3) and Prp46p (A3), as well as with Cef1p (A1) and Syf2p (A4), as bait proteins (Ben-Yehuda et al. 2000; Table 1 ▶). Pkc1p has serine/threonine protein kinase activity, is required for signal transduction function in response to cell stress, and, like several of the splicing factors in this interaction network, is required to promote progression through the cell cycle from G2 to mitosis (Levin et al. 1992). Interestingly, the A. thaliana homolog of Prp46p, PRL1, acts as a heterologous receptor in vitro specifically for the protein kinase C-β II isoform (Nemeth et al. 1998). The yeast Prp46p and Prp45p sequences contain 14 or 7 potential Pkc-phosphorylation sites, respectively, as determined in a ScanProsite search, and when Neubauer et al. (1998) identified SKIP protein in assembled human splicing complexes, the protein separated into three distinct spots after two-dimensional gel electrophoresis, suggesting that SKIP might be subject to phosphorylation or other posttranslational modifications. It will therefore be interesting to determine whether the yeast Pkc1p interactions with spliceosome components represents a potential mechanism for regulating splicing through protein phosphorylation, with Prp46p and Prp45p being potential targets, or whether Prp46p simply acts as a facilitator for protein–protein interactions.
MATERIALS AND METHODS
Yeast growth media
Yeast minimal medium (YMM): 0.67% (w/v) Yeast nitrogen base without amino acids, 2% (w/v) glucose was supplemented with all amino acids except as indicated for auxotrophic selection. YMGRS: 0.67% (w/v) Yeast nitrogen base without amino acids, 2% (w/v) galactose, 2% (w/v) raffinose, 2% (w/v) sucrose was used for induction of the GAL1 promoter. YPDA: 1% (w/v) Yeast extract, 2% (w/v) Bacto-peptone, 2% (w/v) glucose, 0.003% (w/v) adenine sulfate.
Strains
Yeast strains used in this work (diploid markers are homozygous unless indicated otherwise): BMA38 (MATa/α, his3Δ200, leu2-3,112, ura3-1, trp1Δ1, ade2-1, can1-100), BMA38a (MATa, his3Δ200, leu2-3,112, ura3-1, trp1Δ1, ade2-1, can1-100), BMA64α (MATα, his3-11,15, leu2-3,112, ura3-1, trp1Δ1, ade2-1) and the Clontech strains CG1945, L40, and Y187 for two-hybrid screens. Strain YMA45/1 (MATa/α, his3Δ200, leu2-3,112, ura3-1, trp1Δ1, ade2-1, can1-100, PRP45/HIS3-PGAL1-ProtA:PRP45) was made from BMA38 by integration of a linear DNA cassette encoding the HIS3 gene, GAL1 promotor, and two IgG binding sites of the Staphylococcus aureus protein A directly upstream of the PRP45 open reading frame. The cassette was generated by PCR from plasmid pTL27 (Lafontaine and Tollervey 1996) using oligonucleotides PFUN-1 and PFUN-2. Correct integration of the cassette was checked by PCR and inducible production of protein A-tagged Prp45p was monitored by Western blotting. Sporulation of YMA45/1 and subsequent selection for histidine prototrophic spores produced YMA45/2 (MATa, his3Δ200, leu2-3,112, ura3-1, trp1Δ1, ade2-1, can1-100, HIS3-PGAL1-ProtA:PRP45). BMA38a carrying pNOPPATAIL (gift from E. Hurt, Biochemie-Zentrum Heidelberg (BZH), Heidelberg, Germany), which encodes the double protein A epitope, was used as a control.
Strain YMA151KO1 (MATa/α, his3Δ200, leu2-3,112, ura3-1, trp1Δ1, ade2-1, can1-100, PRP46/prp46Δ::HIS3) was constructed from BMA38 by complete replacement of the PRP46 open reading frame with a HIS3 cassette generated using oligonucleotides PLKO-1 and PLKO-2 as described in Baudin et al. (1993).
Strain YMA151/2 (MATα, his3-11,15, leu2-3,112, ura3-1, trp1Δ1, ade2-1, TRP1-PMET3-HA2:PRP46) was generated from BMA64α by targeted integration of a linear cassette containing the TRP1 gene, the MET3-promoter and a double HA-epitope directly upstream of the PRP46 coding sequence. The cassette was produced by PCR from plasmid pUC19-HA55 (van Nues and Beggs 2001) with oligonucleotides 151MetA and 151MetB.
Oligonucleotides
PFun-1: 5′-TTACCTTAACGTATTATTGTAATTCTTCACGAAT TTGATTCTCTTGGCCTCCTCTAGT-3′
PFun-2: 5′-CTTGAGAATGTTTTGGAGGTGGTAGTCTGTTAC TAAACATATTCGCGTCTACTTTCGG-3′
Func1: 5′-TATGAATTCATGTTTAGTAACAGAC-3′
Func2: 5′-TATGTCGACCTAGGCGCCATAGTTATCC-3′
PLKO-1: 5′-GAGGATGCAGACACTGTGTTACATGGAGATTA GTGAGACTCTTGGCCTCCTCTAG-3′
PLKO-2: 5′-CACGTATACAGGGTACGTACTTTTTCCATCTAC TCCCATCGTTCAGAATGACACG-3′
151MetA: 5′-CCACACAAATCCACGATGACCTAAGAACATTC GTTCGCTTAAATCCTTAAATAAATACTACTC-3′
151MetB: 5′-TGTCTACATCTCCTAAATTTTCGACTTTGTGAT CATTTCCGTCCATACGAGCTCCAGCGTAATCTGGAA-3′
151-PR2: 5′-GTATAAAGCCGAAGTCC-3′
YPL151A: 5′-AAGGATCCTAGTGAGAATGGACGG-3′
YPL151B: 5′-AAGGATCCTTCCATCTACTCCCAC-3′
Met3: 5′-TTTAGCTTGTGATCTC-3′
PGK: 5′-ACCGTTTGGTCTACCCAAGTGAGAAGCCAAGACA-3′
RP28A: 5′-TCGTACTGATGCTCCATTC-3′
RP28B: 5′-TGAAACCCTTAGATCTTC-3′
anti-U1: 5′-CACGCCTTCCGCGCCGT-3′
anti-U2: 5′-CTACACTTGATCTAAGCCAAAAG-3′
anti-U4: 5′-CCGTGCATAAGGAT-3′
anti-U5: 5′-AATATGGCAAGCCC-3′
anti-U6: 5′-TC(A/T)TCTCTGTATTG-3′
Plasmids
To make the Gal4:Prp22p two-hybrid bait construct, plasmid pBSKS22 (gift from B. Schwer, Cornell University, NY) was digested with KpnI and blunt-ended by T4 polymerase treatment. After NdeI cleavage, the PRP22-containing fragment was ligated into NdeI/SmaI-digested pAS2ΔΔ (Fromont-Racine et al. 1997). To construct the LexA:Prp45p two-hybrid bait plasmid, the PRP45 ORF was PCR-amplified from yeast genomic DNA using oligonucleotides Func1 and Func2. The PCR product was cleaved with EcoRI, SalI and ligated in frame into EcoRI/SalI-digested pBTM116 (Vojtek et al. 1993).
The LexA:PRP46 two-hybrid bait plasmid was constructed by PCR amplification of the YPL151c ORF using oligonucleotides YPL151A and YPL151B as primers and cloning into BamHI-cleaved pBTM116, in frame with the preceding LEXA sequence.
All constructs were verified by DNA sequencing, and protein expression was tested by Western analysis using anti-Gal4-DNA binding domain antibodies (Santa Cruz Biotechnology) and/or by complementation of a yeast strain bearing a mutation in the corresponding genomic sequence. To make pETMA45 (for His6:Prp45p production in E. coli), the PRP45 ORF was isolated from pMA45 as an EcoRI-SalI fragment, pET19b was digested with NdeI, and both were treated with Klenow and ligated together.
Two-hybrid screens
Two-hybrid screens were performed as described in Fromont-Racine et al. (1997). In the Prp22p screen, positives were selected for growth on YMM-LWH agar and tested for β-galactosidase activity in an X-gal agar overlay assay (Fromont-Racine et al. 1997). In addition, relative strength of reporter gene activation was determined in a β-galactosidase filterlift assay (Transy and Legrain 1995). In the Prp45p two-hybrid screen, positives were selected first on YMM-LWH agar plates, then suspended in microtiterplates and replicated onto YMM-LWH agar containing 20 mM 3-aminotriazole (3AT; which increases the stringency of the His+ selection). One hundred fifteen clones that grew on 20 mM 3AT were analyzed further. The Prp46p screen was as for Prp45p but using 10 mM 3AT, and 34 clones were obtained.
Monitoring yeast cell growth
Cultures were grown to mid-log phase under permissive conditions (galactose-based medium or medium lacking methionine), then used to inoculate media providing either permissive or repressing (glucose or presence of methionine) conditions to an O.D.600 of 0.05–0.1 and grown at 30°C. Maintenance of logarithmic growth was ensured by diluting the cultures with prewarmed medium to keep the O.D.600 readings below 0.8.
Primer extension assay
Yeast total RNA was prepared using the method of Schmitt et al. (1990). Oligonucleotide primers were labeled with T4 polynucleotide kinase, and primer extension was performed and the products analyzed on a 6% (w/v) polyacrylamide gel as described (Beltrame and Tollervey 1992).
Splicing extract preparation and in vitro splicing reactions
Preparation of yeast whole-cell extracts and in vitro splicing reactions were performed as described (Lin et al. 1985). Plasmid p283, which contains an AluI fragment of the yeast actin gene, was transcribed in vitro with T7 RNA polymerase and [α-32P]UTP to produce uniformly labeled splicing substrate (O’Keefe et al. 1996). Splicing reactions (5 μL for splicing activity or 50 μL for immunoprecipitation) were incubated at 23°C for 20 min, and the reaction products were fractionated on a 6% (w/v) polyacrylamide-8 M urea gel and visualized by autoradiography.
Western blot analysis
Proteins were fractionated by SDS-PAGE and detected using peroxidase-anti-peroxidase (PAP; Sigma) and ECL (Amersham) according to the manufacturer’s instructions.
Immunoprecipitations
Immunoprecipitations of snRNPs and spliceosomes were performed as described (Teigelkamp et al. 1995a), using IgG-agarose (Sigma) or anti-Prp8 polyclonal antibodies (anti-8.6), and washes containing 150 mM NaCl (or, when indicated, 75 mM NaCl).
Northern analysis of snRNAs
Immunoprecipitates obtained using IgG-agarose (Sigma) or anti-Prp8 polyclonal antibodies (anti-8.6; Teigelkamp et al. 1995a) were washed, deproteinized by SDS/proteinase K treatment followed by phenol:chloroform:isoamylalcohol extraction, and the RNA was fractionated on a 6% polyacrylamide gel. The snRNAs were detected by Northern analysis using end-labeled oligonucleotides complementary to the U1, U2, U4, U5, or U6 snRNAs (Cooper et al. 1995).
Protein production in bacteria
Proteins were produced in E. coli BL21(DE3)pLysS cells grown at 23–30°C to an O.D.600 of 0.3–0.8 (optimal conditions determined for each protein). IPTG was added to 0.5–1 mM and cultures were incubated for 1–3 h to induce expression. Cell pellets were resuspended in chilled lysis buffer A (50 mM Tris-HCl at pH 7.5, 250 mM NaCl, 10% [w/v] sucrose) containing Sigma P8749 protease inhibitor cocktail, lysozyme was added to 0.2 mg/mL, and the extract was stirred on ice for 45 min. After addition of Triton-X-100 to 0.1% [v/v] and stirring on ice for 5 min, the lysate was centrifuged at 17,000 rpm at 4°C for 45 min and the supernate was stored at −70°C until metal affinity purification of the His-tagged proteins with Ni-NTA agarose (Qiagen).
In vitro synthesis of 35S-labeled proteins
PCR-generated templates were used in an in vitro transcription/translation system (Promega TNT T7 Quick for PCR) according to the manufacturer’s instructions, using 0.37 MBq 35S-methionine (Amersham) for labeling.
Coimmunoprecipitation of proteins
TNT reaction (6 μL) was mixed with 40 μL His-tagged Prp45 protein or BC100 (controls; 20 mM Tris-HCl at pH 7.9, 100 mM KCl, 20% [v/v] glycerol) and incubated at room temperature for 10 min followed by 1 h on ice. Antipentahistidine antibodies (Qiagen) bound to protein A-Sepharose beads were added, or beads alone as a control, and samples incubated at 4°C for 2 h with mixing. Supernates were removed and beads were washed three times with IP300 (6 mM HEPES, 300 mM NaCl, 2.5 mM MgCl2, 0.05% [v/v] Nonidet P40) and suspended in 25 μL 2× SDS buffer (125 mM Tris at pH6.8, 4% SDS, 200 mM DTT, 40% glycerol, 0.02% bromophenol blue). Samples were heated to 70°C and analyzed by SDS-PAGE (Nu-PAGE 4–12% Bis-Tris in MOPS buffer). Gels were dried before autoradiography.
For immunoprecipitation experiments using yeast extract, 25 μL of HA2-tagged PRP46 splicing extract was depleted of ATP by adding 2 mM glucose and incubating at 24°C for 20 min. 35S-labeled PRP45 polypeptides (5 μL TNT reaction) and 4 μL of anti-HA antibodies (12CA5, Roche) were added and the NaCl adjusted to 150 mM with IP300. The mixture was incubated on ice for 1 h before adding protein A-Sepharose beads. The samples were rotated at 4°C for 2 h, then washed three times with ice-cold IP150 (as IP300 but 150 mM NaCl). The pellets and supernates (1.6%) were suspended in 1× SDS-loading buffer and analyzed by SDS-PAGE as above.
Acknowledgments
We thank Michael Mellor-Clark and Alejandra M. Clark for their contributions to constructing the Prp46 bait plasmid and performing the Prp46p two-hybrid screen, and Samantha Kaye for bioinformatic support. We also benefited from interactions with partners, especially Pierre Legrain and Micheline Fromont-Racine, in the EU-funded TAPIR network (Biotech 95007) and with the Edinburgh Protein Interaction Centre. This work was funded by studentships from The Darwin Trust of Edinburgh to M.A. and M.M. and by Wellcome Trust Grant 047685.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2119903.
REFERENCES
- Ajuh, P., Kuster, B., Panov, K., Zomerdijk, J.C.B.M., Mann, M., and Lamond, A.I. 2000. Functional analysis of the human CDC5L complex and identification of its components by mass spectrometry. EMBO J. 19: 6569–6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajuh, P., Sleeman, J., Chusainow, J., and Lamond, A.I. 2001. A direct interaction between the carboxyl-terminal region of CDC5L and the WD40 domain of PLRG1 is essential for pre-mRNA splicing. J. Biol. Chem. 276: 42370–42381. [DOI] [PubMed] [Google Scholar]
- Ambrozkova, M., Puta, F., Fukova, I., Skruzny, M., Brabek, J., and Folk, P. 2001. The fission yeast ortholog of the coregulator SKIP interacts with the small subunit of U2AF. Biochem. Biophys. Res. Commun. 284: 1148–1154. [DOI] [PubMed] [Google Scholar]
- Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F., and Cullin, C. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21: 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudino, T.A., Kraichely, D.M., Jefcoat, Jr., F.C., Winchester, S.K., Partridge, N.C., and MacDonald, P.N. 1998. Isolation and characterization of a novel coactivator protein, NCoA-62, involved in vitamin D-mediated transcription. J. Biol. Chem. 273: 16434– 16441. [DOI] [PubMed] [Google Scholar]
- Beggs, J.D. 1995. Yeast splicing factors and genetic strategies for their analysis. In Pre-mRNA processing (ed. A.I. Lamond), pp. 79–95. R.G. Landes Company, Dallas, TX.
- Beltrame, M. and Tollervey, D. 1992. Identification and functional analysis of two U3 binding sites on yeast preribosomal RNA. EMBO J. 11: 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda, S., Dix, I., Russell, C.S., Levy, S., Beggs, J.D., and Kupiec, M. 2000. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics 156: 1503–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C.B., Tuschl, T., and Sharp, P.A. 1999. Splicing of precursors to mRNAs by the spliceosome. In The RNA world, 2nd ed. (eds. R.F. Gesteland, T.R. Cech, and J.F. Atkins), pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- Caceres, J.F. and Krainer, A.R. 1997. Mammalian pre-mRNA splicing factors. In Eukaryotic mRNA processing (ed. A.R. Krainer), pp. 174–212. IRL Press, Oxford.
- Chen, C.H., Yu, W.C., Tsao, T.Y., Wang, L.Y., Chen, H.R., Lin, J.Y., Tsai, W.Y., and Cheng, S.C. 2002. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 30: 1029–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S., McLean, M.R., and Rymond, B.C. 1999. Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, T.A., Sugnet, C.W., and Ares, Jr., M. 2002. Genomewide analysis of mRNA precessing in yeast using splicing-specific microarrays. Science 296: 907–910. [DOI] [PubMed] [Google Scholar]
- Company, M., Arenas, J., and Abelson, J. 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349: 487–493. [DOI] [PubMed] [Google Scholar]
- Cooper, M., Parkes, V., Johnston, L.H., and Beggs, J.D. 1995. Identification and characterization of Uss1p (Sdb23p): A novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 14: 2066–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl, R., Wani, B., and Hayman, M.J. 1998. The Ski oncoprotein interacts with Skip, the human homolog of Drosophila Bx42. Oncogene 16: 1579–1586. [DOI] [PubMed] [Google Scholar]
- Dix, I., Russell, C.S., Ben-Yehuda, S., Kupiec, M., and Beggs, J.D. 1999. The identification and characterization of a novel splicing protein, Isy1p, of Saccharomyces cerevisiae. RNA 5: 360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk, P., Puta, F., Krpejsova, L., Blahuskova, A., Markos, A., Rabino, M., and Dottin, R.P. 1996. The homolog of chromatin binding protein Bx42 identified in Dictyostelium. Gene 181: 229–231. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Rain, J.-C., and Legrain, P. 1997. Towards a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16: 277–282. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147. [DOI] [PubMed] [Google Scholar]
- Harris, S.D., Cheng, J., Pygh, T.A., and Pringle, J.R. 1992. Molecular analysis of Saccharomyces cerevisiae chromosome I. On the number of genes and the identification number of essential genes using temperature-sensitive-lethal mutations. J. Mol. Biol. 225: 53–65. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D. and Tollervey, D. 1996. One step PCR mediated strategy for the construction of the conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24: 3469–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong, G.M., Subramaniam, N., Figueroa, J., Flanagan, J.L., Hayman, M.J., Eisman, J.A., and Kouzmenko, A.P. 2001. Ski-interacting protein interacts with Smad proteins to augment transforming growth factor-dependent transcription. J. Biol. Chem. 276: 18243–18248. [DOI] [PubMed] [Google Scholar]
- Levin, D.E. and Bertlett-Heubusch, E. 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell. Biol. 116: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R.-J., Newman, A.J., Cheng, S.-C., and Abelson, J. 1985. Yeast mRNA splicing in vitro. J. Biol. Chem. 260: 14780–14792. [PubMed] [Google Scholar]
- McDonald, W.H., Ohi, H.R., Smelkova, N., Frendewey, D., and Gould, K.L. 1999. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol. Cell. Biol. 19: 5352–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, T. and Ogawa, H. 1999. The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase-like protein, is required for crossover control in meiosis. EMBO J. 18: 5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth, K., Salchert, K., Putnoky, P., Bhalerao, R., Koncz-Kalman, Z., Stankovic-Stangeland, B., Bako, L., Mathur, J., Okresz, L., Stabel, S., et al. 1998. Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes & Dev. 12: 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer, G., Gottschalk, A., Fabrizio, P., Séraphin, B., Lührmann, R., and Mann, M. 1997. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc. Natl. Acad. Sci. USA 94: 385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer, G., King, A., Rappsilber, J., Calvio, S.C., Watson, M., Ajuh, P., Sleeman, J., Lamond, A., and Mann, M. 1998. Mass spectrometry and EST database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20: 46–50. [DOI] [PubMed] [Google Scholar]
- Nilsen, T.W. 1998. RNA–RNA interactions in nuclear pre-mRNA splicing. In RNA structure and function. (eds. R. Simons and M. Grunberg-Manago), pp. 279–307. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- ———. 2002. The spliceosome: No assembly required? Mol. Cell 9: 8–9. [DOI] [PubMed] [Google Scholar]
- O’Keefe, R.T., Norman, C., and Newman, A.J. 1996. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell 86: 679–689. [DOI] [PubMed] [Google Scholar]
- Ohi, M.D. and Gould, K.L. 2002. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8: 798–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi, M.D., Link, A.J., Jennings, J.L., McDonald, W.H., Ren, L., and Gould, K.L. 2002. Proteomics analysis reveals stable multi-protein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol. Cell. Biol. 22: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumpton, M., McGarvey, M., and Beggs, J.D. 1994. A dominant negative mutation in the conserved RNA helicase motif SAT causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 13: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potashkin, J., Kim, D., Fons, M., Humphrey, T., and Frendeway, D. 1998. Cell division cycle defects associated with fission yeast pre-mRNA splicing mutants. Curr. Genet. 34: 153–163. [DOI] [PubMed] [Google Scholar]
- Russell, C.S., Ben-Yehuda, S., Dix, I., Kupiec, M., and Beggs, J.D. 2000. Functional analyses of interacting factors involved in both pre-mRNA splicing and cell cycle progresion in Saccharomyces cerevisiae. RNA 6: 1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, M.E., Brown, T.A., and Trumpower, B.L. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18: 3091–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B. and Gross, C.H. 1998. Prp22, a DExH box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17: 2085–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T.F., Gaitatzes, C., Saxena, K., and Neer, E.J. 1999. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185. [DOI] [PubMed] [Google Scholar]
- Stevens, S.W. and Abelson, J. 1999. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl. Acad. Sci. USA 96: 7226–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, S.W., Ryan, D.E., Ge, H.Y., Moore, R.E., Young, M.K., Lee, T.D., and Abelson, J. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9: 31–44. [DOI] [PubMed] [Google Scholar]
- Teigelkamp, S., Newman, A.J., and Beggs, J.D. 1995a. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J. 14: 2602–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp, S., Whittaker, E., and Beggs, J.D. 1995b. Interaction of the yeast splicing factor PRP8 with substrate RNA during both steps of splicing. Nucleic Acids Res. 23: 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Transy, C. and Legrain, P. 1995. The two-hybrid: An in vivo protein–protein interaction assay. Mol. Biol. Rep. 21: 119–127. [DOI] [PubMed] [Google Scholar]
- van Nues, R.W. and Beggs, J.D. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes. Genetics 157: 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek, A.B., Hollenberg, S.M., and Cooper, J.A. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74: 205–214. [DOI] [PubMed] [Google Scholar]
- Wieland, C., Mann, S., von Besser, H., and Saumweber, H. 1992. The Drosophila nuclear protein Bx42, which is found in many puffs on polytene chromosomes, is highly charged. Chromosoma 101: 517–525. [DOI] [PubMed] [Google Scholar]
- Will, C.L. and Lührmann, R. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell. Biol. 13: 290–301. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Nouraini, S., Field, D., Tang, S.J., and Friesen, J.D. 1996. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature 381: 709–713. [DOI] [PubMed] [Google Scholar]
- Zhang, C., Baudino, T.A., Dowd, D.R., Tokumaru, H., Wang, W., and MacDonald, P.N. 2001. Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription. J. Biol. Chem. 276: 40414–40620. [DOI] [PubMed] [Google Scholar]
- Zhou, S., Fujimoro, M., Hsieh, J.J., Chen, L., Miyamoto, A., Weinmaster, G., and Hayward, D.S. 2000. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Biol. Cell 20: 2400–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]