Abstract
Polydnaviruses, obligatorily associated with endoparasitoid wasps, are unique in that their segmented genome is composed of multiple double-stranded DNA circles. We present here the first cytological evidence that virus segments are integrated in the wasp genome, obtained by using in situ hybridization of virus probes with viral sequences in the chromosomes of a wasp from the braconid family of hymenopterans.
Viruses are generally considered as molecular parasites that are pathological for the organisms in which they replicate. They might be neutral but are not expected to provide a selective advantage to the infected organism. However, there is a major exception: the case of polydnaviruses (PDVs), obligatorily associated with thousands of species of parasitoid hymenopterans which develop within lepidopteran host insects. Polydnavirus particles produced in the female wasp's ovaries are injected into host larvae during oviposition. PDV gene expression in host cells protects the parasitoid progeny by inducing both an inhibition of the immune defenses and a developmental arrest of the host (1, 4, 10, 17, 19).
Polydnaviruses have a segmented genome composed of dozens of double-stranded DNA (dsDNA) circles (4, 19). The sequence of each circular molecule is also present in the wasp in a linear form integrated into high-molecular-weight DNA (5, 7, 15, 22). The results of molecular analyses of two PDV segments in different species have demonstrated that sequences flanking virus segments were not included in the virus particles (5, 15). Thus, it was assumed that this DNA corresponded to the wasp chromosomal sequences, indicating that virus segments were integrated as proviruses in the wasp genome. However, this interpretation has not been verified to date through a cytological approach. Here we present the first visualization of PDV sequences integrated within a wasp genome using in situ hybridization of viral probes on C. congregata (Braconidae, Microgastrinae) chromosomes and in addition demonstrate that multiple probes bind to a common site on chromosome 5, suggesting that the PDV genome is organized as a macrolocus on this chromosome.
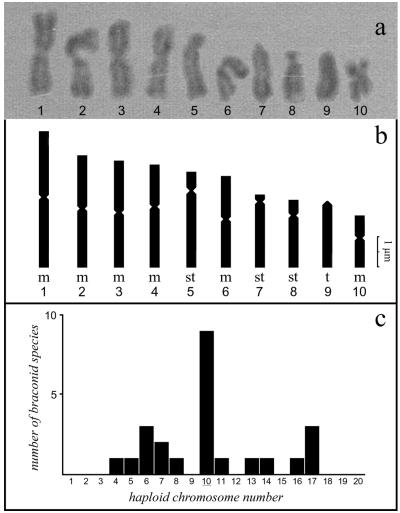
C. congregata karyotype.
Metaphase spreads were prepared as previously described for Drosophila melanogaster (11) using developing brain and gonad cells from wasp larvae at the prepupal stage (13). The complementary sex determination model applies for sex determination in many hymenopteran species (12). In this model, individuals heterozygous at the sex locus develop as females, while hemizygous and homozygous individuals develop as haploid and diploid males, respectively. The C. congregata haploid set of chromosomes (n = 10) was identified using haploid males that constitute the sole progeny of virgin females. As previously suggested (16), diploid males (2 n = 20) were regularly observed in the progeny of mated females in addition to haploid males and diploid females, indicative of a low level of polymorphism at the sex locus in the laboratory strain.
The C. congregata karyogramme shown in Fig. 1 was established from the pictures of 10 metaphase spreads (5 haploid males and 5 diploid females). The chromosome number determined corresponds to the number mode of chromosome distribution in the few braconid species analyzed to date (6) (Fig. 1c). The C. congregata karyotype is comprised of six metacentric (1, 2, 3, 4, 6, and 10), three subtelocentric (5, 7, and 8) and one acrocentric chromosome (9). The size and organization of chromosomes 2, 3, 4, and 6 are roughly similar, while the other chromosomes can be clearly distinguished on the basis of their morphology (see Fig. 1a and 1b).
FIG. 1.
(a) C. congregata karyotype. (b) Schematic diagram of the haploid chromosome set, composed of metacentric (m), subtelocentric (st), and telocentric (t) chromosomes. (c) Bar graph depicting the distribution of chromosome numbers previously determined in braconid species of hymenopterans.
In situ hybridization of rDNA probes to C. congregata chromosomes.
To set up in situ hybridization on C. congregata chromosomes, a fragment corresponding to rDNA sequence, highly conserved among insects, was used as a probe. The rDNA probe was Py12 cloned in pBr322 (13). It contains the Drosophila melanogaster 18S, 5.8S, and 28S genes plus intergenic spacer sequences. Methods for hybridization followed the procedure used before (14). The sites of hybridization were detected by an immunocytological reaction between biotinylated DNA probe and enzyme-conjugated avidin. The peroxidase is revealed by diaminobenzidine, and the brownish reaction product is observed on the Giemsa-stained chromosomes.
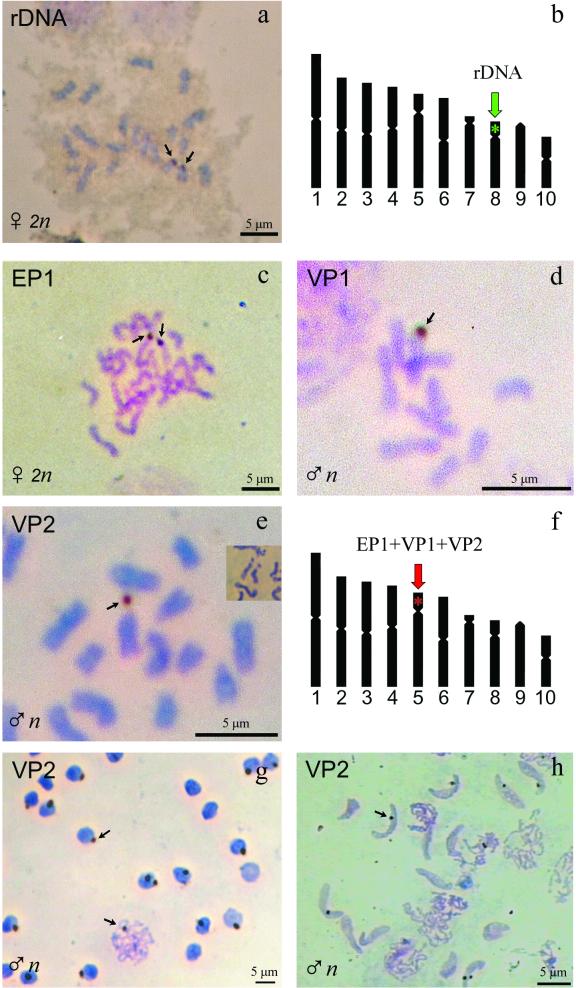
A unique hybridization signal was identified with the rDNA probe on metaphase spreads from haploid larvae cells, located on the dot-shaped short arm of chromosome 8, the smallest subtelocentric chromosome. On metaphase spreads from diploid prepupal cells, two signals were observed in the same region of both chromosome 8 homologues (Fig. 2a). Interphase nuclei were also hybridized with the probe and showed generally one signal when male cells were analyzed and two spots for females. The data obtained indicate that the experimental conditions used are suitable for in situ hybridization of DNA probes on C. congregata chromosomes.
FIG. 2.
(a) In situ hybridization of C. congregata chromosomes with the rDNA probe in developing brain of prepupal stage wasps. (b) Summary of the results, showing that the location of hybridization (arrows) corresponds to the short arm of chromosome 8. (c, d, and e) In situ hybridization of C. congregata chromosomes with the C. congregata PDV probes EP1, VP1, and VP2, respectively, corresponding to different virus segments. Developing brain cells were used in panels c and e, and developing gonads were analyzed in panel d. In panel e, the inset shows that the primary constriction of chromosome 5 can be elastic, so that the dot-shaped arm of chromosome 5 occasionally appears as separated from the centromere, as in the VP2 hybridization picture. (f) Summary of the results, showing that the location of hybridization (arrows) is common to all the virus sequences used as probes and corresponds to the short arm of chromosome 5 (red star). (g) Hybridization of the VP2 probe on interphase nuclei and on meiotic chromosomes from haploid male cells. (h) Hybridization of the same VP2 probe on chromosomal DNA in nuclei from spermatids, where the virus sequences appear to have a common location within the condensed chromatin.
Visualization of EP1 polydnavirus sequences on C. congregata chromosome 5.
To analyze polydnavirus sequences integrated in C. congregata chromosomes, we have a fragment of the cDNA of the early protein 1 (EP1) gene, located on the smallest segment of the virus genome. This gene encodes a virus protein which is expressed abundantly in parasitized host hemolymph in tobacco hornworm larvae and other permissive hosts (8, 9). In Southern blot analysis using male wasp DNA, the EP1 probe gives a hybridization pattern resembling that of a single-copy chromosomal gene (15). Accordingly, metaphase spreads from haploid males cells showed a unique hybridization signal located on the short arm of the subtelocentric chromosome 5.
Chromosome 5 can be readily distinguished because it is much larger than the other subtelocentric chromosomes, chromosome 7 (30% larger) and chromosome 8 (40% larger). Moreover, chromosome 5 has a small but distinct short arm, while chromosome 7 is nearly acrocentric. On metaphase spreads from diploid cells, the signals were observed on both chromosome 5 homologues (Fig. 2c). Interphase nuclei were also hybridized with the EP1 probe and showed generally one signal when haploid male cells were analyzed and two spots for diploid female prepupal cells. Thus, the data indicate unambiguously that the EP1 segment is integrated into chromosomal DNA and that it is localized on the short arm of C. congregata chromosome 5.
Common localization of multiple viral probes suggests a single integration event.
To confirm that the integration of C. congregata PDV segments into C. congregata chromosomes is not restricted to the EP1 sequence, we performed in situ hybridization using two other virus probes. This analysis also addressed the question of the overall organization of the different segments within the wasp genome and permitted the identification of the location of the different viral sequences, which could be either localized at a single chromosomal locus or scattered throughout the wasp chromosomes.
The probes VP1 and VP2 were randomly cloned from XhoI restriction digest fragments of C. congregata PDV DNA which had been purified as previously described (2). Southern blot analysis of undigested purified virus DNA indicated that the VP1 and VP2 probes are specific for two different large virus segments. Strikingly, in situ hybridization of the VP1 and VP2 probes on C. congregata chromosomes gave results similar to those obtained using the EP1 probe. A unique hybridization signal was present on most metaphase spreads from haploid larvae cells, located on the short arm of chromosome 5 (Fig. 2d and e). In metaphase spreads from diploid larvae cells, two signals were generally observed on the chromosome 5 homologues. Interphase nuclei showed one signal when haploid cells were analyzed (Fig. 2g), and two spots were visualized in diploid female cells.
Spermatid nuclei were hybridized with the VP2 probe, and surprisingly, the signals were always localized at the same relative position between the extremities of the nucleus, on the same side and at the periphery (Fig. 2h). Previous studies on human sperm nuclei have yielded a model for sperm nucleus organization in which the centromeres are grouped into a “chromocenter” central inside the sperm nucleus and the telomeres are associated in doublets at the periphery (23). If this model is applicable to C. congregata spermatids, the pattern of hybridization observed indicates a telomeric location of PDV sequences. The precise localization of the signal also suggests that C. congregata chromosomes are arranged in a defined order in the spermatid nucleus, as described for the monotrene mammalian sperm nucleus (18).
We present here the first cytological evidence to demonstrate PDV sequence integration in wasp chromosomal DNA. The C. congregata segments analyzed were all visualized at the same chromosomal location, indicating that they are clustered on chromosome 5 (Fig. 2f). Recent phylogenetic studies indicate that all the wasp species of the braconid family of hymenopterans harboring polydnaviruses have a monophyletic origin (3, 20). Thus, more than 17,000 species conceivably originated from a single event, estimated to have occurred 60 million years ago: the integration of a PDV ancestor as a provirus into the chromosomes of an ancestral wasp species (21).
The PDV would have driven the diversification of this group by facilitating the adaptation of the parasitoid wasps to the physiological contexts of thousands of lepidopteran hosts. Given the common localization of multiple probes derived from different genomic segments, we suggest that the virus genome might be organized as a macrolocus on chromosome 5. Such an organization would support the hypothesis of the unique origin of the braconid wasp-PDV association, since a single integration event would have been sufficient to integrate all the virus sequences in the ancestor wasp genome.
Acknowledgments
This work was supported in France (E.B., J.R., M.P., F.L., and J.-M.D.) by the CNRS grants “Interactions Durables,” “GDR Éléments Transposables,” and “IFR Biologie des Transposons et des Virus” and in the United States by the NSF and the University of California—Riverside (N.E.B.).
REFERENCES
- 1.Beckage, N. E. 1998. Modulation of immune responses to parasitoids by polydnaviruses. Parasitology 116:57-64. [DOI] [PubMed] [Google Scholar]
- 2.Beckage, N. E., F. F. Tan, K. W. Schleifer, R. D. Lane, and L. L. Cherubin. 1994. Characterisation and biological effects of C. congregata polydnavirus on host larvae of the tobacco hornworm, Manduca sexta. Arch. Insect Biochem. Physiol. 26:165-195. [Google Scholar]
- 3.Dowton, M., and A. D. Austin. 1998. Phylogenetic relationships among the microgastroid (Hymenoptera: Braconidae): combined analysis of 16S and 28S rDNA genes, and morphological data. Mol. Phylogenet. Evol. 10:354-366. [DOI] [PubMed] [Google Scholar]
- 4.Drezen, J.-M., E. Huguet, C. Dupuy, and, M. Poirié. 2000. Polydnaviruses: how viruses evolve after domestication, p. 45-77. In Global Research Network (ed.), Research advances in virology, vol. 1. Global Research Network, Puthen Chalai, India. [Google Scholar]
- 5.Fleming, J. G. W., and M. D. Summers. 1991. Polydnavirus DNA is integrated in the DNA of its parasitoid wasp host. Proc. Natl. Acad. Sci. USA 88:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gokhman, V. E., and D. L. J. Quicke. 1995. The last twenty years of parasitic hymenoptera karyology: an update and phylogenetic implications. J. Hymenoptera Res. 4:41-63. [Google Scholar]
- 7.Gruber, A., P. Stettler, P. Heiniger, D. Schumperli, and B. Lanzrein. 1996. Polydnavirus DNA of the braconid wasp Chelonus inanitus is integrated in the wasp genome and excised only in later pupal and adult stages of the female. J. Gen. Virol. 77:2873-2879. [DOI] [PubMed] [Google Scholar]
- 8.Harwood, S. H., and N. E. Beckage. 1994. Purification and characterization of an early-expressed polydnavirus induced protein from the hemolymph of Manduca sexta larvae parasitized by C. congregata. Insect Biochem. Mol. Biol. 24:685-698. [Google Scholar]
- 9.Harwood, S. H., A. J. Grosovsky, E. A. Cowles, J. W. Davis, and N. E. Beckage. 1994. An abundantly expressed hemolymph glycoprotein isolated from newly parasitized Manduca sexta larvae is a polydnavirus gene product. Virology 205:381-392. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence, P. O., and B. Lanzrein. 1993. Hormonal interactions between insect endoparasites and their host insects, p. 59-85. In N. E. Beckage, S. N. Thompson, and B. A. Federici (ed.), Parasites and pathogens of insects, vol. 1. Academic Press, New York, N.Y. [Google Scholar]
- 11.Lemeunier, F., and M. Ashburner. 1984. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). IV. The chromosomes of two new species. Chromosoma 89:343-351. [Google Scholar]
- 12.Poirié, M., G. Periquet, and L. Beukeboom. 1992. The hymenopteran way of determining sex. Semin. Dev. Biol. 3:357-361. [Google Scholar]
- 13.Rousselet, J., C. Géri, G. M. Hewitt, and F. Lemeunier. 1998. The chromosomes of Diprion pini and D. similis (Hym.: Diprionidae): implications for karyotype evolution. Heredity 81:573-578. [DOI] [PubMed] [Google Scholar]
- 14.Rousselet, J., L. Monti, M.-A. Auger-Rozenberg, J. S. Parker, and F. Lemeunier. 2000. Chromosome fission associated growth of ribosomal DNA in Neodiprion abietis (Hymenoptera, Diprionidae). Proc. R. Soc. Lond. B 267:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savary, S., N. E. Beckage, F. Tan, G. Periquet, and J. M. Drezen. 1997. Excision of the polydnavirus chromosomal integrated EP1 sequence of the parasitoid wasp C. congregata (Braconidae, Microgastrinae) at potential recombinase binding sites. J. Gen. Virol. 78:3125-3134. [DOI] [PubMed] [Google Scholar]
- 16.Savary, S., J. M. Drezen, F. Tan, N. E. Beckage, and G. Periquet. 1999. The excision of polydnavirus sequences from the genome of the wasp C. congregata (Braconidae, Microgastrinae) is developmentally regulated but not strictly restricted to the ovaries in the adult. Insect Mol. Biol. 8:319-327. [DOI] [PubMed] [Google Scholar]
- 17.Shelby, K. S., and B. A. Webb. 1999. Polydnavirus-mediated suppression of insect immunity. J. Insect Physiol. 45:507-514. [DOI] [PubMed] [Google Scholar]
- 18.Watson, J. M., J. Meyne, and J. A. Marshall Graves. 1996. Ordered tandem arrangement of chromosomes in the sperm heads of monotreme mammals. Proc. Natl. Acad. Sci. USA 93:10200-10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb, B. A. 1998. Polydnavirus biology, genome structure, and evolution, p. 105-139. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing Corporation, New York, N.Y.
- 20.Whitfield, J. 2000. Phylogeny of microgastroid braconid wasps, and what it tells us about polydnavirus evolution. In A. D. Austin and M. Downton (ed.), The hymenoptera: evolution, biodiversity and biological control. CSIRO Publishing, Melbourne, Australia.
- 21.Whitfield, J. B. 1997. Molecular and morphological data suggest a single origin of the polydnaviruses among braconid wasps. Naturwissenschaften 84:502-507. [Google Scholar]
- 22.Xu, D., and D. B. Stoltz. 1991. Evidence for a chromosomal location of polydnavirus DNA in the ichneumonid parasitoid Hyposoter fugitivus. J. Virol. 65:6693-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zalensky, A. O., M. J. Allen, A. Kobayashi, B. R. Brinkley, and E. M. Bradbury. 1995. Well-defined genome architecture in the human sperm nucleus. Chromosoma 103:577-590. [DOI] [PubMed] [Google Scholar]