Abstract
Human cytomegalovirus (HCMV) expresses a large number of membrane proteins with unknown functions. One class of these membrane proteins apparently acts to allow HCMV to escape detection by the immune system. The best characterized of these are the glycoproteins encoded within the US2 to US11 region of the HCMV genome that mediate resistance to CD8+ and CD4+ T cells. US2, US3, US6, and US11 block various aspects of the major histocompatibility complex (MHC) class I and class II antigen presentation pathways, functioning in cytoplasmic membranes to cause retention, degradation, or mislocalization of MHC proteins. Distantly homologous genes in this region, US7, US8, US9, and US10, are not well characterized. Here, we report expression of the glycoproteins encoded by US7 to US10 by using replication-defective adenovirus (Ad) vectors. US7, US9, and US10 remained sensitive to endoglycosidase H and were exclusively or largely present in the endoplasmic reticulum (ER) as determined by confocal microscopy. US8 reached the Golgi apparatus and trans-Golgi network and was more quickly degraded. Previous studies suggested that US9 could localize to cell junctions and mediate cell-to-cell spread in ARPE-19 retinal epithelial cells. We found no evidence of US9 at cell junctions of HEC-1A epithelial cells. HCMV recombinants lacking US9 produced smaller plaques on ARPE-19 cell monolayers but also exhibited defects in virus replication compared with wild-type HCMV in these cells. Other HCMV recombinants constructed in a similar fashion that were able to express US9 also produced small plaques and some of these exhibited defects in production of infectious progeny in ARPE-19 cells. Thus, there was no correlation between defects in cell-to-cell spread (plaque size) and loss of expression of US9, and it is possible that US9− mutants produce smaller plaques because they produce fewer progeny. Together, our results do not support the hypothesis that US9 plays a direct role in HCMV cell-to-cell spread.
The unique short (US) component of the human cytomegalovirus (HCMV) genome contains a region of genes, US2 to US11, which is predicted to encode at least eight glycoproteins showing limited homology to one another. All of these glycoproteins are predicted to be type 1 membrane glycoproteins with molecular masses of 20 to 38 kDa and may have been derived by duplication and divergence from a common ancestor or ancestors (2, 16). The US2 to US11 glycoproteins are not required for growth of HCMV in cultured human fibroblasts, suggesting that they do not perform functions essential for virus replication in fibroblasts (16, 17). At least four of these glycoproteins act to protect HCMV-infected cells from CD8+ and CD4+ T lymphocytes by blocking the major histocompatibility (MHC) class I and class II antigen presentation pathways (reviewed in references 11, 13, and 24). US2 binds to class I and II proteins and US11 binds class I proteins, in both cases hastening a cellular pathway that normally leads to degradation of misfolded proteins by proteasomes. US6 inhibits the transporter for antigen processing (TAP), blocking import of peptides into the endoplasmic reticulum (ER) for loading onto MHC class I complexes. US3 binds to class I proteins in the ER, causing them to be retained or to accumulate in the ER. Recently, we have shown that US3 can also bind class II protein complexes and inhibit their movement to peptide loading compartments (N. R. Hegde, R. A. Tomazin, T. W. Wisner, C. Dunn, J. Boname, D. M. Lewinsohn, and D. C. Johnson, submitted for publication).
The functions of other glycoproteins in this region, specifically US7 to US10, are not well characterized, at least in terms of immune evasion. Given that four of the eight glycoproteins in the US2 to US11 region can block the MHC class I and class II antigen presentation pathways, it appeared possible that US7, US8, US9, or US10 might also act by inhibiting cell surface expression of MHC or MHC-like proteins. However, efforts to demonstrate inhibition of the MHC class I or class II pathways, by examining either intracellular transport (1) or recognition by CD8+ and CD4+ T cells (Hegde et al., submitted), have, to date, produced no evidence that US7 to US10 can inhibit classical class I and II proteins. There are also numerous nonclassical MHC and MHC-like proteins that can signal to lymphocytes, and these are also potential targets for US2 to US11 proteins (3, 6).
There have been reports that US9 can mediate cell-to-cell spread of HCMV in epithelial cells. HCMV recombinants in which the US9 gene alone or both the US8 and US9 genes were replaced with a β-glucuronidase gene cassette (16, 17) produced smaller plaques in human retinal epithelial ARPE-19 cells than did wild-type HCMV (19, 22). This work leads to the suggestion that US9 might disrupt cell junctions, facilitating cell-to-cell transmission of viruses (18). Replication of these US9− mutants in human fibroblasts was not reduced compared with wild-type HCMV (16). In transfected MDCK (canine epithelial) cells, US9 colocalized with ER and Golgi markers but was also observed at lateral surfaces of cells, colocalizing with E-cadherin and actin (18). It was suggested that US9 might alter the association of cell adhesion molecules with actin, thereby promoting cell-to-cell spread (18, 19, 22). Moreover, US9 was compared to herpes simplex virus (HSV) gE, which is known to function in cell-to-cell spread (reviewed in reference 12), because both glycoproteins are encoded in the S component and they show some very limited sequence homology, and it was said that both glycoproteins accumulate at cell junctions (19).
To investigate further how the US7 to US10 glycoproteins might function during the course of HCMV infection, we expressed the glycoproteins in cells using replication-defective adenovirus (Ad) vectors and characterized intracellular transport and subcellular localization in human epithelial cells. These studies indicated that US7, US9, and US10 remain largely or exclusively in the ER while US8 is transported to the Golgi apparatus. We found no evidence that substantial quantities of any of these glycoproteins, including US9, reached the cell surface or cell junctions or disrupted these junctions. Moreover, a reexamination of several HCMV mutants unable to express US9 suggested that several of them were defective for the production of infectious progeny in ARPE-19 cells. Other HCMV recombinants that expressed US9 also displayed small plaques. These results were not consistent with the conclusion that US9 is involved in HCMV cell-to-cell spread.
MATERIALS AND METHODS
Cells and viruses.
HEC-1A cells and ARPE-19 cells were grown as previously described (27). Normal human diploid fibroblasts (NHDF) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and penicillin-streptomycin-glutamine. Wild-type HCMV strain AD169 was from the American Tissue Culture Collection. Recombinant viruses RV35 (ΔUS6-10), RV47 (ΔUS2-3), RV61 (ΔUS9), RV80 (ΔUS8-9), and RV134 (β-glucuronidase insertion between US9 and US10) have been described previously and were propagated on NHDF (15-17). NHDF were infected with HCMV at 0.1 PFU/cell, and cell culture supernatants were harvested when all the cells were extensively rounded, 6 to 7 days after infection. Supernatant virus stocks were centrifuged at 2,500 rpm for 10 min to remove insoluble debris, and titers were determined by plaque assay on NHDF cells as previously described (21). Construction of Ad vectors expressing US7, US9, or US10 by using an inducible promoter was similar to construction of other vectors expressing US2 and US3 that have been described elsewhere (20, 23; Hegde et al., submitted). Briefly, the relevant genes were amplified by PCR from HCMV AD169 DNA, inserted into plasmid pDE1sp1BTet, and sequenced in both directions. 293 cells were cotransfected with each of these plasmids and with plasmid pJM17 that contains the entire Ad-5 genome (9). Recombinant Ad vectors were screened by PCR and for expression of the relevant glycoproteins by coinfecting cells with a second virus, Adtet-trans (using 20 to 30% of the amount of AdtetUS virus), which expresses the tetracycline transactivator protein. AdCMVUS8 contains the HCMV immediate-early promoter coupled to the HCMV US8 gene and was constructed by using plasmids pCA4 and pJM17 as previously described (9). After their construction, all Ad vectors were propagated and their titers were determined on human 293 cells as previously described (9).
Antibodies.
Rabbit polyclonal sera specific for US7, US8, US9, or US10 were generated largely as described previously (23; Hegde et al., submitted). Briefly, the peptides, the N termini of US8 (NPYEDDDYYYYREDEPRC), of US9 (RSKESLRLS), and of US10 (TDLSSPEGNRPRNYSATC) and the C terminus of US7 (CRFTGKPTYNLLTYPVKG), were synthesized (with an additional cysteine residue) by PeptidoGenic (Berkeley, Calif.); they were coupled to keyhole limpet hemocyanin (KLH) or bovine serum albumin (BSA) bu using m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS; Sigma, St. Louis, Mo.) according to a standard protocol (7a). They were then mixed with Freund's adjuvant and injected into rabbits. A second anti-US9 serum was produced by immunizing rabbits with a fusion protein consisting of C-terminal US9 residues (WIRPWVRGRHRATGRTSREEEAK) fused to the C terminus of glutathione S-transferase. Sheep antiserum specific for TGN46 (Serotec, Oxford, United Kingdom), monoclonal antibodies (MAbs) to β-catenin, calnexin, GM-130, and p115 (Transduction Laboratories, Lexington, Ky.), and protein disulfide isomerase (PDI) (Stressgen, Victoria, British Columbia, Canada) were purchased from commercial sources. A rabbit anti-HCMV serum was purchased from Lee Biomolecular Research Inc. (San Diego, Calif.). Mouse MAbs specific for HCMV gB (residues 27 to 156), pp65 (28 to 19), and pp28 (41 to 8), gifts of Bill Britt (University of Alabama), and a rabbit anti-IE86 serum (R638), a gift of Jay Nelson (Oregon Health Sciences University), were pooled together and diluted 1:200.
Cell-to-cell spread of HCMV recombinants on ARPE-19 cells.
ARPE-19 cells were plated on glass coverslips in 12-well dishes and grown for 6 to 8 days, until approximately 3 days after the cells reached confluency, with changes of cell culture mediim every 2 to 3 days. The cells were infected with diluted stocks of wild-type HCMV or HCMV recombinants for 2 h, the virus inoculum was removed, and the cells were incubated in mediim supplemented with 2% FBS and 0.2% (wt/vol) pooled human gamma globulin (a source of anti-HCMV antibodies) and refed every 2 days. After 7, 14, or 21 days, the cells were washed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 30 min, washed, and permeabilized with 0.2% Triton X-100 for 5 min. The cells were washed and incubated with PBS containing 0.1% Tween 20, 2% normal goat serum, and 2% BSA at 4°C overnight. The cells were stained with a cocktail of anti-HCMV antibodies (see above) and then washed and stained with pooled Alexa594-conjugated goat anti-mouse and anti-rabbit immunoglobulin G (IgG). The numbers of infected cells per plaque were counted by using a Leica fluorescence microscope.
Production of infectious HCMV recombinants on ARPE-19 cells.
ARPE-19 cells were plated into 24-well dishes (approximately 1.8 × 104 cells/well) and incubated overnight. Cells were then treated with 35 mU of neuraminidase type III from Vibrio cholerae (Sigma)/well for 2 h at 37°C, and then the neuraminidase was removed and cells were washed extensively with PBS. Each well was infected with 3 × 106 PFU (titers were derived from fibroblast plaque assays) of wild-type HCMV AD169 or HCMV recombinant for 2 h, and then the dishes were centrifuged at 2,500 rpm for 2 h at 24°C. The virus inoculum was removed, medium containing 5% FBS was added, and cells were incubated at 37°C for 2 to 12 days. At each time point, the cells and cell culture supernatant were harvested and sonicated briefly and infectious HCMV was assayed by plaque formation on human fibroblasts.
Pulse-chase labeling, immunoprecipitation, and endoglycosidase H digestion of proteins.
HEC-1A cells were infected with AdUS8, using 100 PFU/cell (based on titers in 293 cells), or with AdUS7, AdUS9, or AdUS10 (100 293 cell PFU/cell) and, in addition, Adtettrans, an Ad vector expressing the tetracycline transactivator (using 30 293 cell PFU/cell), and then were incubated for 18 h. The cells were incubated in medium lacking methionine and cysteine for 1 h and then with this medium containing [35S]methionine-cysteine (100 μCi/ml; Amersham) for 30 to 60 min as previously described (20). The radiolabel was chased by incubating cells in medium containing a 10-fold excess unlabeled methionine and cysteine for 1, 2, or 4 h. Cell extracts were made using NP-40/DOC lysis buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 2 mg of BSA/ml, and 1 mM phenylmethylsulfonyl fluoride). Cell extracts were immunoprecipitated as described previously (23). Endoglycosidase H (endo H) digestions were carried out using enzyme preparations and protocols supplied by New England Biolabs (Boston, Mass.). Protein samples were subjected to electrophoresis using 12% polyacrylamide gels, and the gels were fixed and incubated with Enlightning (New England Nuclear, Beverly, Mass.), dried, and then exposed to X-ray film.
Confocal immunofluorescence microscopy.
HEC-1A cells (2 × 104/well) were plated in eight-well Lab-Tek plastic microscope slides (Nunc, Naperville, Ill.) for 2 to 4 days (until extensive junctions were formed between cells), and then the cells were infected with recombinant Ad vectors expressing US7 to US10 glycoproteins as described above. The cells were fixed with 4% paraformaldehyde, permeabilized using 0.2% Triton X-100 for 5 min, and incubated overnight at 4°C with PBS containing 0.02% Tween 20, 1% fish gelatin, and 2% normal goat serum. The cells were incubated with rabbit anti-US7, -US8, -US9, or -US10 antibodies and simultaneously with mouse MAb or sheep antibodies specific for cellular marker proteins. The cells were washed and simultaneously incubated with Alexa488-conjugated goat anti-mouse or Alexa488-conjugated donkey anti-sheep IgG and Alexa594-conjugated goat anti-rabbit IgG antibodies. The cells were washed and mounted on microscope slides by using Prolong (Molecular Probes, Eugene, Oreg.) and then visualized using a Bio-Rad 1024 ES laser scanning confocal microscope on a Nikon Eclipse TE300 inverted fluorescent microscope.
RESULTS
Expression and intracellular transport of HCMV US7, US8, US9, and US10 using Ad vectors.
Previously, we have used replication-defective (E1−) Ad vectors to express and characterize several herpesvirus glycoproteins, including HCMV US2, US3, and US11 and HSV gE/gI (4, 20, 23; Hegde et al., submitted). This system allows a relatively high level of expression of HCMV proteins, a condition that is difficult to attain following HCMV infection, especially in diverse cell types. In this case, individual glycoproteins can be characterized in cells without the confounding effects of other HCMV proteins, and Ad proteins are not substantially expressed. Ad vectors that are empty or expressing other glycoproteins can be used to control for the low levels of Ad proteins expressed, and unlike transfection, there is no requirement for cells to grow for many divisions with often toxic viral glycoproteins present. Of course, this may not always accurately reflect properties of these proteins produced in HCMV-infected cells, as other proteins may modify or alter functions or subcellular distribution. There were three reasons that prompted us to study US7, US8, US9, and US10 by using Ad vectors rather than in HCMV-infected cells. First, it is often difficult or impossible to accurately characterize biochemical aspects of these membrane proteins in a number of cultured cell types infected with HCMV. HCMV infects fibroblasts relatively well, but even in these cells the levels of expression of US9 and US10 are relatively low, often too low to accurately assess subcellular localization until relatively late in the infection. Supporting this, we were unable to detect US9 in HCMV-infected fibroblasts by immunofluorescence microscopy, despite extensive efforts (see below). In other cells, such as the ARPE-19 and MDCK epithelial cells used in previous studies (18), infection of cells by HCMV is very inefficient, and the level of expression of viral proteins is lower than in fibroblasts. Therefore, analyses of the localization of US7 to US10 by immunofluorescence or biochemical techniques is unlikely to be successful and may, in fact, be impossible. Second, several of the US2 to US11 glycoproteins act early after HCMV infection, and thus, if these glycoproteins could be detected by immunofluorescence microscopy late after HCMV infection, this might not reflect important sites of action of the proteins. Third, several of the US2 to US11 glycoproteins, specifically US2, US3, US6, and US11, can function autonomously, and all accumulate largely or exclusively in the ER (7, 8, 23, 26; Hegde et al., submitted). Moreover, there is no published evidence that any of the US2 to US11 glycoproteins interact directly with one another or other viral proteins, and our recent efforts to detect interactions with other viral proteins have all been unsuccessful.
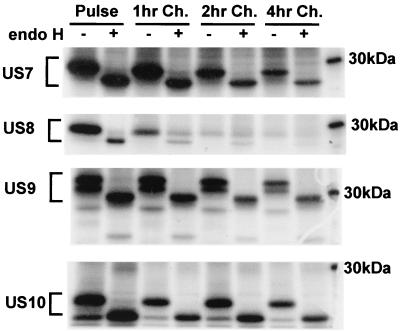
Ad vectors expressing US7, US9, or US10 were constructed by coupling the HCMV genes to a tetracycline-regulated promoter that is induced by coinfecting cells with a second Ad vector expressing a tetracycline transactivator protein, as previously described (20, 23; Hegde et al., submitted). AdUS8 expresses US8 from the constitutive HCMV immediate-early promoter. The products of the US8 and US9 genes were previously described and are denoted gpUS8 and gpUS9, respectively (18); however, for consistency with previous descriptions of US2, US3, US6, and US11 (7, 15, 25, 26; Hegde et al., submitted), we will refer to these glycoproteins as US8 and US9. The glycoproteins were labeled in a pulse-chase format and immunoprecipitated with rabbit polyclonal antibodies, and glycoproteins were treated with endo H. All four glycoproteins were readily detected in HEC-1A human endometrial epithelial cells following shorter label periods of 0.5 to 1.0 h, and these immature glycoproteins were sensitive to endo H as expected (Fig. 1). US9 and US10 were expressed as a doublet, which was converted to a single protein species by endo H. This suggests that a fraction of US9 and US10 was not fully glycosylated, as is the case with US2 (23, 26). US7, US9, and US10 remained largely or exclusively endo H sensitive during subsequent chase periods of up to 4 h. By contrast, US8 attained a significant level of endo H resistance following 1- or 2-h chase periods and was gradually degraded, so that by 4 h there was little US8 remaining. There was also a loss of US7 and US9 during the longer chase periods, and the level of stability of these glycoproteins was similar to that of US2, US3, and US11 observed in other experiments (data not shown). However, US7, US9, and US10 remained entirely endo H sensitive, suggesting that they remained in the ER and did not reach the Golgi apparatus in HEC-1A cells. By contrast, US8 apparently moves to the Golgi apparatus and then likely travels to lysosomes for degradation, as is the case with US3 (7). In another nonepithelial cell type, U373 microglial cells, US7, US9, and US10 also remained endoH sensitive, and US8 attained endo H resistance (R. Tomazin, unpublished results). We have obtained no evidence that pairs of US2 to US11 proteins could interact or promote transport to the cell surface (J. Boname, R. Tomazin, and D. C. Johnson, unpublished data).
FIG. 1.
Radiolabeling and endo H resistance of US7, US8, US9, and US10. HEC-1A cells were grown until confluent and then were infected with Ad vectors expressing US7, US8, US9, or US10 and, in the case of US7, US9, and US10, simultaneously with Adtettrans. After 20 h, the cells were incubated in medium lacking methionine and cysteine for 1 h and then were radiolabeled with [35S]methionine-cysteine for 1 h. The cells were immediately lysed (Pulse) or were incubated for an additional 1, 2, or 4 h in medium containing excess unlabeled methionine and cysteine (1, 2, and 4 hr Ch., respectively), and then cell extracts were made. US7, US8, US9, and US10 were immunoprecipitated using rabbit polyclonal antibodies and digested (+) or not digested (−) with endo H. A 30-kDa marker protein is indicated.
Subcellular distribution of HCMV US7, US8, US9, and US10.
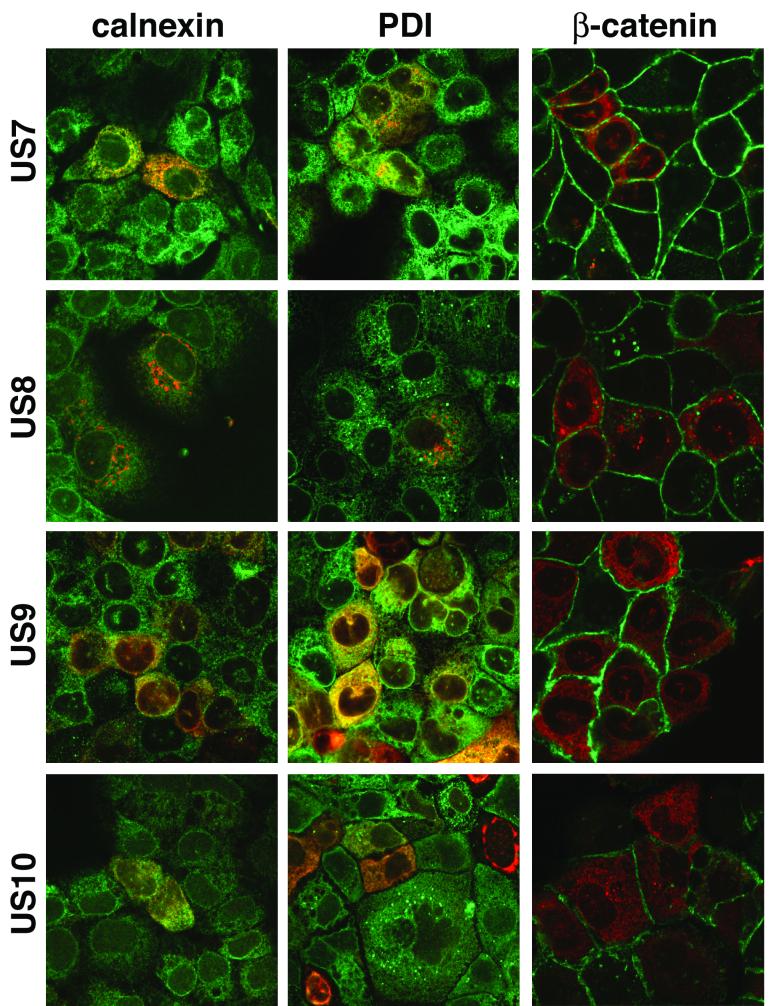
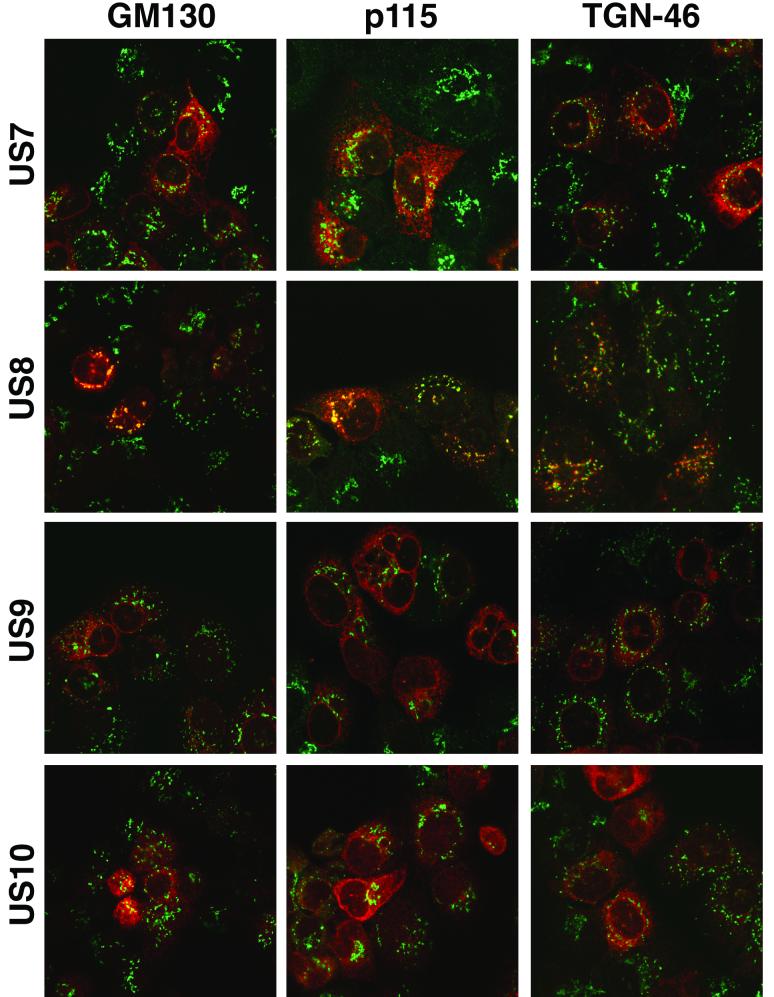
To examine these HCMV glycoproteins further, HEC-1A cells were infected with recombinant Ad vectors and then stained using polyclonal antibodies, and images were analyzed by confocal microscopy. US7, US9, and US10 accumulated in a diffuse cytoplasmic fashion and extensively colocalized with two ER-resident cellular proteins, calnexin and protein disulfide isomerase (Fig. 2). By contrast, US8 was found in more punctate, perinuclear vesicles and did not colocalize with these ER markers (Fig. 2). Instead, US8 was found extensively in the Golgi apparatus, colocalizing with two markers, GM130 and p115, and in the trans-Golgi network (TGN), colocalizing with TGN46 (Fig. 3). Vt1a, a third marker for the Golgi apparatus, also extensively colocalized with US8 but did not colocalize with US7, US9, and US10 (data not shown).
FIG. 2.
Confocal immunofluorescence microscopy of cells expressing US7, US8, US9, or US10 and stained for ER and cell junction markers. HEC-1A cells plated on plastic microscope slides were infected with Ad vectors expressing US7, US8, US9, or US10 and simultaneously with Adtettrans. After 24 h, the cells were fixed, permeabilized, and stained simultaneously with antibodies specific for US7, US8, US9, or US10 and with antibodies specific for calnexin or protein disulfide isomerase (PDI), markers for the ER, or β-catenin, a marker for cell junctions. In every case, the green fluorescence emanates from the cellular marker and the red fluorescence is from US7 to US10.
FIG. 3.
Confocal immunofluorescence microscopy of cells expressing US7, US8, US9, or US10, stained for Golgi and TGN markers. HEC-1A cells were infected with recombinant Ad vectors as in Fig. 2. The cells were fixed, permeabilized, and stained simultaneously with antibodies specific for US7, US8, US9, or US10 and with antibodies specific for GM130 or p115, markers for the Golgi apparatus, or TGN-46, a marker for the TGN. The green fluorescence emanates from the cellular marker, and the red fluorescence is from US7-US10.
HEC-1A cells form extensive cell junctions that can be stained by using antibodies specific for tight junctions (ZO-1) and adherens junctions proteins (E-cadherin or β-catenin) (4, 20). We observed no colocalization of US7, US8, US9, or US10 with the cell junction marker β-catenin (Fig. 2). One could argue that there were small amounts of one of these glycoproteins on the surfaces of cells, especially when the cells were costained with β-catenin antibodies, which might obscure cell surface US7 to US10. However, the presence of US7 to US10 glycoproteins on the surfaces of cells would not be obscured in cell monolayers costained with anticalnexin or anti-PDI antibodies (Fig. 2). Although it is impossible to show multiple images, there was no cell surface expression of US7, US8, US9, or US10 in a large number of separate fields of cells. In limited experiments, immunofluorescence microscopy of nonpermeabilized cells indicated no staining above background with anti-US7, anti-US8, anti-US9, or anti-US10 antibodies, although in these experiments antibodies had no access to lateral surfaces (data not shown). There was also no evidence that cell junctions were altered by morphological criteria, even after long-term, high-level expression of these glycoproteins. We concluded that US7, US9, and US10 are ER-resident glycoproteins whereas US8 moves to the Golgi apparatus and TGN. No obvious fraction of any of these glycoproteins was detected on the surfaces of HEC-1A cells in any instance.
Repeated efforts to detect US9 in HCMV-infected normal human fibroblasts and telomerase-transfected fibroblasts by using immunofluorescence microscopy and two different rabbit anti-US9 polyclonal sera proved futile. In these experiments, the levels of staining of cells infected with wild-type HCMV with anti-US9 antibodies were similar to that observed when cells were infected with mutant RV80, which does not express US9 (data not shown). Other experiments, in which US9 was radiolabeled in HCMV-infected fibroblasts and immunoprecipitated, supported the notion that US9 is expressed at low levels in the cells compared with AdUS9-infected fibroblasts (data not shown). Therefore, the expression of US9 in fibroblasts was insufficient to provide details of the subcellular localization of US9 in HCMV-infected cells. Other cells that can be cultured in the laboratory are not likely to produce higher levels of US9 when infected with HCMV.
HCMV recombinant viruses lacking US9 and other US2 to US11 genes produce smaller plaques on retinal epithelial cells.
Our observations that US9 was exclusively found in the ER and not at cell junctions in HEC-1A cells infected with AdtetUS9 differed from previous observations that a fraction of US9 accumulates at cell junctions after transfection of MDCK cells (18). We obtained no evidence that high-level expression of US9 altered the morphology of cell junctions or the distribution of several components of cell junctions. This prompted a reexamination of the proposal that US9 facilitated cell-to-cell spread of HCMV and disrupted cell junctions. ARPE-19 cells were plated for 3 to 4 days, until the cells attained a cobblestone morphology, and were then infected with HCMV recombinants lacking US9 or other US2 to US11 glycoproteins for 7 to 21 days. The cells were fixed, permeabilized, and stained with a cocktail of anti-HCMV MAbs, polyclonal antibodies, and secondary fluorescent antibodies, and the numbers of infected cells were determined. All of the HCMV recombinants, including a mutant lacking US2 and US3, produced smaller plaques than those made by wild-type AD169 (Table 1). At early (7 days) and late (21 days) times postinfection, both wild-type HCMV and mutants produced both multicell plaques and single-infected ARPE-19 cells that clearly expressed HCMV antigens. We assume that virus replication was aborted in the singly infected cells or the progeny produced were defective for spread or replication in surrounding cells. The wild-type HCMV multicell plaques consisted of an average of 10.5 cells per plaque after 21 days, whereas mutants lacking US2 and US3 (RV47), US6 to US10 (RV35), US8 and US9 (RV80), or US9 (RV61) produced multicell plaques that were substantially smaller (3.0 to 4.6 cells per plaque) (Table 1). It is important to note that there was a broad range of numbers of cells per plaque with both wild-type and mutant HCMV. This is related to the fact that HCMV replication in ARPE-19 cells is relatively slow and inefficient. For example, at 21 days, plaques formed by wild-type HCMV consisted of between 2 and 37 cells. The range was narrower for the mutants, e.g., 3 to 13 cells/plaque for RV35 and 2 to 4 cells/plaque for RV61. Thus, instead of presenting representative photographs of these plaques, we summarized the data by presenting the mean numbers of cells per plaque. The variation in sizes of the plaques meant that standard deviations, especially for wild-type HCMV, were large and were not shown.
TABLE 1.
Plaques formed by wild-type HCMV and US2-US11 mutants on ARPE-19 cells
| Protein and days of infection | No. of single-cell foci | No. of multicell foci | Total no. of foci | Avg no. of cells/ multicell foci |
|---|---|---|---|---|
| AD169 (wild type) | ||||
| 7 days | 357 | 8 | 365 | 2.1 |
| 14 days | 128 | 73 | 201 | 4.8 |
| 21 days | 98 | 108 | 206 | 10.4 |
| RV47 (ΔUS2,3) | ||||
| 7 days | 231 | 12 | 243 | 2 |
| 14 days | 128 | 26 | 154 | 2.8 |
| 21 days | 65 | 37 | 102 | 4 |
| RV35 (ΔUS6-10) | ||||
| 7 days | 306 | 24 | 330 | 2.1 |
| 14 days | 173 | 145 | 318 | 3.3 |
| 21 days | 57 | 65 | 122 | 4.6 |
| RV80 (ΔUS8,9) | ||||
| 7 days | 250 | 15 | 265 | 2 |
| 14 days | 148 | 50 | 198 | 2.8 |
| 21 days | 94 | 63 | 157 | 3.9 |
| RV61 (ΔUS9) | ||||
| 7 days | 252 | 0 | 252 | |
| 14 days | 130 | 24 | 154 | 2.5 |
| 21 days | 40 | 4 | 44 | 3.5 |
| RV134 (intergenic US9/US10) | ||||
| 7 days | 644 | 5 | 649 | 2.0 |
| 14 days | 304 | 50 | 354 | 2.6 |
| 21 days | 41 | 40 | 81 | 3.0 |
RV61, the US9− recombinant, produced fewer multicell plaques than did wild-type HCMV; after 7 days there were no RV61 multicell plaques, and after 21 days there were only rare multicell plaques, composed of on average 3.5 cells (Table 1). By contrast, wild-type HCMV produced small multicell plaques at 7 days, and after 21 days about half the plaques were multicellular and on average were composed of 10.4 cells. Two other US9− mutants, RV80, lacking both US8 and US9, and RV35, lacking US6 to US11, produced more multicell plaques than RV61, and these tended to be somewhat larger than RV61 plaques, though smaller than plaques produced by wild-type HCMV at 21 days (Table 1). RV134, a recombinant with an intergenic insertion of the β-glucuronidase gene between the US9 and US10 genes, also produced smaller plaques at 21 days than did wild-type HCMV. The US9 and US10 open reading frames of RV134 are intact, and it had previously been shown that RV134 expresses US10 (17); also, we found that RV134 expresses wild-type levels of US9 (T. Wisner and D. C. Johnson, unpublished results). In addition, RV47, a recombinant lacking the US2 and US3 genes, also formed smaller plaques. Therefore, mutants unable to express US9, as well as other mutants expressing US9, produced smaller plaques on ARPE-19 cells.
In the previous studies of these recombinants, it was reported that ARPE-19 cells infected with wild-type HCMV displayed altered cell junctions and exhibited increased transepithelial permeability to [3H]inulin, whereas infection with RV61, RV 35, or RV80 did not lead as extensively to these changes. This suggested that US9− mutants were less able to disrupt junctions and that this was connected to defects in cell-to-cell spread. Related to this observation, the morphology of infected cells in plaques produced on ARPE-19 cells was not different from that of cells in plaques produced by US9− mutants RV61, RV80, and RV35. There were fewer infected cells in plaques formed by the mutants than with wild-type HCMV (Table 1), but the morphologies of the infected cells were similar; in both cases, cells were rounded and there was generalized loss of cell adhesion. Since fewer cells were infected with the mutants, we expect that the observed differences in transepithelial permeability in previous studies could be explained by decreased numbers of infected cells.
Reduced replication of HCMV US9 mutants.
Previous characterization of RV47, RV35, RV80, RV61, RV134, and other mutants affecting the US2 to US11 genes indicated that these viruses did not display major defects in virus replication in human fibroblasts (16, 17). US9− mutants produced smaller plaques in ARPE-19 retinal epithelial cells, but there was no characterization of virus replication in these cells (19). In general, HCMV replicates relatively slowly and inefficiently in ARPE-19 cells. Our initial efforts to infect ARPE-19 cells with HCMV AD169 led to infection of only 2 to 10% of the cells, making it difficult to measure a single round of virus replication. However, we found that the cells could be infected more effectively 1 day after the cells were plated, following treatment of the cells with neuraminidase and centrifugation of viruses onto the cells for 2 h. Under these conditions, more than 50% of ARPE-19 cells were infected in every experiment, as judged by expression of immediate-early proteins after 4 to 5 days (data not shown). It should be noted that the cells were plated densely and attained a highly ordered cobblestone morphology.
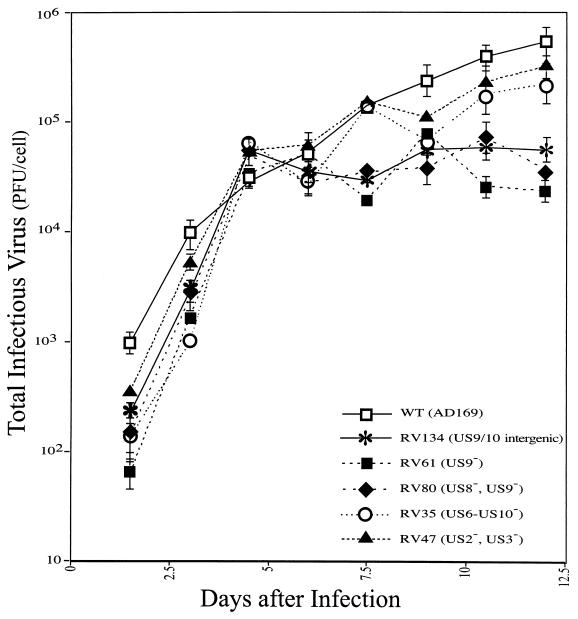
To examine replication of HCMV recombinants, ARPE-19 cells were infected as described above and cell monolayers and culture supernatants were harvested at various times, ranging from 2 to 12 days postinfection. The total amount of virus produced was determined by plaque titrations using human fibroblasts. After 2 days there was significantly less infectivity with many of the virus mutants, e.g., RV61 and RV80, than with wild-type AD169. Since identical amounts of infectious virus were used in every case, based on plaque assays involving human fibroblasts, this appears to reflect early defects in replication, or, more likely, residual input wild-type HCMV was more robust in these cultures and could produce plaques on fibroblasts better than the mutants. After 4.5 and 6 days of infection, there was substantially more virus produced by both mutants and wild-type HCMV than there was at day 2, and there were similar amounts of infectious virus in both cases (Fig. 4). However, from day 6 to day 12, there was an increase in infectious virus produced by wild-type HCMV and certain of the mutants, whereas other mutants, e.g., RV61 (US9−), RV80 (US8/9−), and RV134 (US8/9 intergenic insert), produced less infectious virus. The differences in yields of virus were apparently not due to reduced spread of virus through the monolayer, since the majority of the cells were infected by wild-type as well as recombinant HCMV in the first hours of this experiment, as judged by immediate-early protein expression (data not shown). Moreover, a single round of HCMV replication in these cells required more than 10 days, and quantities of infectious virus did not substantially increase after 12 days (data not shown). The replication curves were steep before day 6 and less steep after day 6 with both wild-type HCMV and recombinants, but there was not a plateau followed by a steep increase, as would be expected if a second round of replication occurred. Curves associated with certain of the recombinants flattened after 6 days more substantially than was observed with wild-type HCMV. Comparing the final yields of virus, RV61 produced the lowest yields of virus, as much as 30-fold lower than was observed with wild-type HCMV (Fig. 4), and this correlated with the observation that RV61 also produced the smallest plaques at early and late times (Table 1). We note that RV61 also produced what appeared to be lower yields of virus when grown on human fibroblasts in previous studies (16) and in our studies (M. Huber, unpublished results). Other recombinants, i.e., RV35, which does not express all of the US6 to US11 genes, including US9, and RV47, a US2− US3− mutant, produced infectious virus in amounts more similar to that produced by wild-type HCMV (Fig. 4). Note that RV47 and RV35 also produced slightly larger plaques than RV61 and RV80, and thus there was a good correlation between production of infectious virus and cell-to-cell spread. These results support the hypothesis that there are defects in the growth of the US9− mutants as well as other recombinants produced by similar methods, and this reduced replication may explain the reduced ability to infect adjacent cells and the smaller plaque sizes.
FIG. 4.
Production of infectious virus by ARPE-19 cells infected with HCMV recombinants lacking US9 or other US2 to US11 genes. ARPE-19 cells were plated in plastic dishes, treated with neuraminidase, and then infected with HCMV recombinants: RV134 (intergenic insert US9/US10), RV61 (ΔUS9), RV80 (ΔUS8, ΔUS9), RV35 (ΔUS6-10), RV47 (ΔUS2, ΔUS3), or wild-type HCMV AD169 in duplicate. After 1.5 to 12 days, the cells and cell culture media were harvested together and sonicated briefly, and infectious virus titers were determined using human fibroblasts.
DISCUSSION
The US2 to US11 region of HCMV includes at least eight membrane glycoproteins that share limited sequence homology, and there is evidence of similar functions for these proteins. Early studies suggested that US6 to US11 comprise a family of genes (5). Our analysis of the sequences of these genes suggests that US8, US9, and US11 display more similarity, as is the case for US2 and US3. However, US2 and US11, at either end of this region and with lower sequence similarity, share the ability to cause proteasome degradation of MHC class I proteins (15, 23, 25, 26; Hegde et al., submitted). Moreover, US2 and US3 share the ability to bind both MHC class I and class II proteins (23; Hegde et al., submitted).
Here, we expressed US7, US8, US9, and US10 glycoproteins in hopes of obtaining clues as to how these glycoproteins function. US7, US9, and US10 remained endo H sensitive in HEC-1A human epithelial cells and other cells. There are examples of herpesvirus glycoproteins that contain some N-linked oligosaccharides which are not processed to complex oligosaccharides during transport through the Golgi apparatus (10). However, we are not aware of examples of glycoproteins which move through the Golgi apparatus yet remain fully sensitive to endo H. Moreover, confocal immunofluorescence microscopy indicated that US7, US9, and US10 were found extensively in the ER and there was little or no glycoprotein detected in the Golgi or TGN or on the cell surface. By contrast, US8 attained endo H resistance and was found in the Golgi apparatus and TGN, but not on the cell surface. The distribution of US8 was reminiscent of US3, which can cause MHC class I proteins to accumulate in the ER for a time, but US3 and a second US3 substrate, MHC class II complexes, leave the ER and move to the Golgi apparatus and TGN (7; Hegde et al., submitted). There is some preliminary evidence that US8, US9, and US10 might bind MHC proteins (M. Chevalier and D. C. Johnson, unpublished results), and efforts are under way to characterize these further.
When we began this work, there were reports that US9 can accumulate at cell junctions. Maidji et al. (18) found that much of the US9 expressed in transfected MDCK cells was observed in the cytoplasm, colocalizing with ER markers, but there was a substantial fraction of US9 along lateral surfaces, colocalizing with cadherins and actin. These authors suggested that US9, and as well HSV gE, might alter cellular junctions, increasing paracellular permeability and allowing virus to move between cells (19, 22). In our studies, US9 expressed using a replication-defective Ad vector was entirely sensitive to endo H and was retained in the ER in several human cell types. Moreover, examination of numerous confocal images did not reveal any substantial fraction of US9 on the lateral surfaces or any plasma membrane domains of HEC-1A epithelial cells and other cells. We found no evidence that expression of US9 could alter cell junctions, though the glycoprotein could be expressed at high levels in cells. Similarly, HSV gE/gI did not decrease transepithelial resistance, cause redistribution of components of cell junctions, or obviously alter the morphology of cells (20; K. Dingwell, unpublished results). These differences may relate to the use of human HEC-1A versus MDCK epithelial cells, the use of Ad vectors versus transfection, or the level of polarization of the cells. While it is possible that US9 or other US7 to US10 glycoproteins may move beyond the ER or Golgi apparatus to the cell surface in HCMV-infected cells, this is difficult to test, as the levels of the glycoproteins produced are low, too low to detect by immunofluorescence microscopy. We attempted to detect US9 in HCMV-infected fibroblasts by immunofluorescence microscopy without success (see above), and ARPE-19 and other epithelial cells expressed much lower levels of US9. This is the major reason for expressing these glycoproteins by using virus vectors here and by transfection in previous studies (18). While it is possible that some other viral protein affects the distribution of US9 or other US7 to US10 glycoproteins, efforts to coprecipitate any of the US2 to US11 glycoproteins with other viral proteins have been entirely negative. Importantly, US2, US3, US6, and US11 all localize to the ER and function autonomously in transfected or virus vector-infected cells, without other HCMV proteins (7, 8, 23, 26; Hegde et al., submitted). Therefore, it is likely, although clearly unproven, that the distribution of US7 to US10 observed here by using virus vectors reflects that in HCMV-infected cells. Regardless of whether this is the case, our results on the subcellular localization of US9 are different from previous observations in which US9 was expressed by transfection and do not support the conclusion that US9 functions in cell-to-cell spread.
On the question of polarization, in our studies, HEC-1A and ARPE-19 epithelial cells were plated on plastic or glass surfaces and for relatively shorter periods than in previous studies. Maidji et al. grew MDCK and ARPE-19 cells on permeable Transwell filters and, in some experiments, for longer periods before HCMV infection, conditions which promote extensive tight junctions and allow measurement of high transepithelial resistances (18, 19). They observed a fraction of US9 at cell junctions in transfected MDCK cells that were highly polarized, but not in less confluent cells. Our previous studies demonstrated that HEC-1A cells grown on permeable filters can attain a fully polarized phenotype as measured by high transepithelial resistances, although these resistances are lower than with MDCK cells. Transepithelial resistances measure only tight junctions, and it appears that HEC-1A cells form less extensive tight junctions but are still highly polarized in terms of assembly of adherens junctions, desmosomes, and gap junctions. HEC-1A cells grown on solid plastic supports attain a similar morphology and establish highly polarized cell junctions compared to cells growing on permeable supports, although one cannot measure transepithelial resistance in this case. Sorting and accumulation of cell junctions proteins E-cadherin, β-catenin, ZO-1, desmosomes, and connexins to sites of cell-cell contact begin within hours of cell plating and were relatively complete with 2 days of contact; i.e., these cellular proteins are found exclusively at cell junctions (4, 20, 27; K. Dingwell and D. C Johnson, unpublished data). Therefore, HEC-1A cells exhibit highly polarized phenotypes early after plating, though tight junctions may not exclude transepithelial movement of small molecules for several more days. We note that HSV membrane proteins, such as gE/gI and HSV particles, are efficiently sorted to cell junctions in HEC-1A cells grown in this fashion (14, 20). However, our studies involved human HEC-1A epithelial cells and those of Maidji et al. involved canine MDCK cells, and the experiments were carried out differently, i.e., transfection versus Ad vectors and permeable versus solid supports, and therefore it is only clear that US9 is not universally delivered to epithelial cell junctions.
Previous conclusions that US9 plays a role in HCMV cell-to-cell spread were based on studies of HCMV mutants lacking US9 that formed smaller plaques in ARPE-19 cells and were less able to disrupt cell junctions and the actin cytoskeleton (19, 22). Earlier characterization of US9− recombinants suggested that the mutants did not have major defects in virus replication in human fibroblasts (16, 17). However, one US9− mutant, RV61 (US9−), produced slightly lower yields on fibroblasts in the previous studies (16), and we also found that RV61 exhibited reduced infectivity (M. Huber, unpublished results). As in the previous studies, two US9− mutants, RV61 (US9−) and RV80 (US8/9−), produced small plaques on ARPE-19 cells. However, yields of the two US9− viruses were reduced by 10- to 30-fold in ARPE-19 cells. It appears plausible that the small-plaque phenotype of both US9− mutants results from reduced yields of infectious virus or slower replication rates in ARPE-19 cells. In the previous work, yields of virus produced by ARPE-19 cells were not determined.
One might argue that the ARPE-19 cells in our studies do not compare exactly with the ARPE-19 cells in previous studies because our cells were not grown on permeable supports. The arguments here are similar to those described above for HEC-1A cells. Our ARPE-19 cells were plated for 6 to 8 days, attaining a highly ordered, cobblestone morphology before being infected with HCMV. Then, plaques required a further 21 days to develop, so the ARPE-19 cells were plated for a period of approximately 28 days in all. These cells faithfully reproduced the small-plaque phenotype of US9− mutants. Moreover, there is no reason to believe that these defects in virus replication would not have occurred no matter how the cells were plated. However, we cannot exclude the possibility that HCMV grows differently in ARPE-19 cells grown on solid than it does in the same cells grown on permeable supports.
Related to the observations that US9− mutants produced smaller plaques and replicated less well on ARPE-19 cells, we observed that several other recombinant HCMV, e.g., RV47 and RV134, also produced smaller plaques. RV47 does not express US2 and US3, and RV134 has a β-glucuronidase insert between US9 and US10 (RV134). Both of these mutants were constructed by similar methods, involving insertion of β-glucuronidase, and both express US9. One simple interpretation of these results is that the process by which the β-glucuronidase gene cassette was inserted into the US2 to US11 region or other aspects of creating these recombinants have caused other mutations that reduce replication and/or plaque size in ARPE-19 cells. In the case of RV134, the reduced plaque size might be explained by reduced replication, as is the case for the two US9− mutants. There is some correlation between yields of infectious virus and plaque size because replication of RV47 (US2− US3−) and RV35 (lacking US6 to US10) was more similar to that of wild-type HCMV and plaques produced by these mutants was more similar to those produced by wild-type HCMV (Fig. 4; Table 1). Perhaps it is not surprising that other mutations that affect cell-to-cell spread or virus replication have accumulated in certain of these recombinants, given the size of the HCMV genome.
To determine more about the relationship between US9 or any of the other US2 to US11 genes and yields of infectious virus in ARPE-19 cells or plaque size, it will be necessary to produce rescued versions of the HCMV recombinants. With HCMV, this is a technically difficult and lengthy process and is beyond the scope of the present study. We note that these US2 to US11 recombinants have been extensively and accurately used to characterize immune evasion, without rescued viruses. Our studies of these HCMV recombinants sought to clarify the conclusion that US9 mediates cell-to-cell spread, a conclusion based on these same HCMV recombinants (18, 19, 22). There are clearly defects in replication of the US9− mutants, at least on ARPE-19 cells, and this may explain small plaques. Moreover, other similar recombinants, on ARPE-19 cells, and other recombinants constructed in a similar fashion and expressing US9 also produce small plaques. Thus, there was no correlation between loss of US9 and the small-plaque phenotype. These results, together with our observations that US9 was found in the ER and not on the cell surface, do not support the hypothesis that US9 promotes HCMV cell-to-cell spread.
Acknowledgments
We are very grateful to Aurelie Snyder for all her hard work and excellent technical assistance with the laser scanning confocal and deconvolution software. We thank Bill Britt and Jay Nelson for anti-HCMV antibodies and Johnan Kaleeba for help making the anti-US9 antibodies. We appreciate Nag Hedge for his advice, analysis of the US2 to US11 genes, and critical review of the manuscript and Tom Jones for critical comments.
This work was supported by grants CA73996 from the National Cancer Institute and EY11245 from the National Eye Institute to D.C.J. and by grant EY07029 from the National Eye Institute to M.T.H.
REFERENCES
- 1.Ahn, K., A. Angulo, P. Ghazal, P. A. Peterson, Y. Yang, and K. Fruh. 1996. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc. Natl. Acad. Sci. USA 93:10990-10995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 3.Cosman, D., J. Mullberg, C. L. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and N. J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 14:123-133. [DOI] [PubMed] [Google Scholar]
- 4.Dingwell, K. S., and D. C. Johnson. 1998. Herpes simplex virus gE/gI facilitates cell-to-cell spread and binds to components of cell junctions. J. Virol. 72:8933-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gretch, D. R., J. F. Bale, Jr., R. C. Gehrz, and M. F. Stinski. 1991. Expression of a human cytomegalovirus glycoprotein multigene family. Virus Genes 5:203-214. [DOI] [PubMed] [Google Scholar]
- 6.Groh, V., R. Rhinehart, J. Randolph-Habecker, M. S. Topp, S. R. Riddell, and T. Spies. 2001. Costimulation of CD8αβ T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat. Immunol. 2:255-260. [DOI] [PubMed] [Google Scholar]
- 7.Gruhler, A., P. A. Peterson, and K. Fruh. 2000. Human cytomegalovirus immediate early glycoprotein US3 retains MHC class I molecules by transient association. Traffic 1:318-325. [DOI] [PubMed] [Google Scholar]
- 7a.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 8.Hengel, H., J. O. Koopmann, T. Flohr, W. Muranyi, E. Goulmy, G. J. Hammerling, U. H. Koszinowski, and F. Momburg. 1997. A viral ER-resident glycoprotein inactivates the MHC-encoded peptide transporter. Immunity 6:623-632. [DOI] [PubMed] [Google Scholar]
- 9.Hitt, M. M., C. L. Addision, and F. L. Graham. 1997. Human adenovirus vectors for gene transfer into mammalian cells. Adv. Pharmacol. 40:137-206. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, D. C., G. Ghosh-Choudhury, J. R. Smiley, L. Fallis, and F. L. Graham. 1988. Abundant expression of herpes simplex virus glycoprotein gB using an adenovirus vector. Virology 164:1-14. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, D. C., and A. B. Hill. 1998. Herpesvirus evasion of the immune system. Curr. Top. Microbiol. Immunol. 232:149-177. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, D. C., and G. McFadden. 2001. Viral immune evasion, p. 357-377. In S. H. E. Kaufmann, A. Sher, and R. Ahmed (ed.), The anti-infective immune response. American Society for Microbiology, Washington, D.C.
- 14.Johnson, D. C., M. Webb, T. W. Wisner, and C. Brunetti. 2001. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J. Virol. 75:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, T. R., L. K. Hanson, L. Sun, J. S. Slater, R. M. Stenberg, and A. E. Campbell. 1995. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J. Virol. 69:4830-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, T. R., and V. P. Muzithras. 1992. A cluster of dispensable genes within the human cytomegalovirus genome short component: IRS1, US1 through US5, and the US6 family. J. Virol. 66:2541-2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, T. R., V. P. Muzithras, and Y. Gluzman. 1991. Replacement mutagenesis of the human cytomegalovirus genome: US10 and US11 gene products are nonessential. J. Virol. 65:5860-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maidji, E., S. Tugizov, G. Abenes, T. Jones, and L. Pereira. 1998. A novel human cytomegalovirus glycoprotein, gpUS9, which promotes cell-to-cell spread in polarized epithelial cells, colocalizes with the cytoskeletal proteins E-cadherin and F-actin. J. Virol. 72:5717-5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maidji, E., S. Tugizov, T. Jones, Z. Zheng, and L. Pereira. 1996. Accessory human cytomegalovirus glycoprotein US9 in the unique short component of the viral genome promotes cell-to-cell transmission of virus in polarized epithelial cells. J. Virol. 70:8402-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McMillan, T. N., and D. C. Johnson. 2001. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J. Virol. 75:1928-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowlin, D. M., N. R. Cooper, and T. Compton. 1991. Expression of a human cytomegalovirus receptor correlates with infectibility of cells. J. Virol. 65:3114-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira, L., E. Maidji, S. Tugizov, and T. Jones. 1995. Deletion mutants in human cytomegalovirus glycoprotein US9 are impaired in cell-cell transmission and in altering tight junctions of polarized human retinal pigment epithelial cells. Scand. J. Infect. Dis. Suppl. 99:82-87. [PubMed] [Google Scholar]
- 23.Tomazin, R., J. Boname, N. R. Hegde, D. M. Lewinsohn, Y. Altschuler, T. R. Jones, P. Cresswell, J. A. Nelson, S. R. Riddell, and D. C. Johnson. 1999. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat. Med. 5:1039-1043. [DOI] [PubMed] [Google Scholar]
- 24.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 25.Wiertz, E. J., T. R. Jones, L. Sun, M. Bogyo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 26.Wiertz, E. J., D. Tortorella, M. Bogyo, J. Yu, W. Mothes, T. R. Jones, T. A. Rapoport, and H. L. Ploegh. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432-438. [DOI] [PubMed] [Google Scholar]
- 27.Wisner, T., C. Brunetti, K. Dingwell, and D. C. Johnson. 2000. The extracellular domain of herpes simplex virus gE is sufficient for accumulation at cell junctions but not for cell-to-cell spread. J. Virol. 74:2278-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]