Abstract
A major pathway of eukaryotic mRNA turnover initiates with deadenylation, which allows a decapping reaction leading to 5′–3′ exonucleolytic degradation. A key control point in this pathway is the decapping of the mRNA. Two proteins, Edc1 and Edc2, were genetically identified previously as enhancers of the decapping reaction. In this work, we demonstrate that Edc1p and Edc2p are RNA-binding proteins. In addition, recombinant Edc1p or Edc2p stimulates mRNA decapping in cell-free extracts or with purified decapping enzyme. These results suggest that Edc1p and Edc2p activate decapping directly by binding to the mRNA substrate and enhancing the activity of the decapping enzyme. Interestingly, edc1Δ strains show defects in utilization of glycerol as a carbon source and misregulation of several mRNAs in response to carbon-source changes. This identifies a critical role for decapping and Edc1p in alterations of gene expression in response to carbon-source changes.
Keywords: Decapping, turnover, yeast, carbon source
INTRODUCTION
The ability to respond to a changing environment by modulating gene expression is crucial to the success of all organisms. In addition to transcription, splicing, and translational control, differential mRNA turnover is important for cells and their ability to respond to various environmental and developmental changes (for review, see Guhaniyogi and Brewer 2001; Wilusz et al. 2001). In yeast, the stability of specific mRNAs can be altered on the basis of the carbon source or the developmental stage of the cell. For example, the SDH2 mRNA is unstable when dextrose is present as the carbon source (t½ <5′), but stable in the presence of glycerol (t½ >30′) (Prieto et al. 2000). Similarly, some mRNAs are unstable during vegetative growth, but become stabilized at specific times in meiosis (Surosky and Esposito 1992). The molecular changes that lead to these specific alterations in mRNA decay are unclear but are likely to involve alterations in both the basal mRNA decay machinery and specific mRNA-binding proteins. A framework for understanding the specific regulation of mRNA turnover has come from the definition of the eukaryotic mRNA decay pathways (for review, see Beelman and Parker 1995). In yeast, both unstable and stable mRNAs are degraded primarily by a general 5′–>3′ mechanism of mRNA turnover. In this pathway, the first step is shortening of the 3′ poly(A) tail (Muhlrad and Parker 1992; Decker and Parker 1993). Once the mRNA has become oligoadenylated (10–12 As), the mRNA becomes a substrate for a decapping reaction (Muhlrad et al. 1994, 1995). After decapping, the mRNA is rapidly degraded in a 5′–>3′ direction by the Xrn1p exoribonuclease (Hsu and Stevens 1993). Differential stabilities among diverse mRNAs are due to differences in the rates of deadenylation and decapping (Muhlrad et al. 1994, 1995). Alternatively, following deadenylation, mRNAs can be degraded in a 3′–5′ direction by the cytoplasmic exosome (Jacobs-Anderson and Parker 1998).
Decapping is a critical control point in the major decay pathway, as it both precedes and permits the degradation of the mRNA body. The decapping reaction requires two interacting proteins, Dcp1p and Dcp2p, which are thought to form the core of a decapping holoenzyme (Beelman et al. 1996; Dunckley and Parker 1999; Steiger et al. 2003). The general activity of this holoenzyme is regulated in both positive and negative ways. For example, the efficiency of the decapping reaction is enhanced by the activity of the Pat1p/Lsm1-7p complex, and Dhh1p, a member of the DEAD box family of RNA helicases (Bonnerot et al. 2000; Bouveret et al. 2000; Tharun et al. 2000; Coller et al. 2001; Fischer and Weis 2002). Although not required for decapping, these factors stimulate the decapping of both stable and unstable mRNAs, suggesting that they are general activators of the decapping reaction. In contrast, proteins that are involved in translation initiation are general inhibitors of decapping. For example, the stability of the interaction between the cytoplasmic cap-binding complex and the mRNA cap structure influences how quickly the mRNA is decapped (Schwartz and Parker 1999, 2000; Vilela et al. 2000; Ramirez et al. 2002). Transcript-specific binding proteins that promote or inhibit the decapping reaction can also modulate the decapping rates of individual mRNAs. For example, Puf3p binds to the 3′UTR of the COX17 mRNA and enhances the rates of both deadenylation and decapping of this transcript (Olivas and Parker 2000).
Two related proteins, Edc1p and Edc2p, were identified as proteins that enhance the rates of decapping in vivo (Dunckley et al. 2001). This was based on their identification as high-copy suppressors of decapping defects in strains containing temperature-sensitive alleles of DCP1 or DCP2. Moreover, in sensitized strains in which an edc1Δ or edc2Δ was combined with partially defective alleles of DCP1 or DCP2, mRNAs were further stabilized. This result indicated that these proteins can play a role in decapping when expressed at their normal level. In addition, Dcp1p and Dcp2p were found to coimmunoprecipitate with a tagged version of Edc1p, indicating that these proteins interact, at least transiently. The biochemical properties of the Edc proteins, the mechanism(s) by which they enhance decapping, and their role in the control of gene expression remains unknown.
In this work, we show that the Edc1p and Edc2p are RNA-binding proteins. Moreover, recombinant Edc1p or Edc2p can stimulate decapping in a purified system, indicating that they play a direct role in decapping. In addition to its role as a general enhancer of decapping, Edc1p is also required for proper alterations in gene expression during a carbon-source switch. These results imply that one role for the Edc proteins may be involved in coordinating changes in mRNA stability in response to environmental cues.
RESULTS
Purification of Edc1p and Edc2p
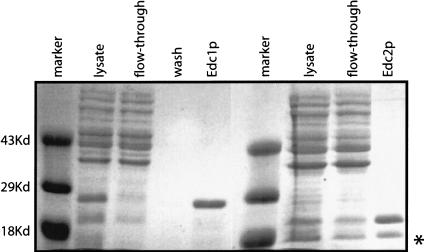
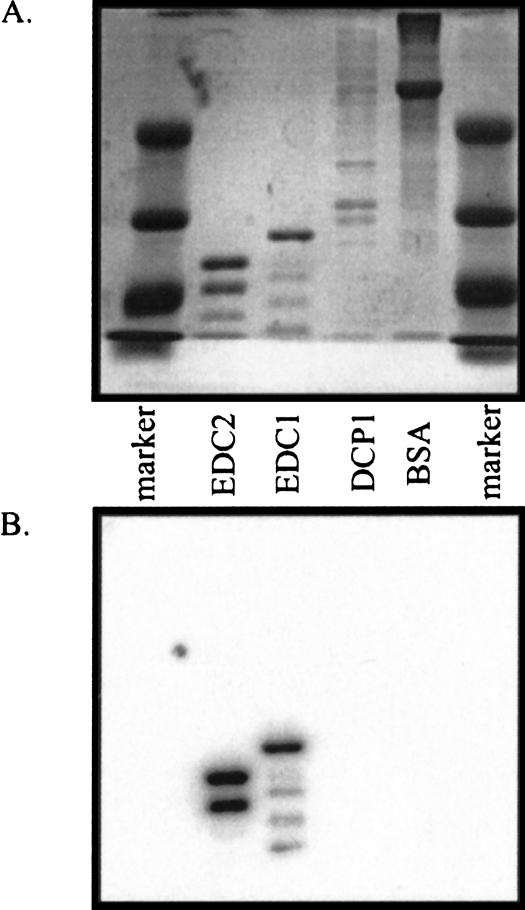
To examine what functional role the Edc proteins might play in decapping, we first purified the two proteins. Both proteins were cloned into plasmid vectors that add a 6× His-tag at the amino terminus of the protein and purified from Escherichia coli to ensure that no other proteins involved in decapping were copurified. In all cases, the preparations were highly purified and free of additional polypeptides as assayed by Coomassie stain (Fig. 1 ▶). We do observe specific breakdown products of Edc1p and Edc2p, although they are always a minority of the preparation (Fig. 1 ▶).
FIGURE 1.
Purification of EDC proteins. Analysis of His-EDC1 and His-EDC2 by Coomassie-stained SDS-PAGE. The proteins were purified from E. coli by use of nickel column chromatography. Shown is a crude lysate, column flow-through, wash, and elution for both EDC1 and EDC2. Protein size markers (in kilodaltons) are shown in the first and sixth lanes. The major breakdown product of Edc2p observed in some preparations is marked with an asterisk.
The Edc proteins can stimulate decapping in yeast cell-free extracts
To determine whether Edc1p and Edc2p directly, or indirectly, affect the decapping reaction, we examined whether Edc1p or Edc2p could affect decapping in vitro. Here, we initially assayed the role of Edc1p and Edc2p in crude extracts, which involves adding an in vitro-transcribed, cap-labeled mRNA to a yeast extract and assaying for the release of 32P-labeled m7GDP by thin-layer chromatography (e.g., Zhang et al. 1999; Dunckley and Parker 2001). Two experiments were performed with these crude extracts.
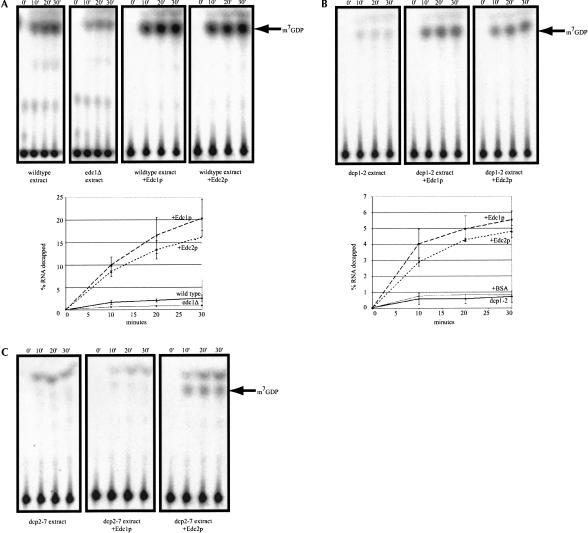
In the first experiment, we asked whether extracts lacking Edc1p or Edc2p showed defects in decapping. Under the conditions used in these experiments, a wild-type yeast extract decaps 2.60% of the substrate in 30 min. In contrast, an edc1Δ extract gives ∼0.81% decapping after 30 min. An extract from an edc2Δ strain decapped similar to the wild-type extract (data not shown). The decrease in decapping seen in the edc1Δ extract is similar to what is seen in extracts from strains that carry conditional lesions in the decapping holoenzyme (dcp1-2 strain [0.90% decapping] or dcp2-7 strain [0.37% decapping], respectively; Fig. 2 ▶). This suggests that Edc1p affects a step in decapping that is rate limiting in crude extracts.
FIGURE 2.
Purified Edc proteins stimulate decapping of yeast extracts. (A) Analysis of decapping activity from a wild-type extract and from wild-type extracts containing either purified Edc1p (26 pmole) or Edc2p (31 pmole). Decapping from an edc1Δ extract is shown for comparison. Decapping was assayed over a 30-min time course with aliquots removed at the designated time points. The products of the reaction were resolved by thin-layer chromatography. (B) Analysis of decapping activity from a dcp1-2 extract and from dcp1-2 extracts containing either purified Edc1p or Edc2p. (C) Analysis of decapping activity from a dcp2-7 extract and from dcp2-7 extracts containing either purified Edc1p or Edc2p. The line graphs show the amount of decapping from multiple experiments over the time course.
In a second experiment, we asked whether recombinant Edc1p or Edc2p could stimulate decapping of the wild-type or mutant decapping enzymes in crude extracts. Addition of purified recombinant Edc1p to a wild-type yeast extract resulted in an eightfold stimulation of decapping (20.4% vs. 2.6%; Fig. 2A ▶). Similarly, addition of Edc2p to a wild-type extract increased the level of decapping to 16.2%. Moreover, addition of either purified Edc1p or Edc2p leads to an approximately eightfold stimulation of the low level of decapping seen in extracts from dcp1-2 mutant strain (Fig. 2B ▶). This is consistent with genetic data that overexpression of Edc1p or Edc2p can suppress the dcp1-2 lesion in vivo (Dunckley et al. 2001). Interestingly, addition of purified Edc2p led to a 6.5-fold increase in decapping activity in the dcp2-7 extract, whereas addition of Edc1p had no effect (Fig. 2C ▶). This result also parallels the specificity of suppression observed previously in vivo, in which overexpression of Edc1p does not suppress the dcp2-7 lesion. This data shows that the EDC proteins purified from E. coli are functional, and that they affect a step in the decapping process that is rate limiting under these conditions in vitro. Moreover, because the interaction in vitro reproduces the specificity of interactions seen in vivo, it argues that the crude extracts are accurately mimicking the in vivo process of decapping, at least for the substep(s) affected by Edc1p and Edc2p.
Edc1p and Edc2p directly affect the Dcp1p/Dcp2p holoenzyme
The ability of Edc1p and Edc2p to stimulate decapping could be either a direct stimulation of the Dcp1p/Dcp2p decapping enzyme, or through indirect effects on other proteins present in crude extracts. To distinguish these two possibilities, we examined whether recombinant Edc1p or Edc2p could enhance the decapping of Dcp1p/Dcp2p enzyme purified from yeast.
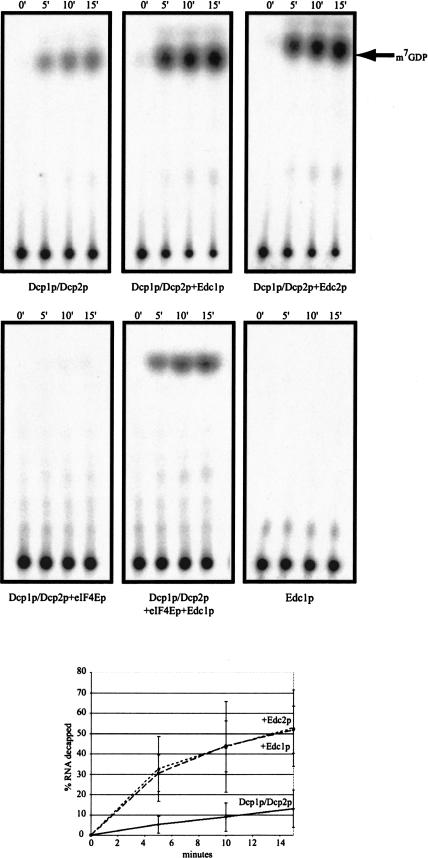
Addition of the Dcp1p/Dcp2p to a cap-labeled MFA2 mRNA leads to decapping of 13% of the substrate after 15 min (Fig. 3 ▶). Addition of either purified Edc1p or Edc2p to the decapping reaction results in an approximately fourfold increase in decapping (52% and 53%, respectively). BSA was added to all reaction mixtures to maintain a constant protein concentration, but BSA alone had no stimulatory effect on the activity of Dcp1p (data not shown). Addition of either Edc1p or Edc2p alone also had no effect on the cap status of the MFA2 substrate (Fig. 3 ▶). These results suggest that the Edc proteins directly affect Dcp1p/Dcp2p function. There was still the possibility that they act indirectly, because, although the preparation of the yeast Dcp1p/Dcp2p was highly purified, it still contained additional proteins (data not shown). However, the Edc proteins also stimulate the activity of recombinant Dcp1p and Dcp2p (Steiger et al. 2003). Therefore, the Edc proteins are able to stimulate decapping of mRNAs in a purified system, indicating that they directly enhance the function of the decapping enzyme.
FIGURE 3.
Purified Edc proteins stimulate decapping in vitro. Analysis of decapping activity by Dcp1p/Dcp2p (left) and Dcp1p/Dcp2p with the addition of either Edc1p (26 pmole), Edc2p (31 pmole), eIF4Ep (21 pmole), Edc1p, and eIF4Ep or Edc1p alone. Decapping by His-Dcp1p/Dcp2p was assayed over a 15-min time course with aliquots removed at the designated time points. The products of the reaction were resolved by thin-layer chromatography. The line graph shows the amounts of decapping from multiple experiments over the time course.
In vivo, the cap-binding complex (eIF4F) and the decapping machinery are thought to be in competition for access to the cap (Schwartz and Parker 2000). If the Edc proteins are increasing the efficiency of decapping, then they may reduce the decapping inhibition observed by the addition of purified eIF4E (cap-binding protein). As reported previously and shown in Figure 3 ▶, the addition of recombinant eIF4E protein alone leads to a substantial loss of decapping in the reconstituted decapping assay. Inhibition by eIF4E is relieved, however, by addition of recombinant Edc proteins (Fig. 3 ▶). Although we believe the major role of the Edc proteins is to enhance decapping activity, this result suggests that in vitro the Edc proteins may enhance the ability of Dcp1p/Dcp2p to compete with eIF4E to stimulate decapping.
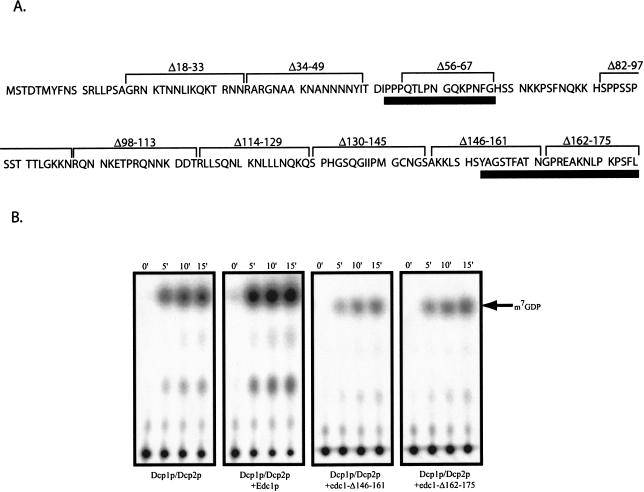
The Edc1 and Edc2 proteins are small basic polypeptides that do not possess any known functional motifs (Dunckley et al. 2001). The two proteins share two small homologous clusters, one internal region and a second at the carboxyl terminus (see Fig. 4A ▶). To identify regions of Edc1p required to stimulate decapping, we created a series of nine edc1 deletion alleles, each removing 13–18 amino acid segments from the coding region of the gene (Fig. 4A ▶). These mutants were expressed in E. coli, purified, and examined for their ability to stimulate decapping in the reconstituted assay. All of the mutant proteins were highly purified as judged by Coomassie staining (data not shown). Two carboxy-terminal deletion mutants (Δ146–161 and Δ162–175) that remove the carboxy-terminal conserved region between Edc1p and Edc2p failed to stimulate decapping (Fig. 4B ▶). In contrast, a deletion of the internal region conserved between Edc1p and Edc2p (Δ56–67) has no effect on stimulation of decapping in vitro (Table 1 ▶). This result demonstrates that the region of the carboxyl terminus conserved between Edc1p and Edc2p contains a domain that is necessary for the stimulation of decapping in vitro.
FIGURE 4.
The carboxyl terminus of EDC1 is required for decapping stimulation. (A) A schematic indicating the regions of the Edc1 protein that were deleted (brackets above sequence) and the regions conserved between Edc1p and Edc2p (black line under sequence). The numbers refer to the amino acids that were removed in each case. (B) Analysis of decapping activity by Dcp1p/Dcp2p (left) and Dcp1p/Dcp2p with either 26 pmole of Edc1p, edc1Δ146–161p, or edc1Δ162–175p. Decapping was assayed over a 15-min time course with aliquots removed at the designated time points. The products of the reaction were resolved by thin-layer chromatography.
TABLE 1.
Phenotype of Edc1p deletions
| EDC1 | Δ18–33 | Δ34–49 | Δ56–67 | Δ82–97 | Δ98–113 | Δ114–129 | Δ130–145 | Δ146–161 | Δ162–175 | EDC2 | |
| RNA binding (Northwestern) | + | “±” | + | + | + | + | + | + | + | + | + |
| RNA binding (Column) | + | + | + | N.D. | N.D. | N.D. | N.D. | N.D. | + | + | + |
| Stimulation of decapping in vitro | + | + | + | + | N.D. | N.D. | N.D. | N.D. | − | − | + |
| Growth on glycerol at 37°C | + | + | + | + | + | + | + | + | “±” | − | + |
To determine whether or not this region of Edc1p was also required for enhancing decapping in vivo, we performed two experiments. First, we asked whether overexpression of the Edc1 protein lacking the carboxyl terminus was able to rescue the growth defect of a dcp1-2 ski8Δ strain. This mutant strain cannot grow at high temperature due to the combination of a defect in decapping resulting from the temperature-sensitive dcp1-2 allele (Tharun and Parker 1999) and a defect in 3′–5′ mRNA decay caused by the absence of Ski8p (Jacobs-Anderson and Parker 1998). Overexpression of wild-type Edc1p allows this strain to grow at high temperature (Dunckley and Parker 2001). In contrast, the Edc1 protein lacking residues 162–175 did not rescue growth (Table 2 ▶). This result indicates that the mutant Edc1p is not functional in vivo, presumably because it is unable to enhance the limited decapping activity that remains at high temperature.
TABLE 2.
Phenotype of edc1Δ162–175 in temperature-sensitive dcp1-2 strains
| Growth | ||
| Strain | 24°C | 33°C |
| dcp1-2 ski8Δ 2μ | + | − |
| dcp1-2 ski8Δ 2μEDC1 | + | + |
| dcp1-2 ski8Δ 2μedc1Δ162–175 | + | − |
| Strain | Fragment/full-lengtha |
| aThe ratio of 5′ to 3′ decay fragment to full-length MFA2pG mRNA. Values are the average of the ratios obtained from two experiments. | |
| wild-type | 1.02 |
| dcp1-2 2μ | 0.13 |
| dcp1-2 2μEDC1 | 0.24 |
| dcp1-2 2μedc1Δ162–175 | 0.11 |
To test directly whether the deletion of the carboxyl terminus prevented Edc1p from promoting decapping in vivo, we examined whether the mutant protein was able to enhance the amount of an mRNA decay fragment that is produced by decapping. Poly(G) tracts inserted into the 3′ end of mRNAs blocks 5′–3′ exonuclease digestion following decapping, allowing the accumulation of 3′ decay fragments (Vreken and Raue 1992; Decker and Parker 1993; Mulhrad et al. 1994). The amount of fragment present relative to the full-length mRNA is a sensitive measure of decapping (Cao and Parker 2001). In dcp1-2 cells, under conditions in which decapping is limited, the level of the 3′ decay fragment of the unstable MFA2 mRNA is reduced relative to the level in wild-type cells (Tharun and Parker 1999). Overexpression of wild-type Edc1p in dcp1-2 cells partially restores the 3′ decay fragment levels (Dunckley and Parker 2001; also see Table 2 ▶). However, overexpression of the Edc1 protein lacking the carboxyl terminus did not result in an increase in the level of the fragment relative to the full-length mRNA (Table 2 ▶). This result indicates that the carboxy-terminal region of Edc1p is required to enhance decapping in vivo as well as in vitro.
Edc1p and Edc2p are a novel class of RNA-binding proteins
The ability of Edc1p and Edc2p to enhance decapping with purified Dcp1p/Dcp2p suggested that these proteins might be RNA-binding proteins that stimulate decapping by binding to both the RNA and the decapping holoenzyme. This is consistent with the observation that the coimmunoprecipitation of Edc1p with Dcp1p and Dcp2p in vivo is partially sensitive to RNase treatment (Dunckley et al. 2001). To test this hypothesis, recombinant Edc1 and Edc2 proteins were first examined for their ability to bind RNA by Northwestern analysis, wherein the proteins are run on a PAGE, transferred to a membrane, and then probed with a radioactive RNA. As shown in Figure 5 ▶, purified recombinant Edc1p and Edc2p bound radiolabeled uncapped MFA2 mRNA, whereas a crude preparation of Dcp1p/Dcp2p, eIF4Ep (data not shown) or BSA did not bind the MFA2 mRNA. This result suggests that Edc1p and Edc2p are RNA-binding proteins.
FIGURE 5.
Edc1p and Edc2p bind RNA. (A) A total of 10 μg of the indicated purified proteins were run by SDS gel electrophoresis on a 12% polyacrylamide gel and stained with Coomassie blue. (B) A gel was run similarly to A, but transferred to nitrocellulose and probed with a radiolabeled uncapped MFA2 RNA. Purified BSA and Dcp1p were run as controls.
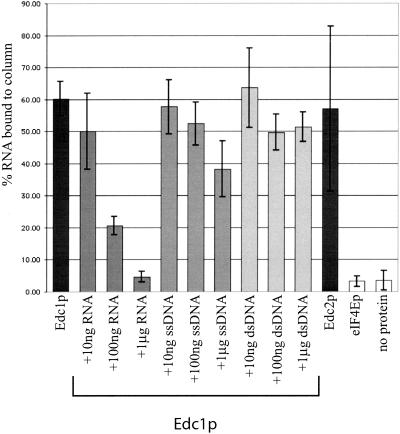
To obtain additional evidence that these proteins bind RNA, and to examine the specificity of the interaction, we bound the His-tagged Edc1p and Edc2p to a nickel column and asked whether radiolabeled RNAs could be specifically retained on the Edc1-His:Ni2+ or Edc2-His:Ni2+ column. Both the Edc1p and the Edc2p columns bound roughly 55% of the input MFA2 mRNA (Fig. 6 ▶). In contrast, <5% of the input mRNA bound to control eIF4E-HisNi2+ or His:Ni2+ columns. This binding was not specific to the MFA2 mRNA, in that several other substrates were retained similarly, including portions of the PGK1 mRNA (data not shown).
FIGURE 6.
Edc1p and Edc2p bind specifically to RNA. A total of 10 μg of purified His-Edc1p, His-Edc2p, or His-eIF4Ep were bound to a nickel column. Radiolabeled MFA2 RNA was added to the column to allow for binding. After washing, the amount of MFA2 RNA remaining bound to the column was quantified using a scintillation counter. Binding was also measured in the presence of the indicated amounts of cold competitor RNA, ssDNA, or dsDNA. Each column represents a minimum of three independent experiments.
To determine whether this binding was specific for RNA, different nucleic acids were used as competitors. Increasing amounts (10- to 1000-fold excess) of cold competitor RNA (capped or uncapped) added to the binding reaction serially reduced the amount of radiolabeled MFA2 mRNA bound by the Edc1-His:Ni2+ column (Fig. 6 ▶). Similar results were observed with the Edc2-His:Ni2+ column (data not shown). In contrast, addition of cold competitor double-stranded or single-stranded DNA (1000-fold excess) did not significantly affect the ability of the Edc proteins to bind the MFA2 RNA. We interpret these observations to indicate that Edc1p and Edc2p are RNA-binding proteins.
The Edc1 and Edc2 proteins do not possess a known RNA-binding motif and, therefore, represent a new class of RNA-binding proteins. To attempt to identify candidate RNA-binding regions within Edc1p, we examined the series of edc1 deletion alleles for their ability to bind RNA. Surprisingly, all of the edc1 deletion mutants were able to bind RNA in the qualitative Northwestern assay (Table 1 ▶). This suggests that the ability of the protein to bind RNA is not limited to any given region, although it is likely that the strength of the RNA–Edc1p interaction may be reduced in some of these alleles. The Edc1p is a particularly basic protein (pI of 11.01), and this feature of the protein may contribute to its general affinity for nucleic acid, but cannot explain its specificity for RNA.
The Edc1p affects growth on glycerol
Both the in vitro experiments described above and the previous in vivo work indicated that Edc1p and Edc2p can enhance decapping rates, but are not required for decapping. This raised the possibility that the Edc proteins could primarily function to regulate the decay of specific mRNAs and/or have a more significant function under different growth/environmental conditions. To predict what growth conditions might reveal a role for the Edc proteins, we examined their expression in a variety of microarray expression databases available over the internet. Interestingly, this revealed that the EDC1 and EDC2 mRNAs are induced during the diauxic shift (DeRisi et al. 1997). This suggests that Edc1p may be required for changes in mRNA turnover that occur following carbon-source shifts (Cereghino et al. 1996). Consistent with this possibility, and the microarray data, we observed that the EDC1 mRNA is induced approximately fourfold following a 15-min shift from glucose to glycerol medium (data not shown).
To determine whether the induction of EDC1 expression following carbon-source shifts was biologically significant, the edc1Δ and edc2Δ strains were tested for their ability to grow on glycerol after the cells were first grown on dextrose-containing medium. This requires the cells to be able to respond rapidly to a change in carbon source and begin aerobic respiration. We found that the edc1Δ strains are unable to grow on glycerol as a carbon source at 37°C (Table 3 ▶). In contrast, edc2Δ strains grew well on glycerol medium at all temperatures tested. This result suggests that Edc1p might be involved in the changes in gene expression necessary for a switch from fermentative to aerobic respiration.
TABLE 3.
Growth in glycerol
| Strain | 24°C | 30°C | 37°C |
| WT | + | + | + |
| dcp1Δ | − | − | − |
| dcp1-2 | + | + | − |
| edc1Δ | + | + | − |
| edc2Δ | + | + | + |
| dcp1-2 edc1Δ | − | − | − |
Three observations suggest that Edc1p’s role in the carbon-source switch is due to its ability to stimulate decapping. First, small deletions in Edc1 protein that inactivate its ability to stimulate decapping in vitro (Fig. 4 ▶) and in vivo (Fig. 5 ▶), also lead to a failure to grow on glycerol at 37°C (Table 1 ▶). In contrast, all of the deletions in Edc1p that allow stimulation of decapping in vitro, support growth on glycerol. Second, when decapping is further compromised in the edc1Δ strain by the addition of the dcp1-2 mutation, the resulting double-mutant strain fails to grow on glycerol at any temperature (Table 3 ▶). The synthetic-lethal phenotype between dcp1-2 and the edc1Δ on glycerol suggests that changes in mRNA turnover rates are an important aspect of regulated gene expression during a carbon-source switch. If decapping of specific messages is necessary for the carbon-source switch, then strains that have no decapping activity should also be unable to grow on glycerol. To test this hypothesis, a dcp1Δ strain was transferred from dextrose to glycerol-containing medium. As with the edc1Δ dcp1-2 strain, the dcp1Δ strain is unable to grow on glycerol at any temperature (Table 3 ▶), indicating that mRNA turnover is an important aspect of the carbon-source switch.
The failure to grow on glycerol can be due to either loss of nuclear gene regulation, or loss of mitochondrial function. To distinguish between these two possibilities, wild-type and edc1Δ strains were crossed to a rhoo tester strain to obtain diploids. Both the resulting diploids and the original haploid parents were tested for their ability to grow on glycerol. The edc1Δ/rhoo diploid is able to grow on glycerol, indicating that the edc1Δ strain has the full mitochondrial genome (data no shown). Therefore, the sensitivity of edc1Δ strains on glycerol is probably due to misregulation of nuclear genes involved in carbon-source regulation.
The Edc1 protein is involved in regulating expression of multiple genes involved in glycolysis and gluconeogenesis
The data above suggested that the edc1Δ was causing defects in the regulation of nuclearly encoded genes whose mRNA levels fluctuate as a result of catabolite repression and/or glycerol metabolism. To test this idea, we examined the expression of several transcripts in a metabolic shift wherein mRNA levels could be followed after glycerol induction and/or catabolite repression. Briefly, cells were grown in dextrose-containing medium to mid-log phase, at which time the dextrose-containing medium was removed and replaced with glycerol-containing medium. After 1 h, the glycerol-containing medium was removed and replaced with dextrose-containing medium. Time points were taken throughout the course of the experiment and levels of specific mRNAs were analyzed.
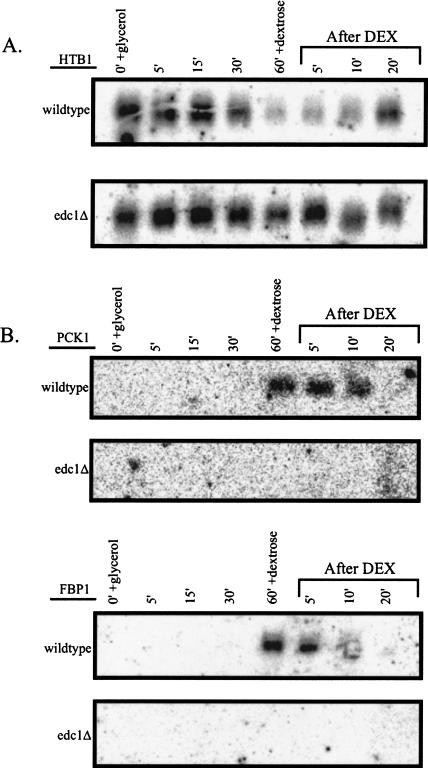
We observed that several mRNAs known to show carbon-source-dependent expression showed altered expression in the edc1Δ strain following changes in carbon source. These changes fell into two different classes. One example was the HTB1 mRNA, which encodes a histone protein. In wild-type cells, the HTB1 mRNA was detected at high levels when grown in dextrose medium (Fig. 7A ▶). After exposure to glycerol, the HTB1 mRNA levels progressively declined over time. Reintroduction of dextrose medium after 60 min in glycerol yielded an increase in the HTB1 mRNA levels. Interestingly, HTB1 mRNA levels do not drop in response to glycerol in the edc1Δ strain. This result suggested that the down-regulation of HTB1 in response to glycerol could be due to more rapid decay of the mRNA. Preliminary results indicate, however, that HTB1 mRNA stability is not altered in response to glycerol or by the deletion of EDC1 (data not shown). Although HTB1 mRNA does not appear to be directly affected by Edc1p, given the relationship between Edc1 protein and the decapping machinery, it is likely that Edc1p does directly regulate the stability of particular mRNAs in response to carbon source.
FIGURE 7.
The Edc1p is required for changes in gene expression following carbon-source changes. Wild-type and edc1Δ strains were grown in dextrose-containing medium to mid-log stage, then grown in glycerol-containing medium for 1 h, then switched back to dextrose-containing medium. The designated mRNAs were examined by Northern analysis. (A) HTB1 mRNA levels do not decline properly in the edc1Δ strain. (B) PCK1 and FBP1 mRNAs are not induced in the edc1Δ strain.
The FBP1 and PCK1 genes represent a second class of transcript. These mRNAs encode gluconeogenic enzymes that are induced by glycerol. In wild-type cells, both FBP1 and PCK1 mRNAs were produced abundantly within 60 min of glycerol addition (Fig. 7B ▶). In contrast, in the edc1Δ strain, these mRNAs were still not induced, even at the end of the glycerol incubation. This class of mRNAs may not be induced properly in the edc1Δ strain for one of two reasons. First, the genes are being transcribed, but the mRNAs are being degraded so rapidly that they do not accumulate to detectable levels. We do not favor this model, as the Edc1p is thought to enhance decapping and, hence, a deletion of this gene would stabilize mRNAs. Secondly, these genes may not be expressed because the proteins and mRNAs involved in catabolite repression are not being turned over properly in the edc1Δ strain, leading to a loss of transcriptional activation of the gluconeogenic mRNAs. We interpret these results to indicate that Edc1p is required to modulate decapping for proper changes in gene expression that occur during shifts from glucose to glycerol carbon sources.
DISCUSSION
Edc1p and Edc2p bind RNA
Several lines of evidence indicate that Edc1p and Edc2p are RNA-binding proteins. First, both proteins bind RNA by Northwestern analysis (Fig. 5 ▶). In addition, both EDC proteins are able to retain RNA in a column-binding assay (Fig. 6 ▶). This binding is specific for RNA because RNA binding can be competed by cold RNA substrates but not by ssDNA or dsDNA substrates. The ability to bind RNA is also consistent with the role of the EDC proteins in enhancing decapping rates both in vivo and in vitro.
Our data imply that Edc1p and Edc2p are able to interact with a wide variety of RNA molecules and, therefore, either have broad RNA-binding specificity or bind to common small motifs. For example, several different RNA molecules can be retained specifically on the Edc1p or Edc2p columns or can function as competitors (Fig. 6 ▶; data not shown). Moreover, in vivo, overexpression of Edc1p or Edc2p restores decapping of both the MFA2 and PGK1 transcripts in a dcp1-2 or dcp2-7 strain, respectively (Dunckley et al. 2001). Additionally, when an edc1Δ or edc2Δ is combined with a dcp1-2 or dcp2-7 mutation, the PGK1 and MFA2 mRNAs are further stabilized. The fact that the Edc proteins affect both MFA2 and PGK1 transcripts, which represent unstable and stable yeast mRNAs, suggests that the Edc proteins can interact with, and affect the decapping of a variety of transcripts. An important issue for future work will be to determine whether or not these proteins do show any specificity in their RNA binding.
Edc1p and Edc2p enhance the activity of the Dcp1p/Dcp2p decapping enzyme
Several observations indicate that the function of the Edc proteins is to directly affect the activity of the Dcp1p/Dcp2p decapping enzyme. For example, addition of purified Edc1p or Edc2p to cell extracts enhanced decapping activity (Fig. 2 ▶). Evidence that this was a direct effect on the decapping enzyme came from the observation that recombinant Edc proteins could also stimulate decapping enzyme purified from yeast (Fig. 3 ▶) as well as recombinant Dcp1/Dcp2 proteins (Steiger et al. 2003).
In principle, the Edc proteins could enhance decapping by one of three mechanisms. First, they could simply bind the RNA and prevent competing secondary structures from forming, which would indirectly stimulate decapping. This is unlikely, as we identified two deletion mutants of Edc1p, which can still bind RNA, but fail to activate decapping in vitro (Fig. 4 ▶). In addition, the nonspecific nucleic acid-binding polyamine, spermidine, inhibits decapping (D. Schwartz and R. Parker, unpubl.). An appealing alternative is that the Edc proteins stimulate decapping by binding both the mRNA substrate and the Dcp1p/Dcp2p decapping enzyme, thereby enhancing the interaction of the enzyme with the substrate and/or the catalytic rate of the enzyme. However, we cannot yet rule out the formal possibility that Edc1p or Edc2p could act as allosteric activators of the decapping enzyme independent of any Edc–RNA interaction, although the RNA-binding properties of the Edc proteins make this appear unlikely.
Implications for control of decapping
The analysis above indicates that the Edc proteins directly affect the actual decapping step in mRNA decay. This conclusion implies that the binding and catalytic steps in decapping are often not the rate-determining steps for decapping rates in vivo. This corollary is based on the fact that loss of Edc1p or Edc2p does not affect the decay of several mRNAs under normal growth conditions (Dunckley et al. 2001). Similarly, conditional mutations in dcp1 or dcp2 do not affect decay rates at the permissive temperature, despite the fact that extracts from these cells are deficient in decapping in vitro at permissive temperatures (Fig. 2 ▶). We interpret these results to suggest that loss of Edc1p or Edc2p, or conditional alleles in dcp1 and dcp2 at the permissive temperatures, each individually slows the rate of the binding/catalytic step in decapping per se in vivo, but other steps in the overall process of mRNA decapping are slower and thereby the edc1Δ, edc2Δ, dcp1-2, or dcp2-7 alleles have no observable affect on overall decapping or decay rates. However, the actual decapping step in decay can be made rate limiting in vivo when lesions are present that severely inhibit this step. Such lesions would include loss of the decapping enzyme (e.g., dcp1Δ or dcp2Δ) or combination of multiple alleles affecting this step, such as edc1Δ dcp1-2 or dcp2-7 edc2Δ.
Additional evidence for multiple distinct substeps in mRNA decapping has come from other observations. First, in genetic experiments, a decapping defect due to a lesion in the decapping enzyme (dcp1-1), but not the decapping defect due to the lsm1Δ or pat1Δ, can be suppressed by a mutation in the cap-binding protein eIF4E (Schwartz and Parker 2000). Second, strains lacking Lsm1p or Pat1p, which show a substantial defect in decapping in vivo (Bonnert et al. 2000; Bouveret et al. 2000; Tharun et al. 2000), produce extracts that have decapping activity similar to wild-type cells (D. Schwartz and R. Parker, unpubl.). Although the precise nature of these other substeps in decapping are not yet known, it is likely that they are related to transitions in mRNP organization and/or translation status, potentially including loss of the cap-binding complex, which serves as an inhibitor of decapping (for review, see Schwartz and Parker 2000; Tharun and Parker 2001). It should also be noted that these observations argue that the decapping rate in our crude cell extracts is predominated by the actual decapping step per se and fails to reproduce significant rate limiting steps of the in vivo process. An important goal for future work will be to create in vitro systems that reproduce more of the rate-determining steps seen in vivo.
The presence of multiple distinct substeps in the decapping process implies that there could be control at each substep either for different mRNAs, and/or under different growth conditions. For example, if the decapping step per se is rate limiting for some mRNAs, then the edc1Δ or the edc2Δ allele might affect decapping of those specific mRNAs. Similarly, if the decapping step per se is rate limiting under some growth condition, then the edc1Δ or dcp1-2 allele might affect decapping rates and/or growth under those conditions (see below).
Edc1p is involved in the dextrose to glycerol carbon-source switch
Several observations argue that Edc1p is required for the switch in gene expression that occurs following a shift from dextrose to glycerol. First, the EDC1 mRNA is induced during this switch (data not shown). Second, the edc1Δ strain is unable to grow on glycerol at 37°C (Table 3 ▶), although this strain is quite healthy at 37°C on dextrose. Examination of the edc1Δ strain on glycerol at lower temperatures shows that the cells are sick and a large percentage of the cells are dead (data not shown). We conclude the edc1Δ strain is dying on glycerol due to a decapping defect, because edc1 alleles that fail to activate decapping in vitro and in vivo (Fig. 4 ▶; Table 2 ▶) also fail to support growth on glycerol (Table 1 ▶), and a dcp1Δ strain is also unable to grow on glycerol (Table 3 ▶), indicating that decapping is required under these conditions. Finally, we observed abnormal expression patterns of several mRNAs following the carbon-source shift in edc1Δ strains (Fig. 7 ▶). These observations indicate that Edc1p is required for the shift in gene expression following this carbon-source shift. The idea that Edc1p is involved in regulating gene expression in response to changes in carbon source is supported by the recent finding that EDC1 is a high-copy suppressor of mutations in PAS kinase, which is involved in the regulation of sugar flux (Rutter et al. 2002).
There are two general manners in which the Edc1p could be required for growth on glycerol. First, it could be that there is a specific subclass of mRNAs, which must be degraded for shifting from dextrose as a carbon source to glycerol, and whose efficient decapping requires the Edc1p. For example, if Edc1p was involved in the rapid decay of transcripts encoding catabolite repressors, then loss of the Edc1 protein would lead to prolonged production of these proteins, possibly leading to lower levels of glycerol-induced mRNAs or the complete absence of glycerol-induced mRNAs as seen with FBP1 and PCK1 (Fig. 7 ▶). An alternative possibility is that the general change in cellular physiology upon a shift to glycerol medium alters the predominant rate-limiting step in decapping to the actual decapping step per se. Under these conditions, Edc1p would be required to allow the cell to continue efficient decapping and proper regulation of a large number of mRNAs, with corresponding significant alterations in gene expression.
MATERIALS AND METHODS
Yeast strains
The genotypes of all of the strains (Saccharomyces cerevisiae) used in this study are listed in Table 4 ▶.
TABLE 4.
Strains used in this study
| Strain | Genotype | Source |
| yRP840 | MAT a his4-539 leu2-3, 112 trp1 ura3-52 cup1::LEU2/PGK1pg/MFA2pG | Hatfield et al. 1996 |
| yRP1340 | MAT a his4-539 leu2-3, 112 lys2-201 trp1 ura3-52 dcp1-2::TRP1 | Dunckley and Parker 1999 |
| yRP1341 | MAT a his4-539 leu2-3, 112 lys2-201 trp1 ura3-52 dcp1-2::TRP1[MFA2pG/LYS2] | Dunckley and Parker 1999 |
| yRP1345 | MAT a his4-539 leu2-3, 112 lys2-201 trp1 ura3-52 dcp1-2::TRP1 ski8::URA3 | Dunckley and Parker 1999 |
| yRP1501 | MAT α leu2-3, 112 lys2-201 trp1 ura3-52 dcp2-7::URA3 | Dunckley and Parker 2001 |
| yRP1503 | MAT a his3-Δ200 ade2-101 leu2-3,112 lys2-201 trp1 ura3-52 edc1::HIS3 cup2::LEU2/PGK1pG/MFA2pG | Dunckley and Parker 2001 |
| yRP1504 | MAT α hist4-539 leu2-3,112 trp1 ura3-52 edc2::NEO cup1::LEU2/PGK1pG/MFA2pG | Dunckley and Parker 2001 |
| yRP1506 | MAT a ade2 leu2-3,112 lys2-201 trp1 ura3-52 dcp1-2::TRP1 edc1::HIS3 cup1::LEU2/PGK1pG/MFA2pG | Dunckley and Parker 2001 |
Vector construction
The His-EDC1 gene was created by PCR of the EDC1 gene from pRP984 with the oligonucleotides CCGCCGCATATGTCGAC GGATACCATGTATTTC (oRP1083) and CGGAAAAGGATCCT TATAAAAAACTTGGTTTGGGCAAG (oRP1084). The His-EDC2 gene was created similarly by using pRP985 with the oligonucleotides CCGCCGCATATGGGTTCCGAGACAAAACATTCTGC (oRP1085) and CGGAAAAGGATCCTCAAACAAAACTAGGTTT GGGTAGCG (oRP1086). The His-edc1-Δ162-175 gene was created similarly by using pRP1056 with the oligonucleotides (oRP1083) and CGGAAAAGGATCCTTAATTAGTGGCGAAAG TGGAGCC (oRP1107). These fragments were digested with NdeI and BamHI and ligated into the E. coli expression vector pET-16b (Novagen) between the NdeI and BamHI sites to yield pRP1054, pRP1055, and pRP1057. These plasmids express either His-EDC1, His-EDC2, or His-edc1-Δ162-175 from the inducible T7 promoter. The additional His-edcΔ plasmids were all made by use of the Quicksite mutagenesis kit from Stratagene using pRP1054 as a template.
The 2μ edc1-Δ162-175 plasmid was created with the oligonucleotides CTTATGCAGGCTCCACTTTCGCCACTAATTAAT CACCCTTTAGAGAGACATTTTTG (oRP1105) and CAAAAAT GTCTCTCTAAAGGGTGATTAATTAGTGGCGAAAGTGGAGCC TGCATAAG (oRP1106) using the Quicksite mutagenesis kit from Stratagene on pRP984 to create pRP1056.
All plasmids were fully sequenced to ensure that all mutations, junctions, and tags were properly generated.
Protein purification
His-EDC1 and His-EDC2 were purified as recommended by the manufacturer (Novagen). Briefly, 100-mL cultures of BL-21(DE3) cells containing either a His-EDC1 (pRP1054) or His-EDC2 (pRP1055) plasmid were grown in Luria broth medium at 37°C to an optical density of 0.6. The cultures were induced with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) for 3 h and then harvested by centrifugation. After elution of the proteins from a nickel-nitrilotriacetic acid column (QIAGEN) with 1× elution buffer, the proteins were dialyzed first into a mixture of 50 mM KCl, 20 mM HEPES (pH 7.5), and 5 mM EDTA to separate the proteins from the nickel, and then into a mixture of 50 mM KCl and 20 mM HEPES (pH 7.5). The proteins were aliquoted and frozen at −80°C.
Preparations of the decapping enzyme containing Dcp1p and Dcp2p were purified from yeast as described previously using a His-Dcp1p (Beelman et al. 1996).
Substrate preparation
Uncapped MFA2 mRNAs lacking poly(A) tails, used for decapping assays, were synthesized by using the Riboprobe in vitro transcription system (Promega). The DNA template used was as described previously (LaGrandeur and Parker 1998). T7 transcription was done as recommended by the manufacturer with 1 to 2 μg of template DNA at 37°C for 2 h. The DNA template was digested with 1 μL of RQ1 DNase. The resulting uncapped transcript was purified by phenol chloroform extraction and Sephadex G-50 chromatography. The samples were precipitated and resuspended in 20 μL of diethyl pyrocarbonate treated water and capped as described previously (LaGrandeur and Parker 1998).
Uncapped MFA2 mRNAs lacking poly(A) tails, used for Northwestern analysis or RNA-binding studies were synthesized as described above using 5′[α-32P]rCTP.
Decapping assays
Decapping reactions with Dcp1p/Dcp2p purified from yeast were assayed at 30°C over a 15-min time course. All experiments were done a minimum of three times. The reaction mixtures generally contained ∼2.3 fmole of m7G[32P]pppMFA2 mRNA, 3.1 pmole of Dcp1p/Dcp2p, 50 mM Tris (pH 7.6), 5 mM MgCl2, 50 mM NH4Cl, 1 mM dithiothreitol (DTT), and 1 μL of RNasin in a volume of 15 μL. Additional proteins were added at the concentrations noted in the figure legends. BSA was added to all reaction mixtures to maintain constant total protein concentrations. Aliquots of the reaction mixture were withdrawn at various time points and decapping was stopped by adding 1 μL of 0.5 M EDTA to the aliquot and placing it on ice. The products of the reaction were separated by polyethyleneimine–cellulose thin-layer chromatography developed in 0.45 M (NH4)2SO4 and detected with a Molecular Dynamics PhosphorImager.
To prepare decapping extracts, a procedure was used from Zhang et al. (1999). Decapping reactions were done similarly to those described above using 20 μg of extract and a 30-minute time course.
Northwestern analysis
Northwesterns were done as described (Bichsel et al. 1997). Briefly, 10 μg of each protein was run by 12% SDS-PAGE, then transferred to nitrocellulose. After renaturing the proteins overnight, a radiolabeled MFA2 mRNA was used as a probe. After washing, RNA binding was detected using a Molecular Dynamics PhosphorImager.
RNA-binding analysis
RNA-binding studies were done using 10 μg of purified His-Edc1p or His-Edc2p bound to a nickel-nitrilotiracetic acid column (QIAGEN). A total of 50,000–100,000 cpm of MFA2 mRNA was added to the column in 100 μL of RNA-binding buffer [50 mM Tris (pH 7.6), 5 mM MgCl2, and 50 mM NH4Cl]. The column was washed two times with RNA-binding buffer. Flow-through material, both washes, and bound material were counted in a scintillation counter to get a measure of the percentage of counts bound to the column.
Suppression of dcp1-2 ski8Δ growth defect
yRP1345 carrying yEP351, pRP984 (Dunckley and Parker 2001), or pRP1056 was grown at 24°C on minimal medium lacking leucine for 4 d, then replica plated and grown for an additional 2 d at 24 or 33°C.
mRNA analysis
To examine the suppression of the dcp1-2 decapping defect, yRP840 and yRP1341 carrying yEP351, pRP984 (Dunckley and Parker 2001), or pRP1056, was grown to mid-log at 24°C in minimal medium lacking leucine and lysine containing 2% galactose. Cultures were then shifted to 33°C for 1 h. Following the temperature shift, cells were harvested and RNA was isolated as described previously (Caponigro et al. 1993). The full-length MFA2pG mRNA and 3′ decay fragment were detected by Northern analysis using oligonucleotide oRP140 (Caponigro and Parker 1995) as a probe. For carbon-shift experiments, cells were grown to mid-log in medium containing dextrose at 30°C, harvested, and resuspended in glycerol-containing medium for 1 h, then harvested again and resuspended in medium containing dextrose. RNA was isolated as described previously. Northern analysis was done using 20 μg of RNA. The amount of RNA loaded in each lane was normalized using the 7S RNA (Decker and Parker 1993).
Acknowledgments
We thank members of the Parker laboratory for comments on the manuscript. An NIH grant (GM45443) and funds from the Howard Hughes Medical Institute supported this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2171203.
REFERENCES
- Beelman, C.A. and Parker, R. 1995. Degradation of mRNA in Eukaryotes. Cell 8: 179–183. [DOI] [PubMed] [Google Scholar]
- Beelman, C.A., Stevens A., Caponigro G., LaGrandeur, T.E., Hatfield L., Fortner, D., and Parker, R. 1996. An essential component of the decapping enzyme required for normal rates of mRNA decay in yeast. Nature 38: 642–646. [DOI] [PubMed] [Google Scholar]
- Bichsel, V.E., Walz, A., and Bickel, M. 1997. Identification of proteins binding specifically to the 3′-untranslated region of granulocyte/macrophage-colony stimulating factor mRNA. Nucleic Acids Res. 25: 2417–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot, C., Boeck, R., and Lapeyre, B. 2000. The two protein Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 20: 5939–5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret, E., Rigaut, G., Shevchenko, A., Wilm, M., and Seraphin, B. 2000. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 19: 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, D. and Parker, R. 2001. Computational modeling of eukaryotic mRNA turnover. RNA 7: 1192–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G. and Parker, R. 1995. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 9: 2421–2432. [DOI] [PubMed] [Google Scholar]
- Caponigro, G., Muhlrad, D., and Parker, R. 1993. A small segment of the MATα1 transcript promotes mRNA decay in yeast: A stimulatory role for rare codons. Mol. Cell. Biol. 13: 5141–5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereghino, G.P. and Scheffler, I.E. 1996. Genetic analysis of glucose regulation in Saccharomyces cerevisiae: Control of transcription versus mRNA turnover. EMBO J. 15: 363–374. [PMC free article] [PubMed] [Google Scholar]
- Coller, J.M., Tucker, M., Sheth, U., Valencia-Sanchez, M.A., and Parker, R. 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7: 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J. and Parker, R. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 7: 1632–1643. [DOI] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278: 680–686. [DOI] [PubMed] [Google Scholar]
- Dunckley, T. and Parker, R. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18: 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2001. Yeast mRNA decapping enzyme. Methods Enzymol. 342: 226–233. [DOI] [PubMed] [Google Scholar]
- Dunckley T., Tucker, M., and Parker, R. 2001. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, N. and Weis, K. 2002. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 21: 2788–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhaniyogi, J. and Brewer, G. 2001. Regulation of mRNA stability in mammalian cells. Gene 265: 11–23. [DOI] [PubMed] [Google Scholar]
- Hatfield, L., Beelman, C.A., Stevens, A., and Parker, R. 1996. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16: 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C.L. and Stevens, A. 1993. Yeast cells lacking 5′ to 3′ exoribonuclease I contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13: 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs-Anderson, J.S. and Parker, R. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur, T.E. and Parker, R. 1998. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17: 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D. and Parker, R. 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes & Dev. 6: 2100–2111. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C., and Parker, R. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′ to 3′ degradation of the transcript. Genes & Dev. 8: 855–866. [DOI] [PubMed] [Google Scholar]
- 1995. Turnover mechanisms of the stable PGK1 mRNA in yeast. Mol. Cell. Biol. 1: 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivas, W. and Parker, R. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 19: 6602–6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto, S., de la Cruz, B.J., and Scheffler, I.E. 2000. Glucose-regulated turnover of mRNA and the influence of poly(A) tail length on half-life. J. Biol. Chem. 275: 14155–14166. [DOI] [PubMed] [Google Scholar]
- Ramirez, C.V., Vilela, C., Berthelot, K., and McCarthy, J.E. 2002. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J. Mol. Biol. 318: 951–962. [DOI] [PubMed] [Google Scholar]
- Rutter, J., Probat, B.L., and McKnight, S.L. 2002. Coordinate regulation of sugar flux and translation by PAS kinase. Cell 111: 17–28. [DOI] [PubMed] [Google Scholar]
- Schwartz, D. and Parker, R. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of yeast mRNAs. Mol. Cell. Biol. 19: 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 20: 7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger, M., Carr-Schmid, A., Schwartz, D.C., Kiledjian, M., and Parker, R. 2003. Analysis of recombinant yeast decapping enzyme. RNA. (this issue). [DOI] [PMC free article] [PubMed]
- Surosky, R.T. and Esposito, R.E. 1992. Early meiotic transcripts are highly unstable in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 3948–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun, S. and Parker, R. 1999. Analysis of mutations in yeast mRNA decapping enzyme. Genetics 151: 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2001. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell 8: 1075–1083. [DOI] [PubMed] [Google Scholar]
- Tharun, S., He, W., Mayes, A.E., Lennertz, P., Beggs, J.D., and Parker, R. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518. [DOI] [PubMed] [Google Scholar]
- Vilela, C., Velasco, C., Ptushkina, M., and McCarthy, J.E. 2000. The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J. 19: 4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken, P. and Raue, H.A. 1992. The rate limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3′ terminal part of the coding region. Mol. Cell Biol. 12: 2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz, C.J., Wormington, M., and Peltz, S.W. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell. Biol. 2: 237–246. [DOI] [PubMed] [Google Scholar]
- Zhang, S., Williams, C.J., Wormington, M., Stevens, A., and Peltz, S.W. 1999. Monitoring mRNA decapping activity. Methods 17: 46–51. [DOI] [PubMed] [Google Scholar]