Abstract
The arginine-rich domains of several RNA-binding proteins have been shown to bind their cognate RNAs with high affinities and specificities as isolated peptides, adopting different conformations within different complexes. The sequence simplicity and structural diversity of the arginine-rich motif has made it a good framework for constructing combinatorial libraries and identifying novel RNA-binding peptides, including those targeted to the HIV Rev response element (RRE). Here we describe a modified transcription antitermination reporter assay engineered with kanamycin resistance that enables larger in vivo screens (∼109 sequences) than previously possible. We show that the assay detects only specific RNA–protein complexes, and that binders are enriched at least 300-fold per round of selection. We screened a large peptide library in which amino acids with charged, polar, and small side chains were randomly distributed within a polyarginine framework and identified a set of high affinity RRE-binding peptides. Most contain glutamine at one particular peptide position, and the best peptides display significantly higher antitermination activities than Rev or other previously described high-affinity RRE-binding peptides. The kanamycin antitermination (KAN) assay should be useful for screening relatively large libraries and thereby facilitate identification of novel RNA binders.
Keywords: Arginine-rich motif, HIV Rev, combinatorial libraries, RNA recognition
INTRODUCTION
Sequence-specific RNA–protein interactions play essential roles in gene expression and often uilize complex three-dimensional RNA structures as surfaces for protein recognition. There have been great advances over the past few years in understanding RNA structure and the principles of recognition, largely from crystallographic and NMR studies of RNAs and RNA–protein complexes, including the two ribosomal subunits (Draper 1999; Perez-Canadillas and Varani 2001; Ramakrishnan 2002). Along with these studies, the identification of several common types of RNA-binding domains, including the RNP (or RRM) domain, the KH domain, and the arginine-rich motif (Draper 1999; Frankel 2000; Perez-Canadillas and Varani 2001), has raised the possibility that proteins with desired binding specificities might be designed or identified from combinatorial libraries. Recently, methods have been developed to screen cDNA or peptide libraries for RNA binders, facilitating the identification of new molecules and detailed structure/function analyses. In vitro screening methods have utilized phage display or mRNA–peptide fusions (Laird-Offringa and Belasco 1995; Barrick et al. 2001), and in vivo methods have utilized reporters in which translation or transcription are altered by protein binding (Harada et al. 1996; Jain and Belasco 1996; Kollmus et al. 1996; SenGupta et al. 1996; Wang et al. 1997; Paraskeva et al. 1998; Tan and Frankel 1998; Landt et al. 2000). Here we describe the construction of a modified transcription antitermination system suitable for high-throughput screening and its use in identifying high-affinity HIV Rev response element (RRE) binding peptides.
The RRE is ∼250 nt in length and contains several RNA hairpins, including the IIB hairpin that forms a high-affinity binding site for the HIV Rev protein. Rev utilizes an α-helical arginine-rich RNA-binding domain to recognize an internal loop region of IIB (Tan et al. 1993; Battiste et al. 1996; Ye et al. 1996) and an additional domain to facilitate viral mRNA export (Pollard and Malim 1998; Hope 1999). Arginine-rich domains are found in a variety of RNA-binding proteins, including the HIV and BIV Tat transcription factors, the bacteriophage antiterminator N proteins, and viral coat proteins, and they often can bind to their RNA sites as short (<20 amino acid) peptides that faithfully mimic many of the binding properties of the intact proteins (Calnan et al. 1991; Weeks and Crothers 1991; Kjems et al. 1992; Tan et al. 1993; Chen and Frankel 1994, 1995; Tan and Frankel 1995; Ansel-McKinney et al. 1996; Cai et al. 1998; Legault et al. 1998; Swanson et al. 1998). In general, the domains share little sequence similarity aside from a preponderance of arginines and show substantial diversity at the structural level (Frankel 2000). For example, an arginine-rich peptide from BIV Tat binds to its TAR RNA site in a β-hairpin conformation, λ and P22 N peptides bind to their boxB hairpins as bent α-helices, and even the HIV Rev peptide, which binds as a single helix to RRE IIB, can adopt an extended conformation in the context of a different RNA site (Puglisi et al. 1995; Ye et al. 1995; Cai et al. 1998; Legault et al. 1998; Ye et al. 1999). Typically, arginine-rich peptides are weakly structured or disordered until bound to RNA (Tan and Frankel 1994; Chen and Frankel 1995; Frankel and Smith 1998; Mogridge et al. 1998; Patel 1999) and utilize just a few types of amino acids other then arginine to make specific RNA contacts (Tan et al. 1993; Chen and Frankel 1995; Tan and Frankel 1998). Because of its structural plasticity and relative sequence simplicity, the arginine-rich motif has proven to be a versatile framework for identifying novel RNA binders from combinatorial libraries (Harada et al. 1996, 1997; Tan and Frankel 1998; Barrick et al. 2001). Such molecules have been useful for defining new RNA recognition strategies, have provided useful reagents for probing RNA function, and may provide starting points for designing therapeutic inhibitors that target key RNA sites, such as the RRE (Hermann 2000; Cheng et al. 2001).
We previously described a bacterial antitermination assay to screen peptide libraries for RNA binders (Harada et al. 1996), based on a bacteriophage λ N protein reporter system (Franklin 1993). In this assay, peptide libraries are fused to the N protein encoded on one plasmid, and those that bind RNA are identified using a second reporter plasmid engineered with an RNA site of interest such that binding of N causes transcription antitermination and LacZ expression (Harada et al. 1996) (see Fig. 1 ▶). High affinity RRE-binding peptides were identified from ∼106 sequences, and additional rounds of mutagenesis and reselection resulted in peptides with enhanced binding affinities (Harada et al. 1996, 1997). Interestingly, the RSG-1.2 peptide, which binds with ∼10-fold higher affinity and specificity than Rev, recognizes the same internal loop of IIB as Rev but adopts a helix-turn-extended conformation in the complex and makes a rather different set of RNA contacts than the helical Rev peptide (Gosser et al. 2001; Zhang et al. 2001).
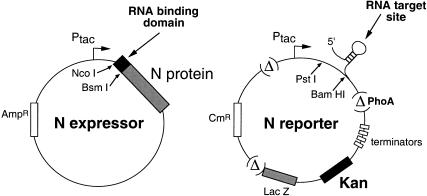
FIGURE 1.
The two-plasmid system for detecting transcription antitermination by heterologous RNA-binding polypeptides fused to the N protein. The reporter plasmid is modified from the previous LacZ reporter (Harada et al. 1996). The transcript, driven by the tac promoter, is arranged with an RNA-binding site upstream of transcription terminators, followed by bicistronic kanamycin resistance and LacZ reporter genes. Three regions of the original reporter have been deleted (indicated by Δ), including the phoA gene and two regions with repeated sequences, as described in the text. The deletions enhanced the antitermination signal and may have increased plasmid stability.
To further improve the RNA-binding properties of selected peptides and to allow for a broader search of peptide sequence space, we have now reengineered the antitermination assay with a kanamycin resistance reporter to allow for selection, rather than visual screening, of binders, with a capacity of ∼109 sequences. By screening a combinatorial library constructed within a polyarginine framework, we have identified a new set of RRE-binding peptides that contain conserved glutamine residues reminiscent of high-affinity binders previously found in a mammalian Tat-hybrid screen (Tan and Frankel 1998). The newly selected peptides show much higher antitermination activities than Rev or the previously identified RSG-1.2 peptide, suggesting that the improved screening capacity of the kanamycin antitermination (KAN) assay will allow for more exhaustive searches of sequence space and further significant binding enhancements.
RESULTS
Kanamycin antitermination (KAN) assay
We previously described an antitermination reporter system that utilizes the bacteriophage λ N protein to monitor RNA–protein interactions (Harada et al. 1996; Wilhelm and Vale 1996; Peled-Zehavi et al. 2000). In λ, N binds to an RNA hairpin (boxB) within the nut (N-utilization) site on the nascent transcript and promotes the assembly of transcription antitermination complexes (Greenblatt et al. 1993). Typically, the host proteins Nus A, Nus G, Nus B, and S10, and another RNA element in nut (box A), are needed for assembly, although under some conditions antitermination complexes can form in the absence of some factors (Rees et al. 1996). To study antitermination in a simplified context, Franklin devised a two-plasmid reporter assay in which N is expressed from one plasmid and LacZ from a second plasmid containing four transcription terminators located downstream of the nut site and upstream of lacZ (Franklin 1993). The system can be modified to study heterologous interactions by fusing an RNA-binding domain to N and replacing boxB with a corresponding RNA-binding site (Fig. 1 ▶) (Harada et al. 1996; Wilhelm and Vale 1996). It has been possible to use the LacZ reporter to identify specific RNA-binding peptides from combinatorial libraries of ∼106 sequences (Harada et al. 1996).
We wished to expand the capacity of the system by replacing lacZ with a selectable marker that, in principle, could allow screening of 109 or more clones. We cloned the neomycin phosphotransferase gene (NPT II), which confers dose-dependent kanamycin resistance, into the N reporter plasmid, retaining lacZ to allow secondary activity assays (Fig. 1 ▶). The ability to select for survival at different kanamycin concentrations was expected to facilitate identification of clones with varying RNA-binding affinities. To further optimize the reporter, we deleted the phoA gene, which initially had been placed upstream of the termination sites to internally control for the levels of transcription initiation (Doelling and Franklin 1989; Franklin 1993). It has been demonstrated that N and NusA alone support partially processive antitermination in vitro provided that the distance between the nut site and terminator is less then a few hundred base pairs (Whalen et al. 1988; Mason et al. 1992). The replacement of boxB in our heterologous system eliminates the NusA-boxB interaction and alters the spatial arrangement of RNA elements, probably reducing the cooperative assembly of antitermination complexes (Chattopadhyay et al. 1995; Mogridge et al. 1995; Rees et al. 1996). Indeed, the activities observed with heterologous interactions are typically lower than with N-boxB (Harada et al. 1996), and thus we deleted phoA to minimize the spacing between the RNA-binding site and improve activity. In addition, two other repeated regions of the reporter were deleted, further reducing the size of the plasmid (see Materials and Methods; Fig. 1 ▶).
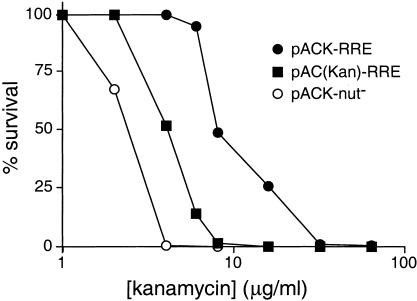
To test the effect of the deletions, we measured kanamycin survival curves using RRE IIB reporters with or without the deletions (Fig. 2 ▶). N567/RRE-reporter cells were transformed with a Rev peptide-N expressor plasmid, incubated at room temperature for 6 h to allow NPT II expression (see Materials and Methods), plated at kanamycin concentrations up to 80 μg/mL, and colonies were counted after 3–5 d. Cells containing the deleted version of the reporter (pACK-RRE) showed 50% survival at 9.2 μg/mL kanamycin, whereas cells containing the undeleted version [pAC(Kan)-RRE] showed 50% survival at 4.4 μg/mL (Fig. 2 ▶). Control cells lacking the RRE (pACK-nut-) survived only at background levels, with 50% survival at 2.5 μg/mL. Thus, the cognate Rev-RRE interaction supports antitermination, as monitored by kanamycin resistance, at levels well above background, and the vector deletions significantly improve activity. We refer to the kanamycin antitermination assay as the KAN assay.
FIGURE 2.
Kanamycin survival curves (KAN assays) with deleted and nondeleted versions of the reporter. N567 cells were transformed with the Rev expressor plasmid and either pAC(Kan)-RRE (nondeleted RRE reporter), pACK-RRE (RRE reporter with deletions), or pACK-nut- (reporter with deletions but lacking the RRE). Percent survival is plotted as a function of kanamycin concentration.
RNA-binding specificity of the KAN assay
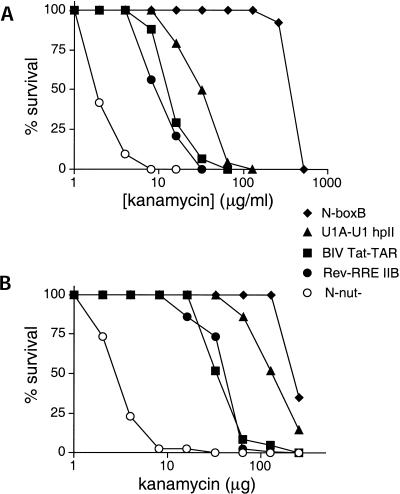
To further characterize the sensitivity, specificity, and dynamic range of the KAN assay, we examined the activities of several other RNA–protein interactions. The BIV Tat-TAR and N-boxB interactions, which also utilize arginine-rich peptides for RNA recognition, were previously characterized using the LacZ reporter assay (Harada et al. 1996). In the KAN assay, the Tat–TAR interaction showed a survival curve similar to that of the Rev–RRE interaction (50% survival at 13 μg/mL for Tat–TAR and 9 μg/mL for Rev–RRE), whereas the N-boxB interaction supported higher levels of antitermination (50% survival at 375 μg/mL) (Fig. 3A ▶), correlating with previous β-galactosidase assays (Harada et al. 1996). To test whether other types of RNA-binding domains that differ in size and structure from arginine-rich peptides can function in this reporter system, we created an expressor in which the first RNP domain of the U1A protein was fused to N (Oubridge et al. 1994) and a reporter with the corresponding hairpin II binding site of U1 snRNA. The U1A–U1 hpII interaction showed resistance at higher levels of kanamycin than the Rev–RRE or Tat–TAR interactions (50% survival at 32 μg/ml), consistent with its known high affinity (Hall 1994) and the high levels of U1A-N expression observed by Western blotting (data not shown).
FIGURE 3.
Kanamycin survival curves for a variety of RNA–protein complexes. (A) Antitermination activities measured by kanamycin resistance (KAN assays). Percent survival is plotted as a function of kanamycin concentration. (B) Antitermination activities measured by the Kirby-Bauer agar diffusion assay. Percent survival is plotted as a function of the total kanamycin amount applied to each filter disc.
We next tested whether a rapid antibiotic resistance assay could be used to monitor antitermination activities. We employed the Kirby-Bauer method in which filter discs saturated with varying amounts of antibiotic are placed onto bacterial lawns and zones of inhibition are measured (Baker and Breach 1980; see Materials and Methods). The results with this assay correlate very well with those of the KAN assay, with activities in the order: N-boxB > U1A-U1 hpII > BIV Tat–TAR ≈ Rev–RRE (Fig. 3B ▶). In both assays, very low antitermination activities were observed using a control reporter with the nut site deleted.
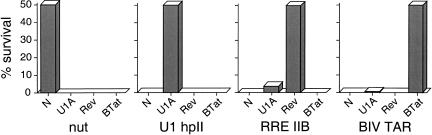
To examine the specificity of the KAN assay, we measured survival curves for N, Rev, BIV Tat, and U1A with cognate and noncognate reporters. In all cases, only the cognate interactions showed significant levels of kanamycin resistance, plotted in Figure 4 ▶ as levels of survival relative to 50% survival with the cognate interaction. U1A showed a slight background with the RRE and BIV TAR reporters, consistent with its nonspecific binding properties (Hall 1994) and activity in β-galactosidase assays (C. Escudé, unpubl.). Thus, the KAN assay can detect a variety of RNA–protein interactions with high sensitivity and specificity, and over a wide dynamic range of activities.
FIGURE 4.
Specificity of the KAN assay. Antitermination activities were measured for cognate and noncognate interactions using N-fusion proteins containing the RNA-binding domains of N, U1A, Rev, or BIV Tat, and reporter plasmids containing nut site boxB, U1 hairpin II, RRE IIB, or BIV TAR. Percent survival at kanamycin concentrations resulting in 50% survival of the cognate pairs is plotted (375 μg/mL for N/boxB, 32 μg/mL for U1A/hpII, 9 μg/mL for Rev/RRE, and 13 μg/mL for BIV Tat/TAR).
Enrichment for RNA binders using the KAN assay
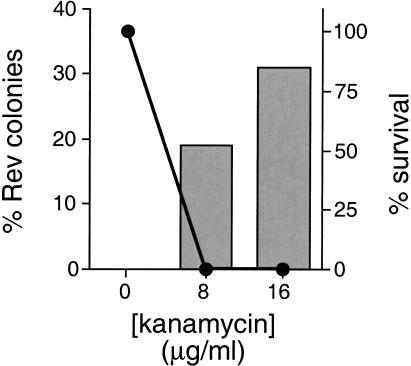
The main purposes for engineering the kanamycin reporter are to enable larger library screens than possible with LacZ and potentially to allow selection of peptides with different affinities by adjusting the kanamycin concentration. Given that LacZ assays show a good correlation between β-galactosidase levels and RNA-binding affinities measured in vitro (Harada et al. 1996), we expected that a KAN screen performed under appropriate conditions should be highly selective for RNA binders. To estimate an enrichment factor, we transformed a ‘‘mock’’ library containing a 1:1000 mixture of Rev-N:wild-type N expressor plasmids into RRE reporter cells, plated at 0, 8, and 16 μg/mL kanamycin, and analyzed surviving colonies by PCR. In the absence of kanamycin, the survival rate was considered to be 100% and 0 of 12 clones analyzed (0%) contained the Rev-N expressor. In contrast, at 8 μg/mL the survival rate was 0.2% and 3 of 16 clones analyzed (19%) contained Rev-N; at 16 μg/mL the survival rate was 0.05% and 5 of 16 clones analyzed (31%) contained Rev-N (Fig. 5 ▶). Thus, we estimate an enrichment factor of ∼300-fold (0.31/0.001) from a single round of selection under these conditions. Actual enrichment factors will depend on the RNA-binding affinities and specificities of library members and the selective conditions used; however, in practice, this value is probably near the maximum enrichment because false positives resulting from reporter plasmid rearrangements arise at a frequency of ∼0.2–0.5% (Harada et al. 1996).
FIGURE 5.
Enrichment for RRE binders from a mock library. A 1:1000 mixture of Rev-N:wild-type N expressor plasmids was transformed into RRE reporter cells and plated at 0, 8, and 16 μg/mL kanamycin. Percent survival is plotted on the right axis (solid circles). Surviving clones were analyzed by PCR to distinguish between the N and Rev expressors, and the percent of Rev expressors is plotted on the left (bars).
Screening a polyarginine-based library for RRE binders using the Lac Z reporter
We next designed a combinatorial peptide library based on the arginine-rich motif and screened for RRE binders, first at a small scale using the original LacZ reporter, and then at a large scale using the new KAN reporter. Previous studies have shown that polyarginine-based libraries can provide a good context for identifying RNA binders in which few amino acids other than arginine are used for specific RNA recognition, as for natural arginine-rich domains (Tan and Frankel 1998; Frankel 2000). We constructed a library within a framework of 15 arginines (see Materials and Methods) in which the first 10 positions randomly encoded charged and polar amino acids to allow potential hydrogen-bonding and electrostatic contacts, and alanine, glycine, and proline to allow conformational diversity. The library was fused to N with a linker of four alanines, potentially biasing the library toward α-helical conformations (Harada and Frankel 1998), as used by Rev for RRE recognition.
The polyarginine-based expressor library was transformed into RRE reporter cells, ∼105 colonies were screened for blue color on X-gal plates, and plasmids from several of the darkest blue colonies (scored as 4+) and slightly lighter colonies (scored as 3+ and comparable to Rev) were sequenced (Table 1 ▶). Clones from the dark blue colonies generally exhibited antitermination activities higher than the Rev peptide and similar to the tight binding RSG-1.2 peptide previously selected from a library (Harada et al. 1997), and all showed good RRE specificity, with low activities on a BIV TAR reporter (Table 1 ▶). Interestingly, all of the peptides contained a glutamine at the C-terminus of the randomized region, reminiscent of the tight RRE-binding glutamine-containing peptides identified in a previous library experiment in mammalian cells (Tan and Frankel 1998).
TABLE 1.
RRE-binding peptides identified from an oligoarginine-based library using the LacZ reporter
| X-gala | ONPGb | ||||||
| Clone # | Peptide sequence | RRE | (BTAR) | RRE | (BTAR) | ||
| Dark Blue (4+) | |||||||
| 1 | MA | PRRDRRRRRQ | RRRRRAAAA | 4 | (0) | 29.3 | (4.2) |
| 2 | MA | DRADRRRRRQ | RRRRRAAAV | 4 | (0) | 28.3 | (3.7) |
| 3 | MA | PRNQRRRRRQ | RRRRRAAAA | 4 | (0) | 28.2 | (3.6) |
| 4 | MA | RRRDRRRERQ | RRRRRAAAA | 3 | (0) | 21.0 | (3.4) |
| Blue (3+) | |||||||
| 5 | MA | RTRRERRHRQ | RRRRRAAAT | 3 | (0) | 18.4 | (3.3) |
| 6 | MA | GRADRRHRRQ | RRRRRAAAA | 3 | (0) | ||
| 7 | MA | PRTRRRRRRQ | RRRRRAAAA | 3 | (0) | ||
| 8 | MA | TRRRRRRNRQ | RRRRRAATS | 3 | (0) | ||
| 9 | MA | GTTQRRRTRQ | RRRRRAAAA | 3 | (0) | 12.2 | (2.8) |
| 10 | MA | TRRDRRRARQ | RRRRRAAAA | 3 | (0) | ||
| Rev | MA | TRQARRNRRRRWRRAAAA | 3 | (0) | 26.2 | (5.0) | |
| RSG-1.2 | MA | RRRRRRGSRPSGAERRRRRAAAA | 4 | (0) | 36.6 | (5.3) | |
| R6QR7c | AAAARRRRRRQRRRRRRRAAAA | ||||||
aX-gal colony assays were performed as described in Peled-Zehavi et al. (2000). Numbers represent plusses used to score blue color intensity. Values in parentheses indicate activities with the nonspecific BIV TAR reporter.
bβ-Galactosidase solution assays were performed as described in Peled-Zehavi et al. (2000). Values in parentheses indicate activities with the nonspecific BIV TAR reporter.
cThe sequence of the R6QR7 peptide identified in a mammalian screening assay (Tan and Frankel 1998) is shown for comparison but experiments with this peptide were not performed in the antitermination assay.
Screening a polyarginine-based library for RRE binders using the KAN assay
To directly compare results with the two reporters and to potentially identify even tighter RRE binders, we used the KAN reporter to more exhaustively screen the same combinatorial library as above. First, library plasmids were transformed into RRE reporter cells, yielding ∼3.2 × 107 total transformants (representing ∼1.1 × 107 independent clones; see Materials and Methods) that were plated at 5 μg/mL kanamycin. A total of 1.7 × 105 colonies (0.5%) survived. Plasmid DNA from the first round survivors was then transformed into RRE reporter cells, and cells were plated at 10 μg/mL kanamycin to further enrich for tight binders and to reduce the background from reporter-related false positives (Peled-Zehavi et al. 2000). Of ∼6.9 × 106 total transformants (representing ∼2.3 × 106 independent clones), 9.6 × 104 colonies (1.4%) survived. Plasmid DNA from the second round survivors was transformed into pAC-RRE reporter cells and plated onto X-gal plates, reasoning that proper antitermination should also result in β-galactosidase expression. Indeed, 100% of colonies that survived two rounds of kanamycin selection were also blue, suggesting that all encoded bona fide antiterminators. Finally, RRE specificity was tested for 46 of the darkest blue colonies, and all showed β-galactosidase expression in colony color assays only with the RRE (and not the BIV TAR) reporter. Sequencing of the 46 clones yielded 23 unique peptides (Table 2 ▶).
TABLE 2.
RRE-binding peptides identified from an oligoarginine-based library using the kanamycin reporter
| X-galc | ONPGd | |||||||
| Clone # | Peptide sequencea | # clonesb | RRE | (BTAR) | RRE | (BTAR) | ||
| 1 | MA | DRRQRRRDRQ | RRRRRAAAA | 3 | 6 | (0) | 153.2 | (9.4) |
| 2 | MA | DRRQRRRRRQ | RRRRRAAAA | 6 | (0) | 74.2 | (7.0) | |
| 3 | MA | DRRERRRTRQ | RRRRRAAAA | 5 | (0) | |||
| 4 | MA | DRRERRRHRQ | RRRRRAAAA | 5 | (0) | |||
| 5 | MA | DRRARRRGRQ | RRRRRAAAT | 6 | (0) | |||
| 6 | MA | NRRTRRRDRQ | RRRRRAAAA | 2 | 5 | (0) | ||
| 7 | MA | NRSARRRDRQ | RRRRRAAAA | 6 | (0) | |||
| 8 | MA | NRKERRRDRQ | RRRRRAATS | 6 | (0) | 142.3 | (10.6) | |
| 9 | MA | NRRLRRRHRQ | RRRRRAAAA | 4 | (0) | |||
| 10 | MA | ERRDRRRRRQ | RRRRRAAAA | 5 | (0) | |||
| 11 | MA | ERNRRRRERQ | RRRRRAAAA | 2 | 4 | (0) | ||
| 12 | MA | SRRDRRRERQ | RRRRRAAAA | 5 | (0) | |||
| 13 | MA | HRKQPRRERQ | RRRRRAAAA | 6 | (0) | 120.1 | (10.6) | |
| 14 | MA | PRHRRRRARQ | RLRRRAAAA | 12 | 5 | (0) | 107.4 | (9.4) |
| 15 | MA | RTNRRRRDRQ | RRRRRAAAA | 2 | 4 | (0) | ||
| 16 | MA | RTTKARRQRQ | RRRRRAAAA | 5 | (0) | 98.5 | (6.3) | |
| 17 | MA | RKERRRERQR | RRRRRAAAA | 4 | (0) | 94.9 | (10.7) | |
| 18 | MA | SQTRTARRNR | RRRRRAAAA | 2 | 5 | (0) | 80.1 | (13.2) |
| 19 | MA | SQRTRRRHRS | RRRRRAAAA | 6 | 5 | (0) | 62.9 | (7.6) |
| 20 | MA | SQRTRRRRRS | RRRRRAAAA | 4 | (0) | 45.2 | (6.8) | |
| 21 | MA | RSRRAGNERR | RRRRRAAAA | 3 | (0) | 39.4 | (4.1) | |
| 22 | MA | NRRRAGNHRR | RRRRRAAAA | 3 | (0) | 41.3 | (6.7) | |
| 23 | MA | RNPRRSGASR | RRRRRAAAA | 4 | (0) | 102.6 | (9.8) | |
| Rev | TRQARRNRRR | RWRRAAAA | 3 | (0) | 63.2 | (9.5) | ||
| RSG-1.2 | 4 | (0) | 75.7 | (7.3) | ||||
| BIV Tat | 0 | (3) | 8.3 | (44.8) | ||||
| N | 0 | (0) | 27.1 | (28.1) | ||||
aNonarginine residues are in bold and arginines with altered codons introduced by the randomized library are underlined.
bNumber of times isolated if more than once.
cNumber of plusses in the colony color assay using RRE (or BTAR) reporter cells. For comparison, N-boxB reporter = 10; Rev-boxB reporter = 0.
dAssayed in RRE (or BTAR) reporter cells. For comparison, N-boxB reporter = 1838; Rev-boxB reporter = 14.2.
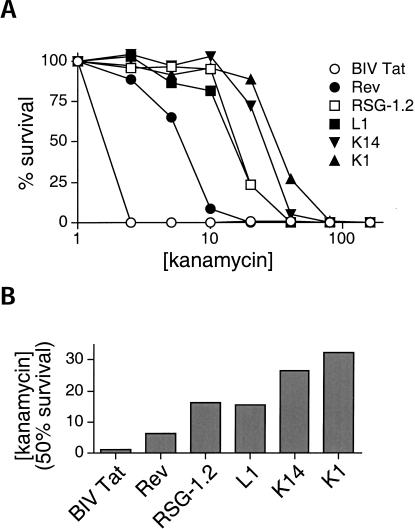
Like the LacZ screen, most of the peptides isolated in the KAN screen contained a glutamine at the C-terminus of the randomized region (clones 1–16, Table 2 ▶), further highlighting the importance of this residue for RRE binding. One sequence (clone 18) resembled Rev, with threonine, alanine, and asparagine residues, and three others (clones 21–23) contained glycines reminiscent of the previously selected RSG peptides (Harada et al. 1996). Levels of activity were quantitated with β-galactosidase colony color and solution assays, and most clones had antitermination activities substantially higher than that of the Rev peptide or even the tight binding RSG-1.2 peptide (Table 2 ▶), and virtually all were higher than the peptides isolated from the LacZ screen (Table 1 ▶). Kanamycin survival curves were measured for the KAN-selected peptide with the highest activity (clone 1) and for one isolated several times from the library (clone 14), and both showed substantially better survival than Rev, RSG-1.2, or the tightest binder selected from the LacZ screen (Fig. 6A ▶). The best peptide (clone 1) showed 50% survival at 32 μg/mL kanamycin, compared to 6 μg/mL for Rev and 16 μg/mL for RSG-1.2 (Fig. 6B ▶). Thus, peptides selected using the KAN reporter system showed high RRE-binding activities and specificities.
FIGURE 6.
KAN assays of selected RRE-binding peptides. (A) Survival curves are shown for N-fusions to Rev, RSG-1.2, L1 (clone #1 from the LacZ screen; Table 1 ▶), K1 and K14 (clones #1 and 14 from the kanamycin screen; Table 2 ▶), and BIV Tat (negative control) in RRE reporter cells. (B) Kanamycin concentrations resulting in 50% survival of the N-fusions in (A) (1 μg/mL for BIV Tat, 6 μg/mL for Rev, 16 μg/mL for RSG-1.2, 15 μg/mL for L1, 27 μg/mL for K14, and 32 μg/mL for K1).
DISCUSSION
We have described the development and utilization of a modified λ N antitermination system to identify RNA-binding peptides from large combinatorial libraries. The use of a kanamycin antitermination reporter (the KAN reporter) has increased screening capacity from ∼106 to ∼109 clones compared to the previous LacZ reporter (Harada et al. 1996), enabling much wider searching of sequence space. With this extra capacity, we have identified new glutamine-containing RRE-binding peptides with substantially higher activities than those isolated from smaller scale screens, even after additional rounds of peptide evolution (Harada et al. 1997). Our libraries so far have been restricted to arginine-rich peptides with 12 types of amino acids that exclude hydrophobic residues (Tan and Frankel 1998), but the KAN reporter now makes less biased screens possible. The KAN assay shows high specificity for cognate RNA–protein interactions, and indeed, all clones isolated from our library after just two rounds of kanamycin selection were RRE-specific. One attractive feature of the system is the ability to adjust kanamycin concentrations during selection, resulting in the identification of higher affinity RRE-binders in the experiment described.
A variety of other methods to screen RNA-binding libraries have been described (Laird-Offringa and Belasco 1995; Jain and Belasco 1996; Kollmus et al. 1996; SenGupta et al. 1996; Wang et al. 1997; Paraskeva et al. 1998; Tan and Frankel 1998; Barrick et al. 2001), each with advantages and disadvantages. The in vitro mRNA–peptide fusion system has been especially useful for studying very large peptide libraries (∼1013 sequences) and has been used to isolate variants of the λ N arginine-rich peptide that bind specifically to boxB loop variants (Barrick et al. 2001). The yeast three-hybrid system is particularly well suited for identifying RNA-binding proteins from cDNA libraries (SenGupta et al. 1996; Bernstein et al. 2002). The KAN system also may be suitable for cDNA screens or analysis of RNA-binding domains other than arginine-rich peptides, as it functions with N fused to the MS2 coat protein (Wilhelm and Vale 1996), zinc fingers (McColl et al. 1999), or the U1A RNP domain (Figs. 3, 4 ▶ ▶). We have not yet found limitations to the length or nature of the fused domains, except that the type of amino acid linker can affect activity in some contexts (Harada and Frankel 1998). The restrictions on the RNA target site may be more severe. Although short hairpin elements generally function well in place of boxB, larger RNAs, such as the full-length 250-nt RRE, or altered arrangements of BIV TAR do not support antitermination (data not shown), perhaps because the spatial arrangement or cooperative assembly of antitermination complexes is compromised (Greenblatt et al. 1993; Mogridge et al. 1995). The spacing between box A and the target site also can affect the level of antitermination (Peled-Zehavi et al. 2000). Despite these limitations, the antitermination system is flexible enough to support a variety of RNA hairpins, and is expected to be useful in further design efforts and structure/function analyses of RNA–protein complexes.
Arginine-rich RNA-binding peptides have provided good model systems for understanding the molecular basis of RNA–protein specificity, and peptides identified by selection have provided interesting contrasts to their natural counterparts, particularly for RRE recognition. The RNA-binding domain of Rev binds to the RRE as a monomeric α-helix, with an asparagine forming hydrogen bonds to a critical G:A base pair and three arginines forming hydrogen bonds to guanines (Battiste et al. 1996; Ye et al. 1996). In contrast, the selected RSG-1.2 peptide, identified from a combinatorial library and evolved through two rounds of mutagenesis and selection in the N system (Harada et al. 1997), uses a rather different recognition strategy. In this case, the RRE adopts a structure similar to the Rev-bound form, but the peptide forms a helix-turn-extended conformation and makes quite different RRE contacts, including hydrophobic and stacking interactions between the RNA and alanine and arginine side chains (Gosser et al. 2001; Zhang et al. 2001). These contacts presumably contribute to the ∼sevenfold higher affinity and ∼15-fold higher specificity of RSG-1.2 compared to Rev. R6QR7, another selected peptide identified from a low-complexity polyarginine-based library, contains a single glutamine residue embedded within a stretch of arginines and binds with ∼fivefold higher affinity than Rev (Tan and Frankel 1998). The structure of the R6QR7–RRE complex is not yet known, but CD data indicate that the peptide, like Rev, binds in a helical conformation. R6QR7 is presumed to make an analogous glutamine-G:A contact as Rev, perhaps with better positioning of the helix within the RNA major groove. We imagine a similar situation for the even higher affinity glutamine-containing peptides identified here. The abundance of acidic residues in these peptides, particularly at the first randomized position (Table 2 ▶), suggests the possibility of salt bridges to arginines that may stabilize the helical conformation (Olson et al. 2001). Structural studies are needed to determine the basis for the enhanced binding affinity.
Our initial results with the KAN system suggest that it will be a powerful addition to the existing methods for screening RNA-binding libraries, particularly for peptides. It will be of great interest to determine what types of RNA targets, beyond the RRE, and what types of peptide motifs, beyond the arginine-rich motif, are amenable to such screens. The diversity of RNA structure and the ability of peptides to adapt to different binding pockets provides an opportunity to explore the evolution of peptide sequences and structures in an RNA context, perhaps as natural evolution did when proteins evolved from an RNA world.
MATERIALS AND METHODS
Reporter and expressor plasmids
The neomycin phosphotransferase gene (NPT II) encoding kanamycin resistance was amplified by PCR from pEGFP-1 (Clontech) and cloned into the unique Bgl II site located between the transcription terminators and lacZ gene of the previously described pAC-nut- reporter plasmid (Franklin 1993; Harada et al. 1996), generating the pAC(kan)-nut- reporter. Forward (5′-GCA TGCAGATCTAAGGAGGCGTTTCGCATGATTGAACAA-3′) and reverse (5′-GCAGTCAGATCTAGTCCCGCTCAGAAGAAC-3′) PCR primers were used, with the Bgl II site underlined.
Three nonessential regions of the pAC(kan)-nut- reporter, or regions that reduced reporter function (see Results), were deleted. First, the phoA gene was removed by digestion with Kpn I and Sph I, replacing the 1.36-kb fragment with a synthetic linker (5′-CCAC ATGGAGAAAATAAAGTGAAACAAAGCACTATTGCACTGGCAT CTCTTACCGTTACTGGCATG-3′). Second, a duplicated segment just downstream of the lacZ gene was removed by digestion with Blp I and Nar I and replaced with a synthetic linker (5′-TGAGCGCCGGTCGCTACCATTACCAGTTGGTCTGGTGTCAA AAATAATAATAACCGGGTAGG-3′) that maintained the C-terminal residues of LacZ. Third, another duplicated segment upstream of the Ptac promoter was removed by digestion with Bgl II and Nde I, further digestion of a subfragment with Apa LI, which excised the duplicated region, and religation of the plasmid. The resulting reporter, pACK-nut-, is nearly 3 kb shorter then the pAC(kan)-nut- plasmid (see Fig. 1 ▶).
To generate the RNA reporters, DNA fragments encoding boxA of the nut site and boxB, RRE IIB, or BIV TAR were cloned into the Pst I and Bam HI sites of pACK-nut- or pAC(kan)-nut- from previously described reporters (Fig. 1 ▶) (Harada et al. 1996). The U1 hairpin II was similarly cloned using a synthetic oligonucleotide (5′-GGTCGACGCTCTTAAAAATTAAGGCCTCATTGCACT CCGAGGCCAGCATTCAAAGCAGG-3′).
Fusions between the arginine-rich domains of HIV Rev or BIV Tat and the λ N protein were generated by cloning oligonucleotides into the Nco I and Bsm I sites of the pBR-based N expressor plasmid (Franklin 1993) as previously described (Fig. 1 ▶) (Harada et al. 1996). A U1A-N fusion was constructed by cloning a corresponding Nco I-Bsm I PCR fragment encoding U1A residues 1–102 from a Tat–U1A expression plasmid (Tan and Frankel 1998).
Kanamycin antitermination (KAN) assay
Competent Escherichia coli N567 host cells (Doelling and Franklin 1989) containing different RNA reporter plasmids were prepared by standard CaCl2 procedures and were transformed with appropriate pBR expressor plasmids (typically 10 ng per 100 μL competent cells) by heat shock. Tryptone medium (1 mL per 100-μL competent cells) was added and cells were allowed to recover by incubating at room temperature for 6 h on a rotating shaker (40 rpm). IPTG was added after 4 h of recovery (to 0.2 mM) to induce expression from the tac promoters. Cells were then plated onto tryptone plates containing ampicillin (50 μg/mL), chloramphenicol (20 μg/mL), IPTG (0.05 mM), and an appropriate amount of kanamycin and plates were incubated at room temperature for 3–5 d. Incubation at room temperature was found to be critical for cell survival. Colonies on low kanamycin concentration plates (0–8 μg/mL) were counted after 3 d, and colonies on high kanamycin concentration plates (16 μg/mL and higher), which typically grow slower, were counted after 5 d. For library screens, expressor plasmids were electroporated into competent N567/pACK-RRE cells as described below, and recovery of cells and plating were performed as above.
Monitoring kanamycin resistance with the Kirby-Bauer filter assay
E. coli N567 cells containing both expressor and reporter plasmids were obtained by heat-shock transformation of competent reporter cells (100 μL) with 10 ng expressor plasmids, incubating at 37°C for 1 h in 1 mL tryptone broth, and plating 100 μL onto tryptone plates containing ampicillin (50 μg/mL) and chloramphenicol (20 μg/mL). When colonies reached ∼2–3 mm in diameter (typically after incubating overnight at 37°C), single colonies were inoculated into 2 mL tryptone/ampicillin/chloramphenicol and cultures were incubated at 37°C until reaching O.D.600 of ∼0.4 (∼4 h). Cultures were then diluted with an additional 3 mL tryptone/ampicillin/chloramphenicol and incubated at room temperature for 6 h on a rotating shaker (reaching O.D.600 of ∼0.8). IPTG was added to cultures after 4 h as described above. Cells were plated on 15-cm diameter tryptone plates containing ampicillin, chloramphenicol, and IPTG (0.05 mM). To ensure constant colony densities, the volumes plated (∼200–250 μL) were adjusted according to culture density (180 ÷ O.D.600 μL). After 10 min, Whatman 3-mm filter discs, cut with a hole punch and impregnated with varying amounts of kanamycin (0, 1, 2, 4, 8, 16, 32, 64, 128, and 256 μg), were placed onto the surface of the plates (Baker and Breach 1980). Plates were incubated at room temperature for 18–20 h, resulting in bacterial lawns with clear zones of inhibition surrounding the kanamycin discs. Diameters of the inhibition zones were measured and total areas were calculated. Kanamycin survival was defined as 100% when total area equaled the area of the filter disc, that is, there was no visible inhibition zone. Percent survival was calculated using a negative exponential equation [% survival = e-(total area − disc area)/disc area * 100], and was plotted as a function of the amount of impregnated kanamycin to generate survival curves.
Combinatorial library design
A combinatorial peptide library was expressed as a fusion to the λ N protein, replacing the RNA-binding domain of N (residues 1–19). An oligonucleotide library was synthesized using a codon-based mutagenesis procedure as previously described (Harada et al. 1997), with a prototype sequence encoding 15 arginines (5′-GA ATCCCCATGGCCCGTCGCCGTCGCCGTCGCCGACGTCGCC GGCGTCGACGTCGCCGTGCAGCTGCTGCGAATGCAGCAAA TCC-3′) and a randomized sequence (5′-GAATCCCCATGGC CVVKVVKVVKVVKVVKVVKVVKVVKVVKVVKCGTCGACGT CGCCGTGCAGCTGCTGCGAATGCAGCAAATCC-3′), where V is an equimolar mixture of A, C, and G, and K is an equimolar mixture of G and T. The VVK codons primarily encode amino acids with charged, polar, and small side chains but no large hydrophobic side chains. The library was synthesized at a 1:1 ratio of arginine:VVK codons and thus is expected to contain an average of ∼5 nonarginine residues per peptide. Assuming a Poisson distribution, it should be possible to sample nearly all sequences with three nonarginine codons by screening 107 clones, ∼10% of sequences with four nonarginine codons, and ∼1% of sequences with five nonarginine codons (Harada and Frankel 1998).
Selection of RRE-binding peptides
Four screening steps were employed to identify RRE-specific binders using the KAN assay. For the primary screen, the synthetic library oligonucleotide was converted to double-stranded DNA using Sequenase 2.0 polymerase (USB), and 0.3 μg was digested with Nco I and Bsm I and ligated into 2.6 μg pBRN-expressor plasmid. Ligation mixtures were phenol extracted, ethanol precipitated using sodium acetate, and redissolved in 8 μL H2O. Plasmids were electroporated into N567/pACK-RRE cells (7 × 80 μL) in 1-mm cuvettes at 2.0 kV using 1 μL of the above solution per electroporation, and 5 mL SOC medium was added immediately after electroporation. Cells were allowed to recover for 6 h at room temperature as described above and were spread onto tryptone plates containing ampicillin, chloramphenicol, IPTG, and kanamycin (5 μg/mL) and incubated at room temperature. For the screen, ∼3.2 × 107 total transformants were plated, representing ∼1.1 × 107 independent clones as estimated by plating cells before the 6 h recovery period on plates lacking kanamycin. After 3 d on the kanamycin plates, surviving colonies were scraped off and plasmid DNA was isolated. Because both the pACK-RRE and pBR library plasmids were present, an EcoR V digestion was performed to selectively digest pACK-RRE. Following phenol extraction and ethanol precipitation, the DNA was redissolved in 10 μL H2O.
For the secondary screen, kanamycin selection was repeated by electroporating 2 μL plasmid DNA (∼80 ng) into 80 μL N567/pACK-RRE cells three times, spreading on 10 μg/mL kanamycin plates, scraping colonies, and isolating plasmid DNA as above. Following EcoR V digestion, the pBR plasmid DNA was redissolved in 200 μL H2O (8 ng/μL).
For the tertiary screen, 1 μL pBR plasmid DNA was electroporated into 80 μL N567/pAC-RRE cells, 5 mL SOC was added, and cultures were incubated in the absence of kanamycin at 37°C for 1 h. A portion of the culture was spread onto X-gal plates, incubated at 37°C for 48 h as previously described (Harada et al. 1996), and the fraction of blue colonies was scored.
For the quaternary screen, RNA-binding specificities of 46 of the darkest blue colonies were assessed. pBR plasmid DNAs were isolated and activities were monitored using the LacZ colony color assay (Harada and Frankel 1998) with N567 reporter cells containing pAC-RRE or pAC-BTAR plasmids. For the tertiary and quaternary screening steps, we used LacZ reporters without the kan gene because the corresponding pACK plasmids typically show slightly higher background levels of β-galactosidase.
Acknowledgments
We thank Colin Smith, Christophe Escudé, Anson Nomura, Bobby Otillar, Shelley Martin, and Lily Chen for help in optimizing the selection conditions and for helpful suggestions; Ami Bhatt for helpful discussions; and Valerie Calabro for comments on the manuscript. This work was supported by grants from the NIH (A.D.F.) and by a Grant-in-Aid for Scientific Research in Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (K.H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2152303.
REFERENCES
- Ansel-McKinney, P., Scott, S.W., Swanson, M., Ge, X., and Gehrke, L. 1996. A plant viral coat protein RNA binding consensus sequence contains a crucial arginine. EMBO J. 15: 5077–5084. [PMC free article] [PubMed] [Google Scholar]
- Baker, F.J. and Breach, M.R. 1980. Medical microbiological techniques. Butterworth Inc., Boston.
- Barrick, J.E., Takahashi, T.T., Ren, J., Xia, T., and Roberts, R.W. 2001. Large libraries reveal diverse solutions to an RNA recognition problem. Proc. Natl. Acad. Sci. 98: 12374–12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste, J.L., Mao, H., Rao, N.S., Tan, R., Muhandiram, D.R., Kay, L.E., Frankel, A.D., and Williamson, J.R. 1996. α helix major groove recognition in an HIV-1 Rev peptide–RRE RNA complex. Science 273: 1547–1551. [DOI] [PubMed] [Google Scholar]
- Bernstein, D.S., Buter, N., Stumpf, C., and Wickens, M. 2002. Analyzing mRNA–protein complexes using a yeast three-hybrid system. Methods 26: 123–141. [DOI] [PubMed] [Google Scholar]
- Cai, Z., Gorin, A., Frederick, R., Ye, X., Hu, W., Majumdar, A., Kettani, A., and Patel, D.J. 1998. Solution structure of P22 transcriptional antitermination N peptide–box B RNA complex. Nat. Struct. Biol. 5: 203–212. [DOI] [PubMed] [Google Scholar]
- Calnan, B.J., Tidor, B., Biancalana, S., Hudson, D., and Frankel, A.D. 1991. Arginine-mediated RNA recognition: The arginine fork. Science 252: 1167–1171. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay, S., Garcia-Mena, J., DeVito, J., Wolska, K., and Das, A. 1995. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage lambda. Proc. Natl. Acad. Sci. 92: 4061–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. and Frankel, A.D. 1994. An RNA-binding peptide from bovine immunodeficiency virus Tat protein recognizes an unusual RNA structure. Biochemistry 33: 2708–2715. [DOI] [PubMed] [Google Scholar]
- ———. 1995. A peptide interaction in the major groove of RNA resembles protein interactions in the minor groove of DNA. Proc. Natl. Acad. Sci. 92: 5077–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, A.C., Calabro, V., and Frankel, A.D. 2001. Design of RNA-binding proteins and ligands. Curr. Opin. Struct. Biol. 11: 478–484. [DOI] [PubMed] [Google Scholar]
- Doelling, J.H. and Franklin, N.C. 1989. Effects of all single base substitutions in the loop of boxB on antitermination of transcription by bacteriophage lambda’s N protein. Nucleic Acids Res. 17: 5565–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, D.E. 1999. Themes in RNA–protein recognition. J. Mol. Biol. 293: 255–270. [DOI] [PubMed] [Google Scholar]
- Frankel, A.D. 2000. Fitting peptides into the RNA world. Curr. Opin. Struct. Biol. 10: 332–340. [DOI] [PubMed] [Google Scholar]
- Frankel, A.D. and Smith, C.A. 1998. Induced folding in RNA–protein recognition: More than a simple molecular handshake. Cell 92: 149–151. [DOI] [PubMed] [Google Scholar]
- Franklin, N.C. 1993. Clustered arginine residues of bacteriophage lambda N protein are essential to antitermination of transcription, but their locale cannot compensate for boxB loop defects. J. Mol. Biol. 231: 343–360. [DOI] [PubMed] [Google Scholar]
- Gosser, Y., Hermann, T., Majumdar, A., Hu, W., Frederick, R., Jiang, F., Xu, W., and Patel, D.J. 2001. Peptide-triggered conformational switch in HIV-1 RRE RNA complexes. Nat. Struct. Biol. 8: 146–150. [DOI] [PubMed] [Google Scholar]
- Greenblatt, J., Nodwell, J.R., and Mason, S.W. 1993. Transcriptional antitermination. Nature 364: 401–406. [DOI] [PubMed] [Google Scholar]
- Hall, K.B. 1994. Interaction of RNA hairpins with the human U1A N-terminal RNA binding domain. Biochemistry 33: 10076–10088. [DOI] [PubMed] [Google Scholar]
- Harada, K. and Frankel, A.D. 1998. In RNA:protein interactions: A practical approach (ed. C.W.J. Smith), pp. 217–236. Oxford University Press, Oxford.
- Harada, K., Martin, S.S., and Frankel, A.D. 1996. Selection of RNA-binding peptides in vivo. Nature 380: 175–179. [DOI] [PubMed] [Google Scholar]
- Harada, K., Martin, S.S., Tan, R., and Frankel, A.D. 1997. Molding a peptide into an RNA site by in vivo peptide evolution. Proc. Natl. Acad. Sci. 94: 11887–11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, T. 2000. Strategies for the design of drugs targeting RNA and RNA–protein complexes. Angew. Chem. Int. Ed. 39: 1890–1904. [DOI] [PubMed] [Google Scholar]
- Hope, T.J. 1999. The ins and outs of Rev. Arch. Biochem. Biophys. 365: 186–191. [DOI] [PubMed] [Google Scholar]
- Jain, C. and Belasco, J.G. 1996. A structural model for the HIV-1 Rev–RRE complex deduced from altered-specificity rev variants isolated by a rapid genetic strategy. Cell 87: 115–125. [DOI] [PubMed] [Google Scholar]
- Kjems, J., Calnan, B.J., Frankel, A.D., and Sharp, P.A. 1992. Specific binding of a basic peptide from HIV-1 Rev. EMBO J. 11: 1119–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmus, H., Hentze, M.W., and Hauser, H. 1996. Regulated ribosomal frameshifting by an RNA–protein interaction. RNA 2: 316–323. [PMC free article] [PubMed] [Google Scholar]
- Laird-Offringa, I.A. and Belasco, J.G. 1995. Analysis of RNA-binding proteins by in vitro genetic selection: Identification of an amino acid residue important for locking U1A onto its RNA target. Proc. Natl. Acad. Sci. 92: 11859–11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt, S.G., Tan, R., and Frankel, A.D. 2000. Screening RNA-binding libraries using Tat-fusion system in mammalian cells. Methods Enzymol. 318: 350–363. [DOI] [PubMed] [Google Scholar]
- Legault, P., Li, J., Mogridge, J., Kay, L.E., and Greenblatt, J. 1998. NMR structure of the bacteriophage lambda N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell 93: 289–299. [DOI] [PubMed] [Google Scholar]
- Mason, S.W., Li, J., and Greenblatt, J. 1992. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J. Biol. Chem. 267: 19418–19426. [PubMed] [Google Scholar]
- McColl, D.J., Honchell, C.D., and Frankel, A.D. 1999. Structure-based design of an RNA-binding zinc finger. Proc. Natl. Acad. Sci. 96: 9521–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogridge, J., Legault, P., Li, J., Van Oene, M.D., Kay, L.E., and Greenblatt, J. 1998. Independent ligand-induced folding of the RNA-binding domain and two functionally distinct antitermination regions in the phage lambda N protein. Mol. Cell 1: 265–275. [DOI] [PubMed] [Google Scholar]
- Mogridge, J., Mah, T.-F., and Greenblatt, J. 1995. A protein–RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the λ N protein. Genes & Dev. 9: 2831–2844. [DOI] [PubMed] [Google Scholar]
- Olson, C.A., Spek, A.J., Shi, Z., Vologodskii, A., and Kallenbach, N.R. 2001. Cooperative helix stabilization by complex Arg-Glu salt bridges. Proteins 44: 123–132. [DOI] [PubMed] [Google Scholar]
- Oubridge, C., Ito, N., Evans, P.R., Teo, C.H., and Nagai, K. 1994. Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372: 432–438. [DOI] [PubMed] [Google Scholar]
- Paraskeva, E., Atzberger, A., and Hentze, M.W. 1998. A translational repression assay procedure (TRAP) for RNA–protein interactions in vivo. Proc. Natl. Acad. Sci. 95: 951–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, D.J. 1999. Adaptive recognition in RNA complexes with peptides and protein modules. Curr. Opin. Struct. Biol. 9: 75–87. [DOI] [PubMed] [Google Scholar]
- Peled-Zehavi, H., Smith, C.A., Harada, K., and Frankel, A.D. 2000. Screening RNA-binding libraries by transcriptional antitermination in bacteria. Methods Enzymol. 318: 297–308. [DOI] [PubMed] [Google Scholar]
- Perez-Canadillas, J.M. and Varani, G. 2001. Recent advances in RNA–protein recognition. Curr. Opin. Struct. Biol. 11: 53–58. [DOI] [PubMed] [Google Scholar]
- Pollard, V.W. and Malim, M.H. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52: 491–532. [DOI] [PubMed] [Google Scholar]
- Puglisi, J.D., Chen, L., Blanchard, S., and Frankel, A.D. 1995. Solution structure of a bovine immunodeficiency virus Tat-TAR peptide–RNA complex. Science 270: 1200–1203. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan, V. 2002. Ribosome structure and the mechanism of translation. Cell 108: 557–572. [DOI] [PubMed] [Google Scholar]
- Rees, W.A., Weitzel, S.E., Yager, T.D., Das, A., and von Hippel, P.H. 1996. Bacteriophage lambda N protein alone can induce transcription antitermination in vitro. Proc. Natl. Acad. Sci. 93: 342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta, D.J., Zhang, B., Kraemer, B., Pochart, P., Fields, S., and Wickens, M. 1996. A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Natl. Acad. Sci. 93: 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, M.M., Ansel-McKinney, P., Houser-Scott, F., Yusibov, V., Loesch-Fries, L.S., and Gehrke, L. 1998. Viral coat protein peptides with limited sequence homology bind similar domains of alfalfa mosaic virus and tobacco streak virus RNAs. J. Virol. 72: 3227–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, R. and Frankel, A.D. 1994. Costabilization of peptide and RNA structure in an HIV Rev peptide–RRE complex. Biochemistry 33: 14579–14585. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Structural variety of arginine-rich RNA-binding peptides. Proc. Natl. Acad. Sci. 92: 5282–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998. A novel glutamine–RNA interaction identified by screening libraries in mammalian cells. Proc. Natl. Acad. Sci. 95: 4247–4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, R., Chen, L., Buettner, J.A., Hudson, D., and Frankel, A.D. 1993. RNA recognition by an isolated α helix. Cell 73: 1031–1040. [DOI] [PubMed] [Google Scholar]
- Wang, S., True, H.L., Seitz, E.M., Bennett, K.A., Fouts, D.E., Gardner, J.F., and Celander, D.W. 1997. Direct genetic selection of two classes of R17/MS2 coat proteins with altered capsid assembly properties and expanded RNA-binding activities. Nucleic Acids Res. 25: 1649–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks, K.M. and Crothers, D.M. 1991. RNA recognition by Tat-derived peptides: Interaction in the major groove? Cell 66: 577–588. [DOI] [PubMed] [Google Scholar]
- Whalen, W., Ghosh, B., and Das, A. 1988. NusA protein is necessary and sufficient in vitro for phage lambda N gene product to suppress a rho-independent terminator placed downstream of nutL. Proc. Natl. Acad. Sci. 85: 2494–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm, J.E. and Vale, R.D. 1996. A one-hybrid system for detecting RNA–protein interactions. Genes Cells 1: 317–323. [DOI] [PubMed] [Google Scholar]
- Ye, X., Kumar, R.A., and Patel, D.J. 1995. Molecular recognition in the bovine immunodeficiency virus Tat peptide–TAR RNA complex. Chem. Biol. 2: 827–840. [DOI] [PubMed] [Google Scholar]
- Ye, X., Gorin, A., Ellington, A.D., and Patel, D.J. 1996. Deep penetration of an α-helix into a widened RNA major groove in the HIV-1 rev peptide–RNA aptamer complex. Nat. Struct. Biol. 3: 1026–1033. [DOI] [PubMed] [Google Scholar]
- Ye, X., Gorin, A., Frederick, R., Hu, W., Majumdar, A., Xu, W., McLendon, G., Ellington, A., and Patel, D.J. 1999. RNA architecture dictates the conformations of a bound peptide. Chem. Biol. 6: 657–669. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Harada, K., Cho, H.S., Frankel, A.D., and Wemmer, D.E. 2001. Structural characterization of the complex of the Rev response element RNA with a selected peptide. Chem. Biol. 8: 511–520. [DOI] [PubMed] [Google Scholar]