Abstract
The basic mechanism of uridine insertion/deletion RNA editing in mitochondria of kinetoplastid protists has been established for some time but the molecular details remained largely unknown. Recently, there has been significant progress in defining the molecular components of the editing reaction. A number of factors have been isolated from trypanosome mitochondria, some of which have been definitely implicated in the uridine insertion/deletion RNA editing reaction and others of which have been circumstantially implicated. Several protein complexes have been isolated which exhibit some editing activities, and the macromolecular organization of these complexes is being analyzed. In addition, there have been several important technical advances in the in vitro analysis of editing. In this review we critically examine the various factors and complexes proposed to be involved in RNA editing.
INTRODUCTION
The RNA modification phenomenon of uridine (U) insertion/deletion RNA editing was discovered in mitochondria of trypanosomatid protists more than 15 years ago (Benne et al. 1986). This was only one of a plethora of RNA modification phenomena that were found to occur in both single-celled and metazoan organisms. These phenomena can be grouped into two basic classes—insertion/deletion editing and substitution editing. The term RNA editing (Benne et al. 1986) has been historically limited to certain RNA nucleotide modifications discovered after 1986, such as C to U editing of the mammalian apoB mRNA and plant mitochondria and chloroplast mRNAs, A to I editing of glutamate receptors mRNAs (Gott and Emerson 2000), C-insertion editing of Physarum mitochondrial mRNAs, and even the cotranscriptional stuttering, leading to insertion of nonencoded G’s in certain negative strand viruses (Vidal et al. 1990), but clearly the term could logically encompass all types of specific nucleotide modifications in RNAs (Rozenski et al. 1999). The issue of terminology is political and not scientific.
The site specificity and the mechanism is not entirely understood in all cases, but in the trypanosomatids it was shown that specificity is determined by complementary guide RNAs acting in trans, or in one case by foldback in cis, to guide enzyme complexes to the site of U-insertion or deletion by base pairing (Blum et al. 1990). Although this specific type of RNA-mediated RNA modification is limited to the kinetoplastid protist lineage, a multitude of unrelated specific nucleotide modifications in RNA molecules have been reported that are mediated by RNA–RNA base pairing either in trans or in cis. For example, specific methylations and pseudouridylations in rRNAs are mediated by complementary snoRNAs guiding the appropriate enzymes to specific nucleotides (Lafontaine and Tollervay 1998), specific A to I modifications in mRNAs by the ADAR family of deaminases are determined by foldback of complementary downstream intronic sequences (Higuchi et al. 1993), and specific cleavages of mRNAs by RNase III-type enzymes, such as Dicer, are mediated by annealing of small siRNAs produced by cleavage of double-stranded RNAs that initiate the process of RNA interference (Bernstein et al. 2001).
Mechanism
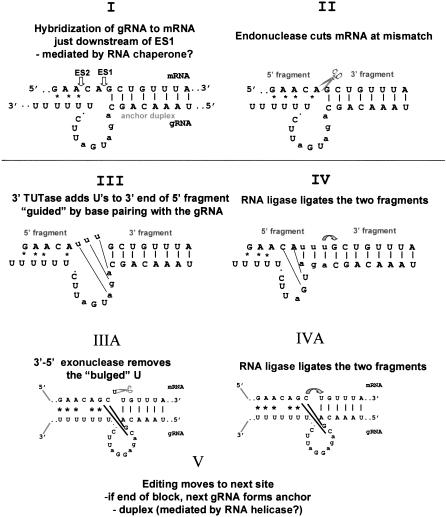
The enzymatic cleavage-ligation mechanism of U-insertion/deletion editing was proposed more than a decade ago (Blum et al. 1990; Fig. 1 ▶), but it took several years to establish the validity of this model by experimental evidence and to disprove the alternative transesterification model (Blum et al. 1991; Cech 1991). This was accomplished by the development of several in vitro editing systems that reproduced portions of the editing process, although very inefficiently, and could test predictions of the model (Seiwert and Stuart 1994, Piller et al. 1995; Byrne et al. 1996; Cruz-Reyes and Sollner-Webb 1996; Kable et al. 1996; Seiwart et al. 1996; Kapushoc and Simpson 1999). Until very recently, however, little was known in detail about the proteins and enzymes involved in this process and their interactions, but this is rapidly changing due to the availability of Leishmania major and Trypanosoma brucei genomic sequences and rapid gene identification techniques such as mass spectroscopy.
FIGURE 1.
The enzyme cascade model for RNA editing. (I–IV) U-insertion editing. (IIIA,IVA) U-deletion editing.
Since our last review of this field in 1999 (Estévez and Simpson 1999), much progress has been made on the mechanism of editing. Several studies raised the intriguing possibility that U-insertion and U-deletion editing may have different regulatory or even enzymatic mechanisms (Cruz-Reyes et al. 1998a, 2002; Huang et al. 2001). The initial observation was that U-deletion editing in vitro using a partially purified editing fraction from T. brucei mitochondria is activated by ATP and ADP, whereas U-insertion in vitro editing is inhibited by these nucleotides (Cruz-Reyes et al. 1998a). The effect appears to be on the initial gRNA-determined cleavage by some type of allosteric mechanism that is not yet understood, and also due to different properties of the two RNA ligases.
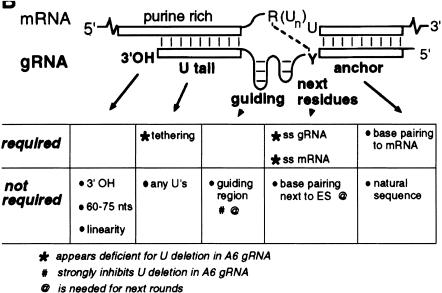
Specific sequence changes in the mRNA and gRNA were also found to differentially affect in vitro U-insertion and U-deletion editing. The gRNA features critical for U-deletion included a single-stranded region of a few adjacent nucleotides and a short upstream tether. These mutations increased the efficiency of in vitro U-deletion activity up to 100-fold (Cruz-Reyes et al. 2001). Requirements for efficient in vitro editing are indicated graphically in Figure 2 ▶.
FIGURE 2.
Guide RNA attributes important for a U-deletion cycle. Characteristic features of gRNAs for U-deletion sites are illustrated. Those shown to be important or not for U deletion are indicated. Modified from Cruz-Reyes et al. (2001).
An important technical advance for in vitro editing was reported, in which the requirement for an initial endonuclease cleavage event was bypassed by supplying an RNA substrate consisting of two mRNA fragments bridged by a gRNA (Igo et al. 2000). The efficiency of correctly edited ligated product in this assay is much greater than that achieved with the editing assay that requires a cleavage. This assay allowed Igo et al. (2002b) to examine the effects of sequence modifications on U-insertion editing. U’s were added to the 5′ cleavage fragment (the upstream fragment of the pre-edited mRNA, which would be produced by the cleavage at the first editing site) adjacent to guiding pyrimidines, but guiding purines were required for efficient ligation. This is not surprising, considering the gRNA-dependent specific addition of U’s by the recombinant 121 kD 3′ TUTase (Aphasizhev et al. 2002b), but it suggests that the specificity of the ligation step may determine the precise number of U’s in the final product by base pairing with guiding A’s or G’s. The propensity of one or both of the RNA ligases to seal nicked, rather than gapped or bulged bridged substrates was shown previously using a partially purified enzyme preparation, probably containing both REL1 and REL2 (Blanc et al. 1999). The advantage of an upstream duplex tether (an RNA–RNA duplex formed by the 3′ portion of the synthetic gRNA and the 5′ portion of the pre-edited mRNA) for efficient U-insertion editing was also demonstrated (Igo et al. 2002b), as had been reported previously for the Leishmania tarentolae system (Kapushoc and Simpson 1999).
An in vivo interaction of the 3′(U) tail of the gRNA with the cognate pre-edited or partially edited mRNA had been predicted as a mechanism to tether the 5′ cleavage fragment in the editing complex. But in vitro studies in which the oligo(U) tail was removed without affecting editing had cast some doubt on this hypothesis. Leung and Koslowsky (2001) attached a 3′ APA photoactivatable group on several Cyb mRNA substrates and cross-linked the APA–gRNA to the mRNA with UV irradiation. The gRNA tail cross-linked to an 11-nucleotide purine-rich sequence that is 5′ of and within the pre-edited region. In addition, chemical and enzymatic structural probing was used to determine single-stranded and duplex regions. When these constraints were placed on the gRNA–mRNA computer folding, similar secondary structures were obtained, each with three duplex regions – the anchor, the guiding nucleotides of the gRNA, and the oligo(U) tail with the mRNA. These folds are remarkably similar to those predicted previously (Blum and Simpson 1990). To reconcile these cross-linking results with the in vitro editing results, it is possible that the formation of a 3′ tether in vivo is mainly due to protein-mediated RNA–RNA interactions, but this remains to be investigated.
Several studies addressed further the L. tarentolae in vitro editing system. It was found previously that this system was much less efficient than the T. brucei one, requiring an RT–PCR step in order to obtain a primer extension signal, and also had several apparent differences such as a gRNA-independent U-insertion activity and a background of 1–15 U-insertions in addition to a major gRNA-determined number of insertions (Byrne et al. 1996). Byrne et al. (1999) and Oppegard et al. (Kabb et al. 2001) reported that a 34-nucleotide AU-rich sequence just upstream of the editing sites in the Cyb mRNA induced gRNA-independent U-insertions to sites both 5′ and 3′ of this sequence, and also inhibited gRNA-directed U-insertions, raising the interesting idea of negative determinants for the specific editing reactions. At least one protein specifically interacted with this sequence and did not react with a mutated sequence that was inactive in stimulating gRNA-independent editing. The protein has not yet been identified.
Complexes
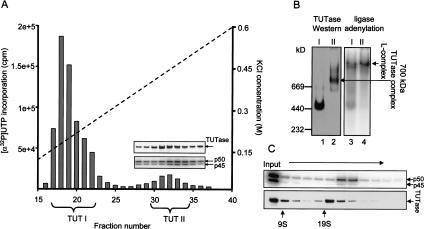
The existence of 3′ TUTase and RNA ligase activities in trypanosomatid mitochondria was actually demonstrated prior to the development of the cleavage-ligation model for editing (White and Borst 1987; Bakalara et al. 1989). The adenylation assay for identification of the two RNA ligases (Sabatini and Jakduk 1995), and the ease of the enzymatic assay for TUTase combined with fractionation of mitochondrial extracts by sedimentation on glycerol gradients rapidly led to the identification of several multiprotein complexes containing these activities. The in vitro editing activities were found to comigrate in a broad region around 19–25 S, together with a peak of TUTase, RNA ligase, gRNA-dependent endoribonuclease, and U-specific 3′–5′ exonuclease activities (Corell et al. 1996; Seiwert et al. 1996; Cruz-Reyes et al. 1998b; Peris et al. 1997; Rusché et al. 1997). A second peak of TUTase and ligase activities was found (Peris et al. 1997) in both L. tarentolae and T. brucei to sediment in the ∼10S region. The absence of a discrete 10S TUTase peak in T. brucei in reports from other laboratories can most likely be attributed to centrifugation conditions. A third variable peak of TUTase activity was reported to sediment at around 35–40S (Pollard et al. 1992; Madison-Antenucci et al. 1998), but this has not been observed by other laboratories, possibly for technical reasons. The two RNA ligases were shown to be components of a high molecular weight complex that could be visualized as an adenylated band in a native gel (Peris et al. 1997; Rusché et al. 1997), which we have termed the L-complex (Aphasizhev et al. 2002b). Isolation of the 121-kD 3′ TUTase led to the finding that this enzyme was present in at least two configurations, one as a possibly tetrameric enzyme, and the other as an ∼700-kD complex, which sedimented in the ∼20S region, but was clearly separable from the L-complex (Aphasizhev et al. 2002b) also found in that region of the gradient (Fig. 3 ▶).
FIGURE 3.
Two forms of TUTase. (A) Two peaks of TUTase activity in L. tarentolae separated by anion exchange chromatography. The presence of TUTase and the LtREL1 (p50) and LtREL2 (p45) RNA ligases in the TUT II peak fractions are shown by Western analysis and by adenylation. (B) Native gel analysis. (I) TUT I peak; (II) TUT II peak. Fractions indicated by brackets were pooled, adenylated, and separated on native gel. (Left) Western using anti-TUTase antiserum; (right) autoradiograph of adenylated L-complex. (C) Glycerol gradient sedimentation of TUT II. Fractions 30–34 from A were concentrated and centrifuged on glycerol gradient. Each fraction was adenylated, and the gel was blotted, exposed to a PhosphorImager screen, and treated with anti-TUTase antiserum. Modified from Aphasizhev et al. (2002b).
Fractionation of editing complexes was also performed by column chromatography. Rusché et al. (1997) fractionated T. brucei mitochondrial extract through Q-Sepharose, followed by DNA cellulose, using an assay for the adenylated REL1 and REL2 ligase proteins. Approximately 8 proteins were visualized in the final preparation, which was enriched around 500-fold for the RNA ligase activity, and sedimented approximately at 20S in glycerol gradients (Fig. 4 ▶). Three of these polypeptides were identified as the REL1 and REL2 proteins. Remarkably, this preparation exhibited a copurification of all four editing activities, a gRNA-directed endonuclease, a 3′ TUTase, a 3′ U-specific exonuclease, and RNA ligase, and also was active in both U-insertion and U-deletion in vitro editing. However, the conclusion that this eight-polypeptide complex represented the complete editing complex turned out to be premature, as all of these genes have now been cloned and the only recognizable enzymes present are the two RNA ligases (Huang et al. 2001; Rusché et al. 2001). It seems clear that additional essential editing components cofractionated with this stable complex.
FIGURE 4.
Protein composition of fractions from DNA-cellulose column fractionation of T. brucei mitochondrial extract. The eight major polypeptides of the purified editing complex isolated by column fractionation of mitochondrial extract are indicated. Protein silver stain (top) or autoradiogram of adenylation reactions showing the two RNA ligases (bottom). Modified from Rusché et al. (1997).
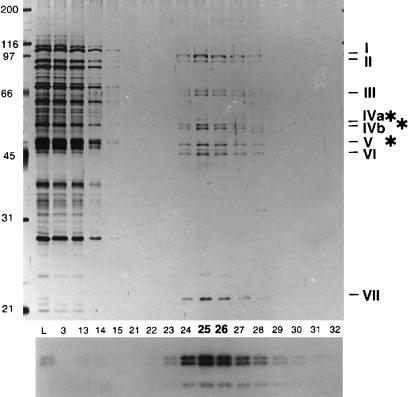
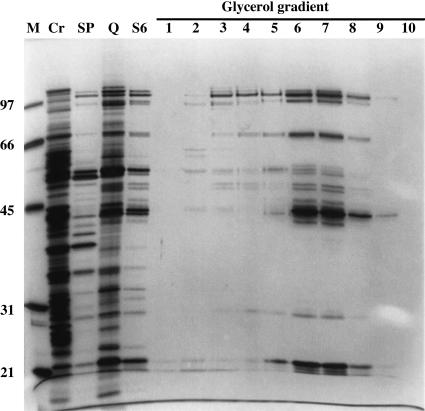
Panigrahi et al. (2001b) performed a similar fractionation of T. brucei mitochondrial extract, using sequential SP-Sepharose, Q-Sepharose, and Superose 6 chromatography and an in vitro U-deletion assay, and recovered a more complex preparation consisting of ∼20 major protein bands, although several appear not to be stoichiometric, and at least 2 bands were found to be common contaminants of mitochondrial extracts, Hsp 70 and glutamate dehydrogenase (Fig. 5 ▶). The activity sedimented at ∼20 S in glycerol gradients. This preparation was used as an immunogen to select 19 monoclonal antibodies directed against 7 different proteins in this preparation. A combination of immunoprecipitation and mass spectrometry of protein bands was used together with T. brucei genomic sequence data to identify a number of the components of this preparation, which are discussed below. See Stuart et al. (2002) for a recent review.
FIGURE 5.
Protein composition of fractions from T. brucei-editing complex purification. The most purified editing activity-positive fraction from the glycerol gradient following the column fractionations shows 20 major polypeptide bands. Protein size standards (M), cleared mitochondrial lysate (Cr), and pooled editing activity-positive fractions from SP Sepharose (SP), Q Sepharose (Q), and Superose 6 (S6) columns, and glycerol gradient are presented. Modified from Panigrahi et al. (2001a).
Madison-Antenucci et al. (1998) fractionated mitochondrial extract from T. brucei through Q-Sepharose, Heparin-agarose, and Phosphocelluose, using enrichment of the adenylated ligases as an assay. The final material contained ∼13 polypeptides, including REL1 and REL2, showed TUTase activity, and contained gRNA. The TUTase activity, however, fractionated on Superose 6 gel filtration slightly ahead of the RNA ligases, as described above for the purified TUT II TUTase and L-complex from L. tarentolae (see Fig. 3 ▶).
Endonuclease
After formation of the gRNA/mRNA anchor duplex, the mRNA is specifically cleaved by a gRNA-dependent riboendonuclease just 5′ of the anchor (Fig. 1 ▶). Several nuclease activities have been described in T. brucei mitochondria (Piller et al. 1997; Salavati et al. 2002). One activity shows the specificity predicted for the editing endonuclease. During glycerol gradient fractionation, this activity cosedimented at ∼20S with other editing enzymatic activities and could be coimmunoprecipitated with the editing complex described by the Stuart laboratory using a anti-TbMP63 antibody (Drozdz et al. 2002). Down-regulation of TbMP63 reduced gRNA-dependent endonuclease activity (Huang et al. 2002). However, recombinant TbMP63 did not show nuclease activity, nor does it contain any known RNase motifs. A possible role for this protein is discussed below. In addition to this activity, an RNase P (Salavati et al. 2002) and at least three gRNA-independent RNase activities that seem to differ in their DTT requirements have been detected (Piller et al. 1997; Salavati et al. 2002). The polypeptides responsible for any of those activities have not yet been identified.
The only riboendonuclease cloned and characterized from kinetoplastid mitochondria so far is LtMAR1 (Alfonzo et al. 1998). This 22-kD protein cleaved edited and pre-edited mRNAs in a gRNA-independent fashion. It is not clear whether MAR1 is involved in RNA editing, or whether the gRNA-independent cleavage observed with the purified enzyme is due to the lack of a specificity factor.
TUTase
In the case of a U-insertion event, the next step in the editing process after cleavage is the addition of a U residue to the 3′-end of the mRNA 5′ cleavage fragment by a 3′-terminal uridylyl transferase (TUTase) (Fig. 1 ▶). A 3′ TUTase activity was first described in whole-cell extracts from T. brucei (White and Borst 1987) and in mitochondrial extracts from L. tarentolae, and a 3′ TUTase has been isolated from mitochondria of L. tarentolae and cloned from both L. tarentolae and T. brucei (Aphasizhev et al. 2002b). The pre-proteins are 121 and 107 kD in size for L. tarentolae and T. brucei, respectively, and contain a mononucleotide binding domain and a poly(A) polymerase-associated domain, characteristic for the pol β superfamily of nucleotidyltransferases. The native and the recombinant enzymes showed a high degree of specificity for UTP. Only limited incorporation of CTP and GTP and, essentially, no incorporation of ATP into substrate RNA was observed. Substitution of Mn++ for Mg++ decreased the specificity substantially.
By use of model pre-cleaved editing substrates, it was shown that the nature of the RNA substrate modulates the activity of the recombinant TUTase from L. tarentolae. The substrates consisted of two short synthetic RNAs resembling the 5′ and the 3′ cleavage fragments of a mRNA and a bridging gRNA that left a gap of variable length. Use of the 5′ fragment alone as substrate led to the addition of a ladder of U’s, the length of which was determined solely by the enzyme and UTP concentrations. Annealing of the 5′ fragment and the bridge RNA significantly decreased the number of U’s added, but annealing the 5′ fragment, the 3′ fragment, and the bridge RNA had a dramatic effect on the number of U’s added. The size of the single-stranded gap affected the number of U’s added to the 5′ fragment. In the case of a 3-nucleotide gap, the predominant species was +3 U’s, whereas the single nucleotide gap showed a predominant species of +1 U. The importance of base pairing of the guiding nucleotides with the added U’s for restricting the number of U’s added was shown by a long ladder obtained with a 3-nucleotide gap containing the nonguiding sequence, CCC. This suggests that the substrate specificity of TUTase combined with that of the ligase(s) represents one of the major factors determining the precise number of U’s added at an editing site, if the identified TUTase proves to be the enzyme that adds U’s at the editing site.
In vivo analysis of TUTase further indicated a role in RNA editing. Down-regulation of TUTase in procyclic T. brucei by RNAi demonstrated that the enzyme is essential for cell viability, and primer extension analysis of transcripts from 6 of the 12 edited genes showed a decrease in the abundance of edited RNA by up to 90%, with no effect on the total amount of mitochondrial RNA (Aphasizhev et al. 2002b).
Immunodepletion of TUTase from mitochondrial extracts in L. tarentolae decreased TUTase activity to near background levels, suggesting that only one TUTase is present in the mitochondrion that is responsible for the formation of the nonencoded oligo(U)-tail of gRNAs and the 3′ addition of U’s to the rRNAs and mRNAs, in addition to the insertion of U’s at an editing site (Aphasizhev et al. 2002b). However, because the anti-TUTase IP also pulls down associated proteins, this result does not eliminate the possibility of the existence of a second TUTase. In fact, Stuart (K. Stuart, pers. comm.), and Aphasizhev and Simpson (R. Aphasizhev and L. Simpson, unpubl.) have very recently detected a second putative 57-kD TUTase as a component of the L-complex in T. brucei and L. tarentolae. However, there is yet no evidence that it is required for RNA editing. The precise roles of the 121-kD TUTase and the 57-kD TUTase are still open questions.
U-specific exonuclease
In the case of a deletion event, the next step in the editing process after cleavage is the removal of U’s from a 3′ overhang of the 5′ cleavage fragment by a U-specific 3′–5′-exonuclease (Fig. 1 ▶). No such enzyme has been cloned yet, but several 3′-exonuclease activities have been partially purified from kinetoplastid mitochondria. In L. tarentolae (Aphasizhev and Simpson 2001), a distributive U-specific 3′–5′-exonuclease activity was purified several thousandfold away from a nonspecific processive 3′-exonuclease and a 3′-phosphate-stimulated exonuclease. This activity was also separated from TUTase, definitely showing that the exonuclease activity is not a reversal of the TUTase activity. In T. brucei (Rusché et al. 1997) a similar 3′-exonuclease activity copurified with an editing complex (Rusché et al. 1997; Lawson et al. 2001; Igo et al. 2002a). In both species, digestion of 3′-terminal U-stretches is single-strand specific and does not proceed past a G, A, or C residue, consistent with its proposed function during editing. The enzyme cascade model (Blum et al. 1990) proposed that guiding A’s and G’s base pair with the added U’s and protect these during U-insertion editing, whereas during deletion editing, the nonbase-paired 3′-terminal U’s are removed. This was confirmed in a pre-cleaved U-deletion editing assay, in which it was shown that the protection from exonucleolytic removal depended on the ability of the 3′ U’s to base pair with guiding nucleotides in the bridge RNA (Aphasizhev and Simpson 2001).
Ligase
After 3′ U residues have been added or removed from the 5′ cleavage fragment, the two fragments are joined by RNA ligase (Fig. 1 ▶). RNA ligase activity copurified with the editing complexes isolated by the Sollner-Webb, Stuart, and Hajduk laboratories, and could be attributed to two auto-adenylatable proteins. Those two proteins, REL1 and REL2, were cloned from both T. brucei (McManus et al. 2001; Panigrahi et al. 2001a; Rusché et al. 2001) and L. tarentolae (G. Gao and L. Simpson, unpubl.). The pre-proteins are 52 kD (REL1) and 48 kD (REL2) in T. brucei, and 53 kD and 48 kD in L. tarentolae. A detailed enzymological study of recombinant ligases has not been reported yet, but an initial analysis showed that recombinant TbREL1 and TbREL2 have RNA ligase activity and can join bridged RNAs, consistent with their proposed function in editing (Schnaufer et al. 2001).
Conditional expression of TbREL1 in bloodstream form T. brucei (Schnaufer et al. 2001) showed that the protein is essential for cell viability, and analysis of mitochondrial RNA from this cell line showed that down-regulation of REL1 was accompanied by a specific reduction of edited RNA in vivo. Surprisingly, the double knockout was lethal for bloodstream trypanosomes, suggesting either that editing is essential also in this stage of the life cycle in spite of the nonmitochondrial metabolism characteristic of the bloodstream stage, or that the ligase plays a vital role outside of the mitochondrion. Support for the former interpretation is provided by the result that RNAi down-regulation of the TbMP81 protein also affects viability of bloodstream trypanosomes (Drozdz et al. 2002)
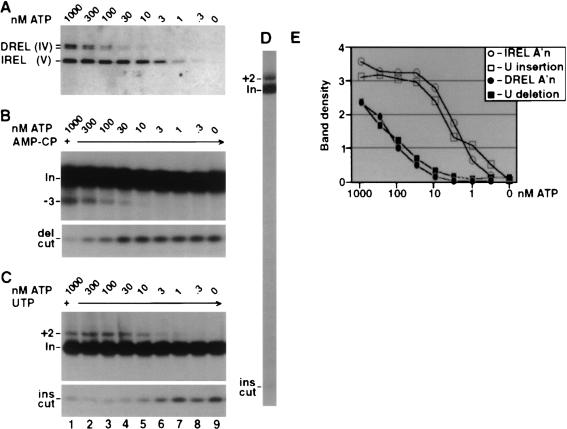
The cloning of the two RNA ligase genes from T. brucei allowed Huang et al. (2001) to produce a conditional dominant-negative REL1 transgene by mutation of the AMP-binding lysine to arginine (K86R). As in the case of the double allele knockout in bloodstream T. brucei, there was a growth phenotype and a decrease in the steady-state abundance of fully edited ND7 mRNA (5′ domain) in procyclic trypanosomes. The authors pointed out that the dominant-negative result provides firm evidence that the REL1 ligase enzymatic activity was required for viability and for editing, in distinction to the above double knockout results (Schnaufer et al. 2001), which could have been due to loss of the entire complex. Poisoned primer extension analysis results of edited and unedited RNAs in the induced cells were interpreted as showing a specific loss of ligation at U-deletion sites. In vitro editing was also examined and the same interpretation made. Cruz-Reyes et al (2002) further examined the differential requirements of REL1 and REL2 for ATP in their purified editing complex (Rusché et al. 1997; Fig. 6 ▶). These results correlated well with the previous data, and this was taken as providing further evidence that REL1 performs ligation at U-deletion sites and REL2 at U-insertion sites. However, the finding that RNAi down-regulation of expression of REL2 in T. brucei produces no phenotype brings into question a role for REL2 in editing at all (Drozdz et al. 2002). This remains an important and open question.
FIGURE 6.
Adenylylation, U-deletion, and U-insertion directed by the editing complex at various ATP concentrations. The purified complex (see Fig. 4 ▶) was first fully decharged with 10 mM PPi, then diluted to 1.5 mM PPi, and supplemented with the indicated ATP. (A) Adenylylation assay. Autoradiogram of protein gel assessing DREL (REL1) and IREL (REL2) activation using [α−32P]ATP. Mixtures that were incubated longer, or that contained the editing RNAs, yielded similar results, and higher ATP concentrations had no additional effect (data not shown). The DREL doublet corresponds to two isoforms. (B,C) U-deletion and U-insertion assays. Electrophoretic analyses show RNA products generated by incubating the treated complex with the indicated amounts of unlabeled ATP and 3′-end-labeled A6 pre-mRNA, preincubated with gRNA D32a (B) or I47G (C). Also, U-deletion reaction mixtures contained 1 mM AMP-CP (B), whereas the U-insertion reaction mixtures contained 150 μM UTP (C). (Top panels) gel region containing input mRNA (In) and the guided (−3) U-deletion product or (+2) U-insertion product; (bottom panels) the gel region containing the fragment remaining from the gRNA-directed cleavage at the U-deletion site (del cut) or at the U-insertion site (ins cut). (D) A larger region of a gel assessing U-insertion, as in C, using 1 mMATP. (E) Quantitation of autoradiograms showing adenylylation of the two ligase proteins in A and the U-deletion and U-insertion products from different exposures of the gels of B and C. Band density is in units of 1000 pixels. Modified from Cruz-Reyes et al. (2002).
RNA helicase
Editing of all except one pre-mRNA involves several gRNAs that overlap and act in a 3′–5′ sequential order. To allow a coordinated exchange of these gRNAs, the participation of an RNA helicase has been proposed. RNA helicase activity has been reported in mitochondrial extracts of T. brucei, and a potential RNA helicase DEAD-box protein, mHEL61, has been cloned (Missel et al. 1997). An involvement of mHEL61 in editing was suggested by an in vivo study. mHEL61 null-mutants were viable, but showed a specific significant reduction of edited mRNAs, a phenotype that was rescued by the expression of an ectopic copy of the gene (Missel et al. 1997).
Proteins of unknown function
Several proteins of unknown function associated with different preparations of editing complexes have been reported (Table 1 ▶). So far, only the sequences of four related proteins have been published. These four proteins are an integral part of both the complex isolated by the Stuart laboratory (Panigrahi et al. 2001b) (TbMP81, TbMP63, TbMP42, and TbMP18) and that isolated by the Sollner-Webb laboratory (Band II, Band III, Band VI, and Band VII) (Rusché et al. 1997). The three larger proteins contain C2H2 zinc-finger motifs. As these motifs are commonly involved in protein–protein or protein–RNA interactions, it has been suggested that these proteins play a role in the structural organization of the editing complex. It has been shown that recombinant TbMP63 binds recombinant REL1 in vitro (Panigrahi et al. 2001a), and this interaction was indirectly confirmed in vivo by showing that down-regulation of TbMP63 by RNAi led to the loss of REL1, but not REL2 (Huang et al. 2002). Glycerol gradient fractionation of TbMP63-depleted mitochondrial extracts further showed that REL2 was not assembled into a high molecular weight complex, thus suggesting that the editing complex could not be assembled in the absence of TbMP63 or REL1. In addition, cells depleted of TbMP63 showed a loss of gRNA-dependent cleavage activity. It is not clear whether the loss of gRNA-dependent cleavage activity is due to TbMP63 itself or due to a coordinated loss of the nuclease or to an effect on the structural integrity of the complex. No evidence so far has been reported that TbMP63 actually is a nuclease.
TABLE 1.
List of cloned genes potentially involved in editing
| Size (kD)b | |||||||
| Namea | Tb | Lt | Cf | Activity/Function | Motifs | KO phenotypea | Ref |
| TUTase | 107 | 121 | — | TUTase activity | Nucleotidyltransferase | GI (RNAi) | (1) |
| TbMP81 | 81 | — | — | May interact with REL2 | zinc finger | GI (RNAi) | (48) |
| RGG1 | — | — | 76 | oligo(U) binding | RGG RNA-binding | — | (72) |
| TbMP63 | 63 | — | — | Interacts with REL1 | zinc finger | GI (RNAi) | (45,49) |
| mHEL61 | 61 | — | — | Putative RNA helicase | DEAD-box | GI (KO) | (42,43,45) |
| REL1 | 52 | 54 | — | RNA ligase activity | ligase | GI (dom. neg.) | (41,53) |
| REL2 | 48 | 48 | — | RNA ligase activity | ligase | NE (RNAi) | (48,a) |
| REAP1 | 45 | — | — | mRNA binding | 21mer-repeat | — | (40,68) |
| TbMP42 | 42 | — | — | Associated with complex | zinc finger | — | (36,57) |
| gBP27 | 25 | 27 | 27 | Annealing, gRNA binding | — | — | (67) |
| gBP21 | 23 | 27 | 29 | Annealing, gRNA binding | — | NE (KO) | (61,62) |
| MAR1 | — | 22 | — | Endonuclease activity | — | — | (50) |
| TbMP18 | 18 | — | — | Associated with complex | — | — | (57) |
| RBP16 | 15 | — | — | RNA binding | Y-box | — | |
(MP81) Band II; (MP63) Band III; (mHEL61) MP61; (REL1) Band IV, TbMP52, p57, p50; (REL2) Band V, TbMP48, p50, p45; (MP42) Band VI; (gBP27) gBP25 (Tb); (gBP21) gBP29 (Cf), (MP18) Band VII.
a(GI) growth inhibition; (NE) No effect. (a). G. Gao and L. Simpson, unpubl.
bApproximate size of the pre-proteins, (Tb) T. brucei; (Lt) L. tarentolae; (Cf) C. fasciculata; (—) no data available.
The second protein analyzed in detail was TbMP81 (Drozdz et al. 2002). In vitro transcribed-translated TbMP81 did not show any exonuclease or endonuclease, TUTase, or ligase activity. Down-regulation by RNAi in both the bloodstream and procyclic forms of T. brucei showed a lethal phenotype and a decrease of edited mRNAs, suggesting a role in editing. Glycerol gradient fractionation of mitochondrial extracts of TbMP81-depleted cells suggested that the ∼20S-editing complex could still assemble. Nevertheless, immunoprecipitation experiments using this extract showed that adenylatable REL2 and TUTase activities were missing from the editing complex. These immunoprecipitates also showed reduced gRNA-directed cleavage activity, a reduced exonuclease activity, and reduced pre-cleaved in vitro deletion and U-insertion editing activities. This rather general effect on a variety of editing activities may suggest a structural role in the editing complex.
RNA-binding proteins
RNA-binding proteins have always been assumed to be a part of the RNA-editing machinery as common sense feature of any RNA-processing reaction taking place in high molecular weight RNP complexes. Several polypeptides capable of binding RNA have been isolated from mitochondrial extracts of T. brucei, L. tarentolae, and Crithidia fasciculata and characterized to different extents. To date, there is no decisive evidence that any RNA-binding protein is essential for U-insertion/deletion editing in trypanosomes. However, the wealth of information that has been accumulated makes it worthwhile to critically analyze some of the most promising candidates, even if they are only guilty by association.
gBP21
The capacity to cross-link synthetic radiolabeled gRNA by UV-irradiation was used extensively to isolate a number of proteins, some of which turned out to be mitochondrial metabolic proteins (Bringaud et al. 1995, 1997; Estévez et al. 1999) that are unlikely to have any part in editing. This approach also resulted in identification (Köller et al. 1994), purification, and cloning (Köller et al. 1997) of the gRNA-binding protein, gBP21, from T. brucei. Database searches revealed neither the presence of known RNA-binding motifs nor homologs beyond trypanosomes. Extensive studies of the RNA-binding properties of the recombinant protein showed that gBP21 binds to synthetic gRNA specifically and with high affinity (kD in the low nanomolar range), although there is also a substantial nonspecific single-stranded RNA-binding activity as well (Köller et al. 1997). Binding of gBP21 did not alter the tertiary structure of gRNA, and most likely, involved ionic contacts. Furthermore, a specific binding site for gBP21 was determined in gND7-506 by RNase footprinting experiments (Hermann et al. 1997). Computer-assisted modeling was used to construct a three-dimensional model of gRNA structure and to propose that a stem/loop structure that can form near the 3′-end of the gRNA may be a binding site for the recombinant gBP21. The 3′ oligo(U) tail was found to be irrelevant for this interaction. The most interesting in vitro activity that has been described for gBP21 is the capacity to promote the hybridization of complementary single-stranded RNAs (Muller et al. 2001). This activity is likely to be achieved by concert action of two RNA-binding sites within gBP21 (Muller et al. 2001). It has been proposed that one binding site is responsible for the formation of a stable gRNA–gBP21 complex by interacting with an irregular hairpin at the 3′-end, whereas the other site causes the unfolding of the 5′-end anchor, thereby facilitating hybridization. In addition, the electrostatic repulsion between two nucleic acids may be mollified by the highly positively charged protein.
The possible involvement of gBP21 in RNA editing has been the subject of several studies. The results suggest that gBP21 in mitochondrial extract is associated with high molecular weight complexes that possess in vitro editing activity (Allen et al. 1998). In addition, polyclonal antibody against gBP21 inhibited editing activity in vitro (Lambert et al. 1999). However, this protein was found not to be essential for cell survival and RNA editing in vivo, but the null/null gBP21 mutant was not able to differentiate from bloodstream to procyclic form. The observed effect of this gene knockout on editing in vivo in bloodstream trypanosomes was somewhat ambiguous; editing patterns of ND7 and A6 mRNAs were largely unaffected, but the abundance of unedited Cyb mRNA and never-edited COI mRNA decreased 70%–80%. No effect on gRNA abundance was detected (Lambert et al. 1999). These findings make the participation of gBP21 in reactions other than RNA editing very likely.
Two gRNA/oligo U-binding proteins (gBP27 and gBP29) with similar RNA-binding properties were purified from C. fasciculata mitochondrial extract by virtue of their ability to cross-link to oligo(U) (Leegwater et al. 1995). Sequence analysis showed a high degree of homology between gBP21 and gBP29, but some probably species-specific differences should be noted. gBP21 was purified as an individual protein (Köller et al. 1997), whereas gBP27 and gBP29 copurified from the extract. Furthermore, orthologs of gBP27 and gBP29 were purified from L. tarentolae as a 100-kD complex, and their direct interaction within a heterotetramer has been demonstrated (Aphasizhev et al. 2002a). Secondly, the 3′ oligo(U) tail was unnecessary for gBP21 binding to gRNA, but was reported to be required for efficient interactions of both gBP27 and gBP29 with RNA (Blom et al. 2001). gBP21 could be coimmunoprecipitated with in vitro editing activity and individual editing enzymes (Allen et al. 1998), but no such interactions were reported for the proteins from C. fasciculata (Blom et al. 2001). However, immunodepletion of either gBP27 or gBP29 resulted in removal of up to 50% of the gRNA and 25% of both edited and unedited mRNAs with little effect on ribosomal RNA. Finally, the demonstration of RNA annealing activity in gBP27 orthologs from T. brucei and L. tarentolae (Aphasizhev et al. 2002a) suggests that this biological function may be redundant in trypanosomes, which would be consistent with a nonessential role of gBP21 in editing.
REAP1
An alternative shotgun-like approach was used to isolate a protein possibly involved in RNA editing in T. brucei, REAP1 (Madison-Antenucci et al. 1998). The experimental strategy involved generation of a panel of monoclonal antibodies against anything that sedimented at 35–40S, the region of the glycerol gradient in which high molecular weight gRNA/mRNA-containing RNP complexes have also been reported (Pollard et al. 1992). An antibody that recognized a protein of ∼45 kD from this region was used to clone the gene from an expression library. It was shown that REAP1 copurified with RNA ligase and TUTase through several conventional chromatography columns, but the absence of quantitative data on the fold purification makes it difficult to establish whether these proteins copurified as a stable complex, or that these activities could be simply detected in the same fractions. Specific immunopurification of REAP1 combined with size-fractionation also showed copurification of RNA ligase and TUTase, but the molecular weight of such a complex was estimated at ∼700 kD, which is consistent with a sedimentation value of ∼20S, not 35–40S. This may suggest that the original definition of REAP1 as a component of a 35–40S-editing complex is in question, or that the stability of the complexes under the experimental conditions of glycerol gradient and affinity/gel filtration fractionations differs substantially.
The final line of evidence presented for an involvement of REAP1 in editing was a dose-dependent inhibition of U-insertion in vitro editing activity up to 50% by the monoclonal antibody against REAP1. The antibody did not inhibit any of the individual enzymatic activities. A poly(G)-binding activity was localized to the amino-terminal third of the protein by a series of carboxy-terminal deletions (Madison-Antenucci and Hajduk 2001). On the basis of this evidence, a model was proposed that REAP-1 acts as a pre-edited region-binding protein of yet unknown function.
RBP16
The presence of a large number of proteins that bind poly(U) or oligo(U) RNA substrates in mitochondrial extracts was one of the earliest observations in the RNA-editing saga (Köller et al. 1994; Leegwater et al. 1995). The rationale behind the purification of poly(U)-binding proteins was their potential propensity to bind the oligo(U) tail of gRNA or U-rich tracks of edited mRNAs. One such protein, RBP16, was purified by positive selection on poly(U) Sepharose and negative selection on poly(A) resin using cross-linking with uniformly radiolabeled gRNA as a purification assay (Hayman and Read 1999). Sequence analysis revealed the presence of a cold-shock domain (CSD), characteristic of the Y-box protein superfamily that had been found responsible for a variety of RNA-binding functions in prokaryotes and eukaryotes (Matsumoto and Wolffe 1998). The most commonly assumed mechanism of CSD function is destabilization of RNA secondary structure (Jiang et al. 1997). Competition experiments with homoribopolymers and recombinant RBP16 in vitro confirmed the poly(U)-binding activity of the isolated protein and also detected a less efficient poly(G) binding. However, the most effective competitor appeared to be a single-stranded oligo[dT] DNA. Immunodepletion of RBP16 led to the removal of equal (∼30%) relative amounts of gRNA and ribosomal RNA. In mitochondrial extracts, RBP16 interacted directly with a 22-kD protein, a homolog of human p32 (Krainer et al. 1991). The biological role of this interaction is yet unclear, as is the possible involvement of RBP16 in RNA editing.
TBRGG1
Another major oligo(U)-dependent cross-link in mitochondrial extract of T. brucei of ∼83 kD (Leegwater et al. 1995) was serendipitously cloned in a search for a trypanosome homolog of nucleolin from higher eukaryotes (Vanhamme et al. 1998). The presence of repeated RGG (Arg-Gly-Gly) tripeptides, found in a class of RNA-binding proteins (Kiledjian and Dreyfuss 1992) suggested an RNA-binding role for TBRGG1. Recombinant protein was able to bind efficiently all homoribopolymers except poly(C) at low-salt concentrations, but under increasing stringency conditions a preference for poly(U) substrates became apparent. No single-stranded DNA binding was detected. Glycerol gradient fractionation of mitochondrial lysates showed that TBRGG1 sedimented in the 30–50S range. However, a peak of in vitro editing activity was also shown to cosediment in these fractions, but this activity was reported previously from several laboratories to sediment in the ∼20S region. The mitochondrial localization, poly(U)-binding activity, and the presence in high molecular weight complexes may argue for some kind of involvement in RNA editing, but further work is clearly required in order to accept or reject this idea.
SUMMARY
A great deal of progress in the identification of specific editing factors has been made in the last few years, and some progress toward understanding the interactions of these factors. The identified complexes fall short of being entitled to be labeled the elusive editosome and, in fact, it appears that there may be several complexes that interact dynamically during the editing reaction. It is premature to construct a detailed model of the editing reaction until more is known about the players in the process and their interactions. The definitive proof of the involvement of any specific factor in editing, of course, would be reconstitution of activities in vitro with recombinant proteins and synthetic RNAs. At the current rate of progress, this may occur in the not-too-distant future.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2178403.
REFERENCES
- Alfonzo, J.D., Thiemann, O.H., and Simpson, L. 1998. Purification and characterization of MAR1 - A mitochondrial associated ribonuclease from Leishmania tarentolae. J. Biol. Chem. 273: 30003–30011. [DOI] [PubMed] [Google Scholar]
- Allen, T.E., Heidmann, S., Reed, R., Myler, P.J., Goringer, H.U., and Stuart, K.D. 1998. Association of guide RNA binding protein gBP21 with active RNA editing complexes in Trypanosoma brucei. Mol. Cell. Biol. 18: 6014–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev, R. and Simpson, L. 2001. Isolation and characterization of a U-specific 3′-5′ exonuclease from mitochondria of Leishmania tarentolae. J. Biol. Chem. 276: 21280–21284. [DOI] [PubMed] [Google Scholar]
- Aphasizhev, R., Aphasizheva, I., Nelson, R.E., and Simpson, L. 2002a. A 100 kDa complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several editing components. RNA (in press). [DOI] [PMC free article] [PubMed]
- Aphasizhev, R., Sbicego, S., Peris, M., Jang, S.H., Aphasizheva, I., Simpson, A.M., Rivlin, A., and Simpson, L. 2002b. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell 108: 637–648. [DOI] [PubMed] [Google Scholar]
- Bakalara, N., Simpson, A.M., and Simpson, L. 1989. The Leishmania kinetoplast-mitochondrion contains terminal uridylyltransferase and RNA ligase activities. J. Biol. Chem. 264: 18679–18686. [PubMed] [Google Scholar]
- Benne, R., Van den Burg, J., Brakenhoff, J., Sloof, P., Van Boom, J., and Tromp, M. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46: 819–826. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Blanc, V., Aphasizhev, R., Alfonzo, J.D., and Simpson, L. 1999. The mitochondrial RNA ligase from Leishmania tarentolae can join RNA molecules bridged by a complementary RNA. J. Biol. Chem. 274: 24289–24296. [DOI] [PubMed] [Google Scholar]
- Blom, D., Burg, J., Breek, C.K., Speijer, D., Muijsers, A.O., and Benne, R. 2001. Cloning and characterization of two guide RNA-binding proteins from mitochondria of Crithidia fasciculata: gBP27, A novel protein, and gBP29, the orthologue of Trypanosoma brucei gBP21. Nucleic Acids Res. 29: 2950–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum, B, and Simpson, L. 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo-(U) tail involved in recognition of the pre-edited region. Cell 62: 391–397. [DOI] [PubMed] [Google Scholar]
- Blum, B., Bakalara, N., and Simpson, L. 1990. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60: 189–198. [DOI] [PubMed] [Google Scholar]
- Blum, B., Sturm, N.R., Simpson, A.M., and Simpson, L. 1991. Chimeric gRNA-mRNA molecules with oligo(U) tails covalently linked at sites of RNA editing suggest that U addition occurs by transesterification. Cell 65: 543–550. [DOI] [PubMed] [Google Scholar]
- Bringaud, F., Peris, M., Zen, K.H., and Simpson, L. 1995. Characterization of two nuclear-encoded protein components of mitochondrial ribonucleoprotein complexes from Leishmania tarentolae. Mol. Biochem. Parasitol. 71: 65–79. [DOI] [PubMed] [Google Scholar]
- Bringaud, F., Stripecke, R., Frech, G.C., Freedland, S., Turck, C., Byrne, E.M., and Simpson, L. 1997. Mitochondrial glutamate dehydrogenase from Leishmania tarentolae is a guide RNA-binding protein. Mol. Cell. Biol. 17: 3915–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, L.M., Burbach, B.J., McKenzie, B.A., and Connell, G.J. 1999. A cis-acting A-U sequence element induces kinetoplastid U-insertions. J. Biol. Chem. 274: 6295–6304. [DOI] [PubMed] [Google Scholar]
- Byrne, E.M., Connell, G.J., and Simpson, L. 1996. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 15: 6758–6765. [PMC free article] [PubMed] [Google Scholar]
- Cech, T.R. 1991. RNA editing: World’s smallest introns. Cell 64: 667–669. [DOI] [PubMed] [Google Scholar]
- Corell, R.A., Read, L.K., Riley, G.R., Nellissery, J.K., Allen, T.E., Kable, M.L., Wachal, M.D., Seiwert, S.D., Myler, P.J., and Stuart, K.D. 1996. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol. Cell. Biol. 16: 1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes, J. and Sollner-Webb, B. 1996. Trypanosome U-deletional RNA editing involves guide RNA-directed endonuclease cleavage, terminal U exonuclease, and RNA ligase activities. Proc. Natl. Acad. Sci. 93: 8901–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Rusché, L., Piller, K.J., and Sollner-Webb, B. 1998a. T. brucei RNA editing: Adenosine nucleotides inversely affect U-deletion and U-insertion reactions at mRNA cleavage. Mol. Cell 1: 401–409. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Rusché, L.N., and Sollner-Webb, B. 1998b. Trypanosoma brucei U insertion and U deletion activities co-purify with an enzymatic editing complex but are differentially optimized. Nucleic Acids Res. 26: 3634–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Zhelonkina, A., Rusche, L., and Sollner-Webb, B. 2001. Trypanosome RNA editing: Simple guide RNA features enhance U deletion 100-fold. Mol. Cell. Biol. 21: 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Reyes, J., Zhelonkina, A.G., Huang, C.E., and Sollner-Webb, B. 2002. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol. Cell. Biol. 22: 4652–4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdz, M., Palazzo, S.S., Salavati, R., O’Rear, J., Clayton, C., and Stuart, K. 2002. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 21: 1791–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez, A.M. and Simpson, L. 1999. Uridine insertion/deletion RNA editing in trypanosome mitochondria - a review. Gene 240: 247–260. [DOI] [PubMed] [Google Scholar]
- Estévez, A.M., Kierszenbaum, F., Wirtz, E., Bringaud, F., Grunstein, J., and Simpson, L. 1999. Knockout of the glutamate dehydrogenase gene in bloodstream Trypanosoma brucei in culture has no effect on editing of mitochondrial mRNAs. Mol. Biochem. Parasitol. 100: 5–17. [DOI] [PubMed] [Google Scholar]
- Gott, J.M. and Emeson, R.B. 2000. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 34: 499–531. [DOI] [PubMed] [Google Scholar]
- Hayman, M.L. and Read, LK. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 274: 12067–12074. [DOI] [PubMed] [Google Scholar]
- Hermann, T., Schmid, B., Heumann, H., and Göringer, H.U. 1997. A three-dimensional working model for a guide RNA from Trypanosoma brucei. Nucleic Acids Res. 25: 2311–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, M., Single, F.N., Köhler, M., Sommer, B., Sprengel, R., and Seeburg, P.H. 1993. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron-exon structure determines position and efficiency. Cell 75: 1361–1370. [DOI] [PubMed] [Google Scholar]
- Huang, C.E., Cruz-Reyes, J., Zhelonkina, A.G., O’Hearn, S., Wirtz, E., and Sollner-Webb, B. 2001. Roles for ligases in the RNA editing complex of Trypanosoma brucei: Band IV is needed for U-deletion and RNA repair. EMBO J. 20: 4694–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.E., O’Hearn, S.F., and Sollner-Webb, B. 2002. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 22: 3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo, R.P., Palazzo, S.S., Burgess, M.L., Panigrahi, A.K., and Stuart, K. 2000. Uridylate addition and RNA ligation contribute to the specificity of kinetoplastid insertion RNA editing. Mol. Cell. Biol. 20: 8447–8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo Jr., R.P., Weston, D.S., Ernst, N.L., Panigrahi, A.K., Salavati, R., and Stuart, K. 2002a. Role of uridylylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukar. Cell 1: 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo Jr., R.P., Lawson, S.D., and Stuart, K. 2002b. RNA sequence and base pairing effects on insertion editing in Trypanosoma brucei. Mol. Cell. Biol. 22: 1567–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W., Hou, Y., and Inouye, M. 1997. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J. Biol. Chem. 272: 196–202. [DOI] [PubMed] [Google Scholar]
- Kabb, A.L., Oppegard, L.M., McKenzie, B.A., and Connell, G.J. 2001. A mRNA determinant of gRNA-directed kinetoplastid editing. Nucleic Acids Res. 29: 2575–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kable, M.L., Seiwert, S.D., Heidmann, S., and Stuart, K. 1996. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science 273: 1189–1195. [DOI] [PubMed] [Google Scholar]
- Kapushoc, S.T. and Simpson, L. 1999. In vitro uridine insertion RNA editing mediated by cis-acting guide RNAs. RNA 5: 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian, M. and Dreyfuss, G. 1992. Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. EMBO J. 11: 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köller, J., Nörskau, G., Paul, A.S., Stuart, K., and Goringer, H.U. 1994. Different Trypanosoma brucei guide RNA molecules associate with an identical complement of mitochondrial proteins in vitro. Nucleic Acids Res. 22: 1988–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köller, J., Muller, U.F., Schmid, B., Missel, A., Kruft, V., Stuart, K., and Goringer, H.U. 1997. Trypanosoma brucei gBP21. An arginine-rich mitochondrial protein that binds to guide RNA with high affinity. J. Biol. Chem. 272: 3749–3757. [DOI] [PubMed] [Google Scholar]
- Krainer, A.R., Mayeda, A., Kozak, D., and Binns, G. 1991. Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66: 383–394. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D.L.J. and Tollervey, D. 1998. Birth of the snoRNPs: The evolution of the modification-guide snoRNAs. Trends Biochem. Sci.. 23: 383–388. [DOI] [PubMed] [Google Scholar]
- Lambert, L., Muller, U.F., Souza, A.E., and Goringer, H.U. 1999. The involvement of gRNA-binding protein gBP21 in RNA editing-an in vitro and in vivo analysis. Nucleic Acids Res. 27: 1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, S.D., Igo Jr., R.P., Salavati, R., and Stuart, K.D. 2001. The specificity of nucleotide removal during RNA editing in Trypanosoma brucei. RNA 7: 1793–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegwater, P., Speijer, D., and Benne, R. 1995. Identification by UV cross-linking of oligo(U)-binding proteins in mitochondria of the insect trypanosomatid Crithidia fasciculata. Eur. J. Biochem. 227: 780–786. [DOI] [PubMed] [Google Scholar]
- Leung, S.S. and Koslowsky, D.J. 2001. Interactions of mRNAs and gRNAs involved in trypanosome mitochondrial RNA editing: Structure probing of an mRNA bound to its cognate gRNA. RNA 7: 1803–1816. [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci, S. and Hajduk, S.L. 2001. RNA editing-associated protein 1 is an RNA binding protein with specificity for preedited mRNA. Mol. Cell 7: 879–886. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Sabatini, R.S., Pollard, V.W., and Hajduk, S.L. 1998. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): A novel editing complex protein with repetitive domains. EMBO J. 17: 6368–6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, K. and Wolffe, A.P. 1998. Gene regulation by Y-box proteins: Coupling control of transcription and translation. Trends Cell. Biol. 8: 318–323. [DOI] [PubMed] [Google Scholar]
- McManus, M.T., Shimamura, M., Grams, J., and Hajduk, S.L. 2001. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA 7: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missel, A., Souza, A.E., Nörskau, G., and Göringer, H.U. 1997. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 17: 4895–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, U,F., Lambert, L., and Goringer, H.U. 2001. Annealing of RNA editing substrates facilitated by guide RNA-binding protein gBP21. EMBO J. 20: 1394–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Gygi, S.P., Ernst, N.L., Igo, R.P., Palazzo, S.S., Schnaufer, A., Weston, D.S., Carmean, N., Salavati, R., Aebersold, R., et al. 2001a. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21: 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Schnaufer, A., Carmean, N., Igo Jr., R.P., Gygi, S.P., Ernst, N.L., Palazzo, S.S., Weston, D.S., Aebersold, R., Salavati, R., et al. 2001b. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21: 6833–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris, M., Simpson, A.M., Grunstein, J., Liliental, J.E., Frech, G.C., and Simpson, L. 1997. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol. Biochem. Parasitol. 85: 9–24. [DOI] [PubMed] [Google Scholar]
- Piller, K.J., Decker, C.J., Rusche, L.N., and Sollner-Webb, B. 1995. Trypanosoma brucei mitochondrial guide RNA-mRNA chimera-forming activity cofractionates with an editing-domain-specific endonuclease and RNA ligase and is mimicked by heterologous nuclease and RNA ligase. Mol. Cell. Biol. 15: 2925–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piller, K.J., Rusche, L.N., Cruz-Reyes, J., and Sollner-Webb, B. 1997. Resolution of the RNA editing gRNA-directed endonuclease from two other endonucleases of Trypanosoma brucei mitochondria. RNA 3: 279–290. [PMC free article] [PubMed] [Google Scholar]
- Pollard, V.W., Harris, M.E., and Hajduk, S.L. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11: 4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenski, J., Crain, P.F., and McCloskey, J.A. 1999. The RNA modification database: 1999 update. Nucleic Acids Res. 27: 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché, L.N., Cruz-Reyes, J., Piller, K.J., and Sollner-Webb, B. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16: 4069–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché, L.N., Huang, C.E., Piller, K.J., Hemann, M., Wirtz, E., and Sollner-Webb, B. 2001. The two RNA ligases of the Trypanosoma brucei RNA editing complex: Cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 21: 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, R. and Hajduk, S.L. 1995. RNA ligase and its involvement in guide RNA/mRNA chimera formation. J. Biol. Chem. 270: 7233–7240. [DOI] [PubMed] [Google Scholar]
- Salavati, R., Panigrahi, A.K., Morach, B.A., Palazzo, S.S., Igo, R.P., and Stuart, K. 2002. Endoribonuclease activities of Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 120: 23–31. [DOI] [PubMed] [Google Scholar]
- Schnaufer, A., Panigrahi, A.K., Panicucci, B., Igo, R.P., Salavati, R., and Stuart, K. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291: 2159–2161. [DOI] [PubMed] [Google Scholar]
- Seiwert, S.D. and Stuart, K. 1994. RNA editing: Transfer of genetic information from gRNA to precursor mRNA in vitro. Science 266: 114–117. [DOI] [PubMed] [Google Scholar]
- Seiwert, S.D., Heidmann, S., and Stuart, K. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84: 831–841. [DOI] [PubMed] [Google Scholar]
- Stuart, K., Panigrahi, A.K., Schnaufer, A., Drozdz, M., Clayton, C., and Salavati, R. 2002. Composition of the editing complex of Trypanosoma brucei. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 357: 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme, L., Perez-Morga, D., Marchal, C., Speijer, D., Lambert, L., Geuskens, M., Alexandre, S., Ismaïli, N., Göringer, U., Benne, R., et al. 1998. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 273: 21825–21833. [DOI] [PubMed] [Google Scholar]
- Vidal, S., Curran, J., and Kolakofsky, D. 1990. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 9: 2017–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, T. and Borst, P. 1987. RNA end-labeling and RNA ligase activities can produce a circular ribosomal RNA in whole cell extracts from trypanosomes. Nucleic Acids Res. 15: 3275–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]