Abstract
We have investigated the role in the fold and RNA-binding properties of the KH modules of a hydrophobic to asparagine mutation of clinical importance in the fragile X syndrome. The mutation involves a well-conserved hydrophobic residue close to the N terminus of the second helix of the KH fold (α2(3) position). The effect of the mutation has been long debated: Although the mutant has been shown to disrupt the three-dimensional fold of several KH domains, the residue seems also to be directly involved in RNA binding, the main function of the KH module. Here we have used the KH3 of Nova-1, whose structure is known both in isolation and in an RNA complex, to study in detail the role of the α2(3) position. A detailed comparison of Nova KH3 structure with its RNA/KH complex and with other KH structures suggests a dual role for the α2(3) residue, which is involved both in stabilizing the hydrophobic core and in RNA contacts. We further show by nuclear magnetic resonance (NMR) studies in solution that L447 of Nova-1 in position α2(3) is in exchange in the absence of RNA, and becomes locked in a more rigid conformation only upon formation of an RNA complex. This implies that position α2(3) functions as a “gate” in the mechanism of RNA recognition of KH motifs based on the rigidification of the fold upon RNA binding.
Keywords: KH modules, Nova, FMR1 fragile X, RNA binding, structure
INTRODUCTION
The fragile X syndrome—the most common form of inherited mental retardation—is a dominant dysfunction associated with the anomalous expansion of a CGG triplet near the promoter of the FMR1 gene (for review, see Kaufmann 2002). In most of the fragile X patients, the expansion leads to silencing of the gene with consequent absence of the FMR1 protein (FMRP) in the affected individuals. However, expression of the protein containing an I304N missense mutation was observed in a single patient with an unusually severe phenotype (De Boulle et al. 1993). The mutation concerns a highly conserved hydrophobic amino acid in the second of two copies of a K-homology (KH) domain, an RNA-binding motif that occurs in more than 100 proteins involved in RNA regulation (Gibson et al. 1993; Siomi et al. 1993).
Understanding the effect of the I304N mutation in the KH modules and which loss of function it might cause in the cell is of high interest both in view of its clinical importance and, more in general, for rationalizing its role in the KH fold. However, although several studies have investigated the behavior of the I304N mutant, an unambiguous comprehension of its importance is still lacking. Mutation of the residue in full-length FMRP was shown to result in complete or partial impairment of the RNA-binding properties of the protein (Siomi et al. 1994; Feng et al. 1997; Brown et al. 1998). The mutant protein no longer associates with translating poly-ribosomes, and becomes part of smaller abnormal mRNP (Feng et al. 1997). Furthermore, translation inhibition of various mRNA is observed in vivo and in vitro with wild-type FMRP but not with the mutant protein (Laggerbauer et al. 2001). The I304N mutant also seems to inhibit oligomerization of FMRP, suggesting that the second KH has a role in homo-association. Introduction of the mutation in isolated KH domains from FMRP and Nova-1 led to an unfolded protein (Musco et al. 1997; Lewis et al. 2000), supporting the hypothesis that the conserved hydrophobic residue is essential for the fold stability of a functionally important domain of FMRP. However, mutation of the corresponding residue in isolated KH of the Splicing factor 1 (Sf1) preserves the fold but impairs RNA binding (Liu et al. 2001). Consistently, the conserved residue makes direct contact with the RNA in all the high-resolution structures of KH–nucleic acid complexes now available (Lewis et al. 2000; Liu et al. 2001; Braddock et al. 2002a, 2002b).
Does the conserved residue have an essential structural role in the domain stability or is it mostly implicated in RNA recognition? In the present article, we have addressed this question first comparing the currently available structures of KH domains, using them to rationalize the functional information. We have then focused on the KH3 from Nova-1 as a paradigm. Nova KH3 is one of the few examples for which the structures of both the isolated domain and of a KH/RNA complex are available (Lewis et al. 1999, 2000). Our choice was also suggested by the observation of an unusual behavior of L447 (the residue corresponding to I304 in FMRP) in the NMR spectrum of Nova-1 KH3: The resonance of the amide proton of L447 could not be identified (Ramos et al. 2002). Here, we show that the apparently contradictory data on the function of the conserved residue stem from its dual role in protein stability and RNA recognition. Based on our NMR evidence, we suggest a mechanism that clarifies the role of this residue in RNA recognition.
RESULTS AND DISCUSSION
Comparison of the known KH structures suggests a dual role of α2(3) position
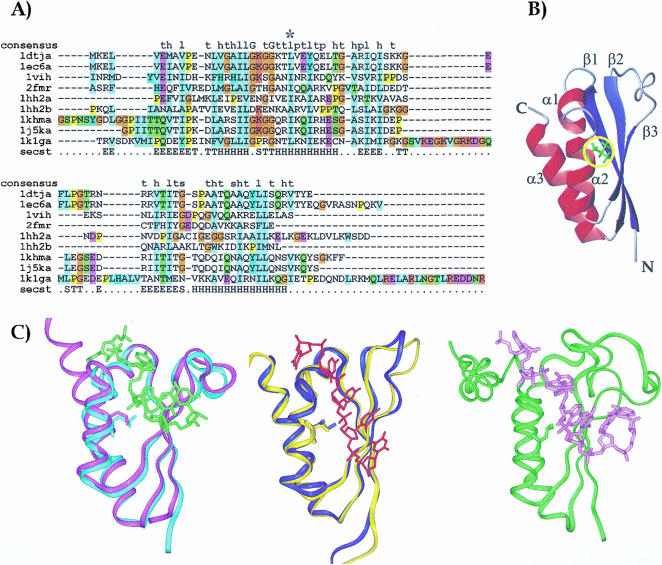
A bulky hydrophobic amino acid (a leucine or an isoleucine in most of the sequences, rarely a short-chain hydrophobic residue, never an aromatic) is present in the whole KH family at a position equivalent to I304 in FMRP (Fig. 1A ▶). Inspection of the currently available structures of KH domains shows that this residue lies in the first turn of helix α2 (position α2(3); Fig. 1B ▶). The side chain is part of the domain hydrophobic core and is completely buried: The exposed surface areas of residues in α2(3) are around 20Å2 with the side chain pointing towards the protein interior. Evidence from our group and from others (Musco et al. 1996; Lewis et al. 1999) conclusively shows that mutation of position α2(3) strongly destabilizes the fold in several KH domains, leading to unfolding of the isolated domain. The only exception is that of the KH domain of Sf1 (Liu et al. 2001). This exception can, however, be rationalised because the fold of the KH domain is most likely stabilized by the presence of the contiguous QUA2 motif, which packs against the KH domain (Fig. 1C ▶). In all the high-resolution KH structures, the amide proton of the residue in α2(3) is hydrogen bonded with the oxygen of the residue in position i-7 [where i is the residue number in α2(3)]. However, the crystallographic B factors of Nova KH3 show that, in the isolated Nova KH3 structure, L447 in α2(3) is significantly more flexible than in the KH3/RNA complex (Lewis et al. 1999, 2000).
FIGURE 1.
(A) Sequence alignment of the KH constructs whose structure is currently known. Sequence alignment was obtained by the Clustalx program (Thompson et al. 1997). The sequences are labeled with the names of the corresponding PDB entries: 1dtj and 1ec6 correspond to the isolated and the complexed Nova KH3 constructs respectively (Lewis et al. 1999, 2000); 1vih, 2fmr, 1hh2-1 and 1hh2-2 are the sequences of vigilin KH6 (Musco et al. 1996), FMRP KH1 (Musco et al. 1997), and of the two KH repeats of NusA (Worbs et al. 2001); 1khm and 1j5k span the sequences of the isolated and complexed hnRNP K KH3, respectively (Baber et al. 1999; Braddock et al. 2002a); 1k1g is the sequence of the complexed Sf1 (Liu et al. 2001). A star indicates position α2(3). The consensus sequence as given by SMART is indicated in the first row (Consensus). The secondary structure of Nova is indicated in the last row (Secst). The DSSP convention was used (Kabsch and Sander 1983). (B) A ribbon representation of the KH fold. The coordinates of Nova KH3 have been used (Lewis et al. 1999). Red ribbons and blue arrows indicate the α helices and the β strands, respectively. The side chain of position α2(3) is shown in green and underlined by a yellow circle. (C) Structure comparison of KH structures. The protein backbones are displayed as a ribbon, and the α2(3) conserved amino acid is displayed in full. The structures were superposed by the Dali server (Holm and Sander 1993) and displayed by InsightII (Accelerys). (Left) Superposition of the free (cyan ribbon) and RNA complex (magenta ribbon with the RNA indicated in green) structures of Nova-2 KH3 (Lewis et al. 1999, 2000). (Middle) Superposition of the free (yellow ribbon) and RNA complex (blue ribbon with the RNA indicated in red) structures of the KH3 of hnRNP-K protein (Baber et al. 1999; Braddock et al. 2002a). (Right) KH-QUA2 structure in complex with RNA (green ribbon with the RNA in pink; Liu et al. 2001).
Because it is inaccessible to the outside, it is difficult to believe that I304 of FMRP KH2 can easily be involved in protein–protein contacts and promote dimerization or self-aggregation directly as suggested by Laggerbauer et al. (2001). A much more likely explanation is that mutation of KH2 causes unfolding of this domain, which leads to a drastic change of the shape of the full-length FMRP molecule, thus preventing self-association of other regions of the protein.
No major structural rearrangement is observed upon RNA complexation (Fig. 1C ▶). The RNA inserts in a groove formed between α2 and β3. The side chain of the leucine/isoleucine in α2(3) packs against an adenine (Lewis et al. 2000; Liu et al. 2001; Braddock et al. 2002a, 2002b). The sequence conservation of all the residues involved in RNA binding indeed suggests that the mechanism of recognition observed in the currently known complexes can be generalized to the whole KH family. Because of the close interaction of the α2(3) hydrophobic residue with RNA in the complex, this position must be very important for RNA binding, so that mutation must reduce significantly the binding affinity. This prediction is consistent with the observation that when the mutation is introduced into the KH-QUA construct of Sf1, the protein is not able to bind to RNA although retaining its fold (Liu et al. 2001).
We can therefore confidently expect that the I304N mutant of FMRP must lose completely the RNA-binding properties associated to KH2, such as polysomal association (Feng et al. 1997). The inability of the I304N mutant to associate with translating ribosomes would explain the increased shuttling activity of the mutant as compared with the wild-type protein (Tamanini et al. 1999). This, however, does not affect the absolute RNA binding capability of the protein (Feng et al. 1997; Brown et al. 1998) as FMRP is a multidomain protein whose individual domains are all able to recognize RNA in vitro (Adinolfi et al. 1999). Other regions can contribute to RNA binding, thus compensating for the loss of function of KH2.
L447 of Nova-1 KH3 is in exchange in the absence of RNA
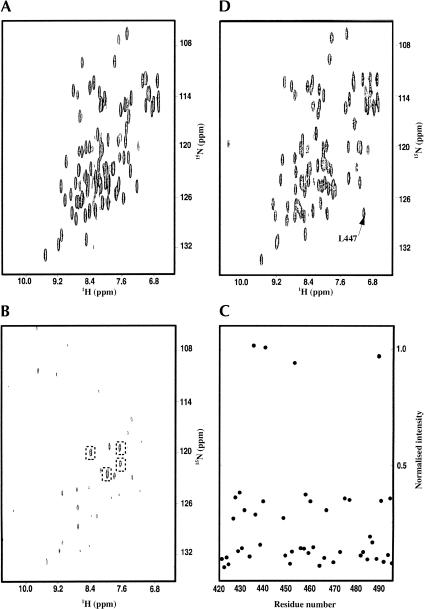
In our previous work on the characterization of Nova-1 KH3 RNA-binding properties (Ramos et al. 2002), we did not succeed in identifying the resonance of the amide proton of L447 in the NMR spectra, even when all distinguishable peaks had been assigned (Fig. 2A ▶). Only 3 backbone amide resonances out of 76 could not be identified, 2 of them (G443 and T480) being in exposed and flexible regions of the molecule (Ramos et al. 2002). In contrast, the amide of L447 is part of a well-defined secondary structure element and the neighboring resonances could be readily identified. We then explored the possible causes of the missing resonance.
FIGURE 2.
Comparison of the spectra of Nova-1 KH3 free and in an RNA complex recorded at 500 MHz and 25°C. (A) 15N HSQC spectrum of the free protein. (B) 15N HSQC of a 15N leucine selectively labeled sample of free Nova-1 KH3. (C) Plot of the normalized (see Materials and Methods) values of peak intensities of the spectrum in A versus the sequence of the protein. The peaks with intensities close to 1 are those boxed in Figure 2B ▶. (D) 15N HSQC spectrum of a 1:1 Nova-1 KH3/RNA1 complex.
The possibility of an unusual chemical shift of the amide proton of this residue in the isolated protein domain seems unlikely, as no aromatic residues are in spatial proximity. Accordingly, the peak could not be identified even at large sweep-widths. Because the chemical shifts of the Hα protons of residues in the position equivalent to L447 in other KH domains are all remarkably similar (3.74 ± 0.07 ppm) irrespective of the amino acid type (leucine or isoleucine), we tried to identify the Hα to side chain connectivities of L447 in the homo-nuclear NOESY and TOCSY spectra (data not shown). However, no spin system seemed to be consistent with the presence of a leucine in the range between 3.65 and 3.88 ppm.
Finally, to discard the possibility that resonance overlap could mask the L447 amide resonance, a selectively labeled sample was produced by adding to the minimal medium a mixture of the 20 natural amino acids where only leucine was 15N labeled. An 15N HSQC spectrum recorded on a sample of purified protein is shown in Figure 2B ▶. The peak intensities were normalized against the intensities of corresponding peaks observed in an 15N HSQC spectrum recorded on an 15N uniformly labeled sample of Nova-1, to correct for line width differences. The results (Fig. 2C ▶) show that the peaks observed in the spectrum of the selectively labeled sample can be divided in three classes based on their normalized intensities. Four peaks have a value between 0.97 and 1.02. These peaks had previously been assigned as four of the five leucine amides in the protein. Fourteen peaks have a value of between 0.38 and 0.27. These peaks correspond to resonances previously assigned as the amides of the 14 isoleucines and valines present in the construct. The remaining peaks, with intensities lower than 0.2, correspond to amides of a number of different amino acids, all previously identified. A degree of labeling of isoleucines and valines and other amino acids is expected, as transamination mechanisms cannot be completely eliminated.
These results clearly indicate that only four leucines are identifiable in the HSQC spectrum of Nova-1 KH3. If the resonance of L447 amide overlapped with one of the peaks observable by transamination in the selectively labeled spectrum, it should have a significantly lower intensity than that of other leucines. A peak broadening to the limit of disappearance strongly suggests that this residue is in chemical or conformational exchange (or in a mixture of both). Varying the temperature (in the range 5°C–65°C) and/or the magnetic field (in the range 500 to 800 MHz), two factors that should influence the exchange properties, did not reveal the appearance of new peaks in the spectra (data not shown).
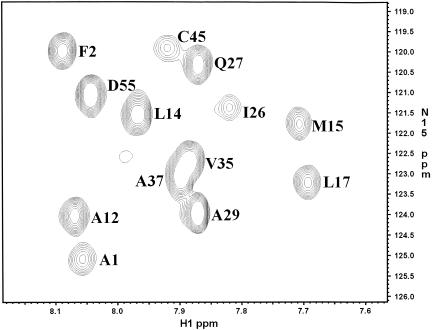
A similar although not so dramatic effect was observed in FMRP KH1 and in Vigilin KH15: resonance broadening in the 1H-dimension (but not of the 15N dimension as demonstrated by the 15N relaxation values) was observed in the homo-nuclear and 15N HSQC spectra (Fig. 3 ▶; Musco et al. 1996, 1997).
FIGURE 3.
Close-up of a 15N HSQC spectrum recorded at 500 MHz and 25°C on a sample of free FMRP KH1. The peaks are labeled according to the assignment of Musco et al. (1997). Position α2(3) is here labeled I26.
α2(3) has a dual role in stabilizing the fold and recognizing RNA
Next, we investigated whether the resonance of the L447 amide is identifiable in the HSQC spectrum of a KH3–RNA complex (Fig. 2D ▶). The RNA sequence used is the same identified as RNA1 and described in Ramos et al. (2002). The backbone assignment of the complex was accomplished using 15N-edited NOESY and HNCA spectra. The amide resonance of L447 is clearly visible at 7.1 ppm and 127.3 ppm (1H and 15N frequencies, respectively); the line width both in the 1H and 15N dimensions is comparable with that of the other amide peaks. 15N T1 (646 ms) and T2 (63.9 ms) and heteronuclear 15N NOE (0.65) values observed at 500 MHz for the L447 amide resonance are close to the average values of Nova-1 KH3 amides in secondary structure elements (Ramos et al. 2002). Thus, L447 is in a stable conformation when the protein is bound to the RNA. The phenomenon leading to the broadening of the resonance observed in the free protein is no longer occurring, suggesting that the residue is locked in a more rigid conformation.
We have previously demonstrated that RNA recognition of Nova-1 KH3 occurs through a rigidification of the overall fold of the protein upon binding, presumably necessary to decrease the entropic step linked to binding (Ramos et al. 2002). Our present observations indicate that L447 is an integral part of this process, suggesting a detailed mechanism of the KH/RNA recognition: The amide proton resonance is in exchange (likely both conformational and chemical) in the absence of RNA to allow insertion of the RNA in the groove between α2 and β3. This local flexibility becomes unnecessary once the complex is formed. Conformational exchange of this residue in the free protein can ensure accessibility of the binding groove without perturbing the protein structure. A local “breathing” of position α2(3) is also supported by the B-factors of this residue in the crystal structure of Nova KH3, which are close to the values observed in loop regions (Lewis et al. 1999, 2000).
In conclusion, the dual function of position α2(3) constitutes yet another example of how RNA binding may be achieved through subtle tuning of the mechanism of recognition.
MATERIALS AND METHODS
RNA and protein preparation
The RNA1 oligonucleotide was synthesized in vitro using T7 polymerase and synthetic DNA templates using established techniques (Varani et al. 1996). The Nova-1 KH3 construct, which spans amino acids 421–496 of the Nova-1 sequence, was expressed in BL21 pLysS cells as His-tagged proteins and isolated using a two-step standard purification procedure as previously described (Ramos et al. 2002). 15N labeled and 15N,13C double-labeled protein was obtained by growing the cells in M9 minimal media supplemented with 15N-labeled ammonium sulphate and 13C-labeled glucose. 15N leucine selectively labeled protein was obtained as described in Biekofsky et al. (2002) but for the use of 15N leucine rather than 15N isoleucine. Briefly, the cells were grown in M9 minimal media supplemented with 13C-labeled glucose and a mixture of 19 unlabeled and 1 (leucine) 15N-labeled amino acids. The large excess of amino acids is known to reduce transaminase expression and therefore scrambling. Expression of the protein was significantly increased by the amino acid cocktail and 12 mg/L of pure protein were obtain from 5 L of cell culture.
Nuclear magnetic resonance spectroscopy and resonance assignment
NMR spectra were recorded in the temperature range between 5°C and 65°C on Varian INOVA and UnityPlus spectrometers operating at 500, 600, and 800 MHz 1H frequency. Full assignment of free and bound Nova KH3 backbone resonances at 25°C was obtained as previously described (Ramos et al. 2002) using standard methods (Marion et al. 1989; Grzesiek and Bax 1992a, 1992b). Water suppression was achieved by the WATERGATE pulse-sequence (Piotto et al. 1992; Sklenar et al. 1993). The spectra were processed and zero-filled to the next power of two using the NMRPIPE program (Delaglio et al. 1995). Baseline correction was applied when necessary. The spectra were analyzed using the Felix (MSI) and XEASY programs (Bartels et al. 1995). HSQC spectra were recorded for uniformly 15N-labeled and 15N-leucine selectively labeled samples of Nova-1 KH3 at 1.3 mM concentration. The peak intensities in the leucine labeled spectra were normalized with respect to the intensity of peaks in the uniformly labeled spectra.
Relaxation data were recorded at 500 MHz on a Varian Unityplus-500 spectrometer using standard sequences (Kay et al. 1992). Peak intensities as a function of delay time were extracted from the spectra and normalized to the intensity of the first time point using the NMRPIPE/NMRDRAW package. The values of T1 and T2 and 15N-1H NOE values were obtained using a standard procedure, as described for Nova-1 KH3 (Ramos et al. 2002).
Acknowledgments
The project was supported by a HFSP grant. We thank P.A. Temussi for critical reading of the manuscript and M. Sattler for helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2168503.
REFERENCES
- Adinolfi, S., Bagni, C., Musco, G., Gibson, T., Mazzarella, L., and Pastore, A. 1999. Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. RNA 5: 1248–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baber, J.L., Libutti, D., Levens, D., and Tjandra, N. 1999. High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor. J. Mol. Biol. 289: 949–962. [DOI] [PubMed] [Google Scholar]
- Bartels, C., Xia, T.H., Billeter, M., Gunter, P., and Wuthrich, K. 1995. The program XEASY for computer-supported NMR spectral-analysis of biological macromolecules. J. Biomol. NMR 6: 1–10. [DOI] [PubMed] [Google Scholar]
- Biekofsky, R.B., Martin, S.R., McCormick, J.E., Masino, L., Fefeu, S., Bayley, P.M., and Feeney, J. 2002. Thermal stability of Calmodulin and M mutants studied by HSQC NMR measureaments of selectively labelled (15N)Ile proteins. Biochemistry 41: 6850–6859. [DOI] [PubMed] [Google Scholar]
- Braddock, D.T., Baber, J.L., Levens, D., and Clore, M. 2002a. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: Solution structure of a complex between hnRNPK KH3 and single-stranded DNA. EMBO J. 13: 3476–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock, D.T., Louis, J.M., Baber, J.L., Levens, D., and Clore, G.M. 2002b. Structure and dynamics of KH domains from FBP bound to single stranded DNA. Nature 415: 1051–1056. [DOI] [PubMed] [Google Scholar]
- Brown, V., Small, K., Lakkis, L., Feng, Y., Gunter, C., Wilkinson, K.D., and Warren, S.T. 1998. Purified recombinant FMRP exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J. Biol. Chem. 273: 15521–15527. [DOI] [PubMed] [Google Scholar]
- De Boulle, K., Ververk, A.J.M.H., Reyniers, E., Vits, L., Hendrickx, J., Van Roy, B., VanDenBos, F., de Graaff, E., Oostra, B.A., and Willems, P.J.A. 1993. Point mutation in the Fmr1 gene associated with fragile X mental retardation. Nat. Genet. 3: 31–35. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.V., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Feng, Y., Absher, D., Eberhart, D.E., Brown, V., Malter, H.E., and Warren, S.T. 1997. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1: 109–118. [DOI] [PubMed] [Google Scholar]
- Gibson, T.J., Thompson, J.D., and Heringa, J. 1993. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 324: 361–366. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S. and Bax, A. 1992a. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J. Magn. Res. 99: 201–207. [Google Scholar]
- Grzesiek, S. and Bax, A. 1992b. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114: 6291–6293. [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123–138. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22: 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kaufmann, W.E. 2002. Neurobiology of Fragile X syndrome: From molecular genetics to neurobehavioral phenotype. Microsc. Res. Tech. 57: 131–134. [DOI] [PubMed] [Google Scholar]
- Kay, L.E., Nicholson, L.K., Delaglio, F., Bax, A., and Torchia, D.A. 1992. Pulse sequences for removal of the effects of cross correlation between dipolar and chemical shift anisotropy relaxation mechanisms on the measureament of heteronuclear T1 and T2 values in proteins. J. Mag. Res. 97: 359–375. [Google Scholar]
- Laggerbauer, B., Ostareck, D., Keidel, E.M., Ostareck-Lederer, A., and Fischer, U. 2001. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 10: 329–338. [DOI] [PubMed] [Google Scholar]
- Lewis, H.A., Chen, H., Edo, C., Buckanovich, R.J., Yang, Y.Y., Musunuru, K., Zhong, R., Darnell, R.B., and Burley, S.K. 1999. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Struct. Fold. Des. 7: 191–203. [DOI] [PubMed] [Google Scholar]
- Lewis, H.A., Musunuru, K., Jensen, K.B., Edo, C., Chen, H., Darnell, R.B., and Burley, S.K. 2000. Sequence-specific RNA binding by a Nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell 100: 323–332. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Luyten, I., Bottomley, M.J., Messias, A.C., Houngniou-Molango, S., Sprangers, R., Zanier, K., Kramer, A., and Sattler, M. 2001. Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science 294: 1098–1102. [DOI] [PubMed] [Google Scholar]
- Marion, D., Driscoll, P.C., Kay, L.E., Wingfield, P.T., Bax, A., Gronenborn, A.M., and Clore, G.M. 1989. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: Application to interleukin 1 β. Biochemistry 28: 6150–6156. [DOI] [PubMed] [Google Scholar]
- Musco, G., Stier, G., Joseph, C., Castiglione Morelli, M.A., Nilges, M., Gibson, T.J., and Pastore, A. 1996. Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell 85: 237–245. [DOI] [PubMed] [Google Scholar]
- Musco, G., Kharrat, A., Stier, G., Fraternali, F., Gibson, T.J., Nilges, M., and Pastore, A. 1997. The solution structure of the first KH domain of Fmr1, the protein responsible for the fragile X syndrome. Nat. Struct. Biol. 4: 712–716. [DOI] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2: 661–665. [DOI] [PubMed] [Google Scholar]
- Ramos, A., Hollingworth, D., Major, S.A., Adinolfi, S., Kelly, G., Muskett, F.W., and Pastore, A. 2002. The role of dimerisation in KH/RNA complexes: The example of Nova KH3. Biochemistry 41: 4193–4201. [DOI] [PubMed] [Google Scholar]
- Siomi, H., Matunis, M.J., Michael, W.M., and Dreyfuss, G. 1993. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 21: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, H., Choi, M.C., Siomi, M.C., Nussbaum, R.L., and Dreyfuss, G. 1994. Essential role for KH domains in RNA binding: Impaired RNA binding by a mutation in the KH domain of Fmr1 that causes fragile X syndrome. Cell 77: 33–39. [DOI] [PubMed] [Google Scholar]
- Sklenar, V., Peterson, R.D., Rejante, M.R., and Feigon, J. 1993. Two- and three-dimensional HCN experiments for correlating base and sugar resonances in 15N, 13C-labeled RNA oligonucleotides. J. Biomol. NMR 3: 721–727. [DOI] [PubMed] [Google Scholar]
- Tamanini, F., Bontekoe, C., Bakker, C.E., van Unen, L., Anar, B., Willemsen, R., Yoshida, M., Galjaard, H., Oostra, B.A., and Hoogeveen, A.T. 1999. Different targets for the fragile X-related proteins revealed by their distinct nuclear localizations. Hum. Mol. Genet. 8: 863–869. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTALX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani, G., Aboul-ela, F., and Allain, H.T. 1996. NMR investigation of RNA structure. Progr. Nucl. Magn. Res. 29: 51–127. [Google Scholar]
- Worbs, M., Bourenkov, G.P., Bartunik, H.D., Huber, R., and Wahl, M.C. 2001. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell 7: 1177–1189. [DOI] [PubMed] [Google Scholar]