Abstract
Adenosine deaminase that acts on RNA, ADAR, catalyzes the conversion of adenosine into inosine within double-stranded RNA. This type of editing has mainly been found in genes involved in neurotransmission. Site-specific A to I modifications often require intronic sequences to create the double-stranded structure necessary for editing. A system was developed to investigate if editing and splicing of pre-mRNA are coordinated. We have focused on a selectively edited site (R/G) in the glutamate receptor subunit B pre-mRNA. This editing site is situated in close proximity to a 5′ splice site. To ensure efficient splicing, the editing site, together with its natural 5′ splice site, was fused to a 3′ splice site of the major late transcript from adenovirus. In vitro, on a premade transcript, ADAR2 editing and splicing were found to interfere with each other. The stable stem-loop required for ADAR2 editing had a negative effect on in vitro splicing, possibly by sequestering the 5′ splice site. Further, RNA helicase A was shown to overcome the splicing inhibition caused by ADAR2. In vivo, allowing cotranscriptional processing, the same construct was found to efficiently edit and splice without interference, suggesting that the two RNA processing events are coordinated.
Keywords: ADAR, GluR-B, RNA editing, RNA processing, pre-mRNA splicing
INTRODUCTION
The surprisingly low number of genes encoded by the human genome suggests that there are alternative procedures to increase the genetic variation. Pre-mRNA processing involves important and intricate mechanisms to regulate and fine-tune gene expression. By regulation of these processing events, multiple isoforms can be generated from the same gene, creating an increased number of gene products for time- and tissue-specific functions.
RNA editing by base modification is a processing event that has the potential to alter single codons. In addition to codon changes, this type of modification has been shown to generate both introduction and removal of stop codons and change of splice sites (reviewed in Maas and Rich 2000; Keegan et al. 2001). RNA editing by ADAR enzymes catalyze adenosine deamination, which results in the formation of inosine within double-stranded or highly structured RNA. Inosine is recognized as guanosine at translation, and can therefore cause a change in codon. Two ADAR enzymes with similar enzymatic activity, but somewhat different substrate specificity, have been characterized in mammals, ADAR1 and ADAR2 (Kim et al. 1994; Maas et al. 1996; Melcher et al. 1996). The ADAR enzymes contain a conserved catalytic deamination domain as well as two or three double-stranded RNA binding motifs (dsRBMs). The ADAR proteins are highly expressed in the mammalian brain (Kim et al. 1994; Melcher et al. 1996), and a majority of the targets for A to I editing have been found in neural receptor pre-mRNA (reviewed in Gerber and Keller 2001). Non-NMDA glutamate receptors, serotonine receptors, and the ADAR2 pre-mRNAs have been shown to be substrates for A to I editing in mammals (Sommer et al. 1991; Burns et al. 1997; Rueter et al. 1999).
One of the most well-studied editing events is the modification of glutamate receptor subunit B pre-mRNA (GluR-B). Two edited sites (Q/R and R/G) are situated in the coding sequence of GluR-B (Fig. 1 ▶). At the Q/R site, located in exon 11, a glutamine codon is changed to an arginine codon upon editing. In the mammalian brain, Q/R is endogenously edited to nearly 100% (Sommer et al. 1991; Higuchi et al. 1993). AMPA receptors assembled with edited GluR-B subunits have reduced permeability for Ca2+ ions (Hume et al. 1991; Burnashev et al. 1992). The R/G site is located in exon 13, where the consequence of editing is a change of an encoded arginine to a glycine. The R/G site displays variable levels of editing during development (reviewed in Seeburg et al. 1998). Receptors assembled with an R/G edited GluR-B subunit has been shown to recover faster from desensitization (Lomeli et al. 1994).
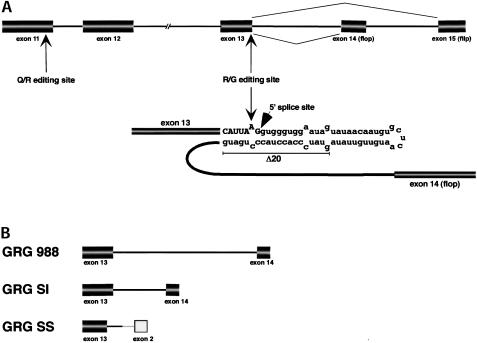
FIGURE 1.
(A) Schematic representation of a part of rat glutamate receptor subunit B pre-mRNA. Exons are represented by boxes and introns by lines, not drawn to scale. The R/G editing site and editing complementary sequence are shown in detail. Exonic sequences are written in capitals and intronic sequences in lowercase. The sequence deleted in the Δ20 mutant is underlined. (B) The editing/splicing reporter constructs used in the study. All constructs contain exon 13 including the R/G editing site and the 5′ part of intron 13. GRG 988 contains full-length intron 13 and exon 14, whereas GRG SI has a shortened intron. GRG SS contains part of an intron, the 3′ splice site and exon from a modified adeno major late transcript.
Both the Q/R and the R/G editing sites are located close to 5′ splice sites, with the R/G site situated only one nucleotide from the intron border. Exonic sequence surrounding the R/G site together with the editing complementary sequence (ECS) in the downstream intron form the double-stranded RNA (dsRNA) structure required for editing (Higuchi et al. 1993). There are several other examples where the ECS is located in an intron, and frequently there is also proximity to splice sites (Burns et al. 1997; Seeburg et al. 1998; Rueter et al. 1999; Reenan et al. 2000). One exception is the antigenome of the hepatitis delta virus that is site selectively edited at the amber/w site, enabling expression of two proteins of different lengths and functions. However, because the ECS is almost exclusively situated in the intron, editing has to happen before splicing; therefore, it is likely that these two RNA processing events interact with each other. One indication that editing and splicing are linked comes from the observation that mice deficient in ADAR2 show a decrease in splicing of GluR-B pre-mRNA (Higuchi et al. 2000). Further, in Drosophila melanogaster, the dsRNA helicase Mle (maleless) has been suggested to link editing and splicing of para pre-mRNA. Mutations in the RNA helicase (mlenapts) result in aberrant editing and exon skipping of the para sodium channel pre-mRNA (Reenan et al. 2000). The mammalian homolog of Mle is RNA helicase A (RHA), a DEAH box RNA helicase that has been shown to unwind dsRNA structures in vitro (Lee and Hurwitz 1992).
In this study we have investigated the relationship between editing and splicing both in vitro and in vivo. We show that in a standard in vitro assay, editing and splicing can be interfering events when their sites of recognition are overlapping. The ADAR2 editing enzyme interfered with splicing of a pre-mRNA containing the R/G editing site. Exposing the transcript with splicing factors prior to addition of ADAR2 abolished editing, which indicated that editing and splicing function as inhibitors upon each other in vitro. RNA helicase A had a positive effect on splicing, possibly by disruption of the stem sequestering the 5′ splice site. By coexpression of ADAR2 and GluR-B pre-mRNA in transfected cell lines, a link between editing and splicing is suggested. We show that there is no interference between editing and splicing of the in vivo transcribed reporter construct. A model for synchronized editing and splicing by the transcriptional machinery is hypothesized.
RESULTS
The R/G stem-loop sequesters the 5′ splice site of GluR-B pre-mRNA
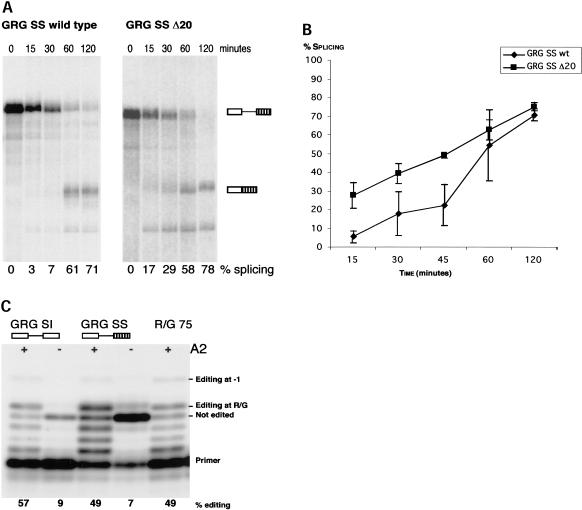
The scope of this study was to analyze the effect editing and splicing has on each other at a site where these RNA processing events coincide. In GluR-B pre-mRNA the R/G editing site is situated one nucleotide from the exon–intron border (Fig. 1A ▶). A GluR-B transcript (GRG 988) containing the R/G editing site in exon 13, intron 13, and exon 14 (flop) was assayed for in vitro splicing. However, splicing of GRG 988 was inefficient, and shortening of the intron by 500 nucleotides (GRG SI) did not increase the efficiency of the in vitro splicing (data not shown). One explanation for the inefficient splicing might be that splicing is complex in this region. The transcript is subjected to alternative splicing by the downstream exon flop or flip, and splicing enhancer sequences might be missing in these constructs. To increase the efficiency of splicing without changing the conditions for editing, the 3′ splice site was changed. Exon 13 and the first part of intron 13 including the editing complementary sequence, was fused to the 3′ splice site of a modified first intron in the adenovirus major late transcript (Fig. 1B ▶) (Ghosh and Garcia-Blanco 2000). In this construct (GRG SS), splicing and R/G editing are efficient (Fig. 2 ▶). After 120 min of standard in vitro splicing reaction 70% of the GRG SS transcript was spliced (Fig. 2A, B ▶). ADAR2 could edit both GRG SI and GRG SS pre-mRNAs in vitro to a similar extent as the R/G 75 stem-loop RNA, previously shown to be a good substrate for ADAR2 editing (Fig. 2C ▶; Ohman et al. 2000). Using this editing/splicing construct the stem-loop structure required for A to I editing was shown to have a negative effect on the splicing efficiency. It has previously been shown that disruption of the double-stranded structure surrounding the R/G site impair site selective editing (Lomeli et al. 1994). Deleting 20 nucleotides of the sequence complementary to the R/G site (see Δ20 in Fig. 1A ▶) increased the rate of splicing, as shown in Figures 2A, 2B, and 3A ▶ ▶. No editing was detected in sequence of GRG SS Δ20 after RT-PCR of the transcript (data not shown). Also noteworthy is that the wild-type GluR-B 3′ splice site of exon 14 also contributes to the inefficient removal of intron 13, as an exchange of the GRG SI 5′ splice site with the strong 5′ splice site of the major late transcript (Glu5′SS) did not enhance in vitro splicing (data not shown). The stable R/G stem functions as a splicing inhibitor, and the 3′ splice site might add to the poor splicing of the wild-type GluR-B constructs in vitro.
FIGURE 2.
Efficient splicing and editing of the GRG SS pre-mRNA. (A) In vitro splicing of GRG SS pre-mRNA and GRG SS pre-mRNA lacking the ECS (Δ20). Splicing products were separated on denaturing polyacrylamide gel. The position of nonspliced pre-mRNA and spliced mRNA is indicated to the right of the gel; exons are represented by boxes and introns by lines. The percentage of splicing is indicated below the gel. (B) Diagram of in vitro splicing efficiency of GRG SS wild type and Δ20 pre-mRNA. Splicing efficiency was plotted against time. Data represents three independent experiments, error bars show standard error of the mean, SEM. (C) GRG SI and GRG SS pre-mRNAs can be edited in vitro by recombinant rADAR2a (A2) as shown by limited primer extension. Position of primer and products corresponding to nonedited, R/G, and -1 edited pre-mRNAs are indicated. Numbers below show the amount of editing in percentage in the different substrates.
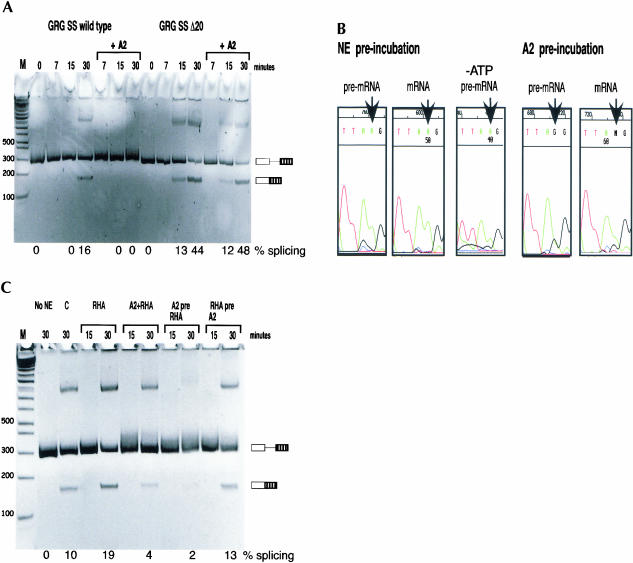
FIGURE 3.
(A) ADAR2 inhibited splicing of GRG SS pre-mRNA. In vitro splicing of GRG SS or GRG SS Δ20 pre-mRNA with or without recombinant rADAR2a (A2) was analyzed using RT-PCR at time points as indicated. Products were separated on a nondenaturing 6% polyacrylamide gel. Additional upper bands have been sequenced and correspond to full-length pre-mRNA. The position of precursor and spliced mRNA are indicated on the right, exons are represented by boxes, and introns by lines. Used for size determination was 1 kb PLUS marker (Life Technologies™). The percentage of splicing is indicated below the gel. (B) Splicing and editing are competing in vitro. In vitro editing/splicing reactions with preincubation of either nuclear extract (NE) or recombinant rADAR2a (A2) for 5 min, as indicated. PCR products corresponding to pre-mRNA and mRNA at 30 min of splicing reaction were sequenced, and the chromatograms with the R/G editing site indicated by an arrow are shown. ATP was omitted (-ATP) in the splicing reaction in the middle panel. (C) RNA helicase A (RHA) enhanced in vitro splicing of GRG SS and counteracted splicing inhibition by ADAR2 (A2). In vitro editing/splicing reactions with preincubation of recombinant rADAR2a and/or RHA, as indicated. Two controls are included: a splicing reaction without nuclear extract (No NE), and a control splicing reaction (C) containing an equal volume of ADAR storage buffer without rADAR2a. The positions of precursor and spliced mRNA are indicated to the right of the gel; exons are represented by boxes and introns by lines. Used for size determination was 1 kb PLUS marker (Life Technologies™). The percentage of splicing is indicated below the gel.
The relation between R/G editing and splicing in vitro
To investigate whether ADAR2 binding has an effect on splicing in vitro, recombinant ADAR2 was added to in vitro splicing reactions of GRG SS transcripts. In the presence of ADAR2, an inhibition of splicing was observed after 30 min of the reaction (Fig. 3A ▶). ADAR2 had little effect on splicing of the GRG SS Δ20 pre-mRNA, lacking the complementary sequence required for editing (Fig. 3A ▶).
To examine if splicing could have a negative effect on editing, the GRG SS transcript was preincubated with nuclear extract for 5 min allowing splicing factors to bind prior to the addition of ADAR2. After 30 min of incubation, no R/G editing was detected, determined by sequencing of products obtained by reverse transcription and polymerase chain reaction (RT-PCR) (Fig. 3B ▶, left). The ability of splicing factors to block editing in vitro was not dependent on ATP, indicating that the initial binding of splice factors to the 5′ splice site is sufficient to inhibit editing (Fig. 3B ▶, middle). As a control, ADAR2 was preincubated with the GRG SS pre-mRNA for 5 min prior to the in vitro splicing assay, resulting in the majority of the transcripts being edited after 30 min (Fig. 3B ▶, right). These results suggest that ADAR2 editing and splicing are competing events in vitro.
The influence of RNA helicase A on GluR-B editing and splicing
RNA helicases have been suggested to be involved in pre-mRNA splicing, unwinding RNA structures, and also possibly disrupting RNA–protein complexes (reviewed in Linder et al. 2001; Schwer 2001). We analyzed whether RNA helicase A (RHA) can promote splicing of a 5′ splice site sequestered in a stable stem that also contains an editing site. Recombinant RHA was added to splicing reactions of the GRG SS transcript. The rate of splicing was enhanced by RHA (Fig. 3C ▶). The helicase was also observed to counteract the splicing inhibitory effect of ADAR2. When the transcript was preincubated with RHA, prior to editing and splicing, ADAR2 had no effect on splicing (Fig. 3C ▶). Further, preincubation with RHA decreased the amount of A to I editing (data not shown). These results suggest that RHA has the potential to disrupt the R/G stem, facilitating the binding of splice factors to the 5′ splice site and thereby preventing ADAR2 binding and editing.
Editing and splicing of GluR-B pre-mRNA transcribed by RNA polymerase II
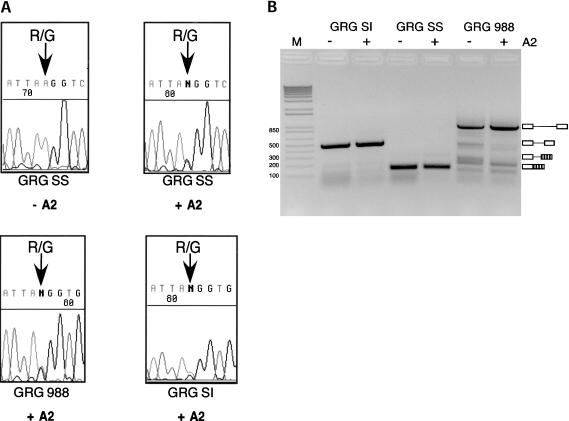
The GluR-B constructs were transfected into a 293 cell line, and editing and splicing of the transcribed GluR-B RNAs were analyzed. Editing and splicing of transcripts derived from the transfected vectors were defined as in vivo RNA processing. The editing/splicing construct GRG SS was cloned into a mammalian expression vector. Approximately 40 h after transfection, total RNA was extracted and the amount of editing was determined by direct sequencing of the RT-PCR products. No endogenous A to I editing of the GRG SS transcript was observed in the 293 cells (Fig. 4A ▶, top left). When an ADAR2 expression plasmid was cotransfected with the editing/splicing reporter construct, efficient R/G editing was observed as a heterogeneous A and G peak in the chromatogram after sequencing (Fig. 4A ▶, top right). As a control, the efficiency of R/G editing in the wild-type GRG 988 and GRG SI constructs was also investigated. Editing of these transcripts was in the same range as the GRG SS transcript (Fig. 4A ▶, bottom panels). In vivo splicing efficiency of the transcripts containing the R/G editing site was determined as described in Materials and Methods. The GRG SS transcript was efficiently spliced, while splicing of the wild-type GluR-B constructs (GRG 988 and SI) could not be detected (Fig. 4B ▶).
FIGURE 4.
(A) ADAR2 edits reporter constructs at the R/G site in vivo. Transfection of 293 cells with editing/splicing reporter constructs with or without ADAR2 expression vector. Total RNA was isolated and RT-PCR products corresponding to either spliced or nonspliced RNA were sequenced. The top left sequence corresponds to a nonedited R/G site, indicated by an arrow. Cotransfection with ADAR2 expression vector results in editing seen as a mixed A and G peak at the R/G editing site in the top right and bottom chromatograms. (B) GRG SI and 988 pre-mRNAs were inefficiently spliced in vivo, whereas GRG SS RNA was only detectable as spliced mRNA. The degree of splicing was not affected by coexpression of ADAR2. RT-PCR products corresponding to pre-mRNA and spliced mRNA were separated on 2% agarose gel. Used for size determination was 1 kb PLUS marker (M) (Life Technologies™).
Splicing of the GRG SS transcript was not inhibited by ADAR2 coexpression (Fig. 4B ▶). Further, ADAR2 had no effect on the processing of the wild-type GRG 988 and shortened intron (GRG SI) transcripts. These results suggest that ADAR2 does not have a negative effect on splicing in vivo. Likewise, the efficient splicing of GRG SS did not significantly influence editing, as GRG SS was edited to the same extent as GRG SI and GRG 988 (Fig. 4A ▶).
DISCUSSION
Competing RNA processing events in vitro
Most A to I editing sites in mammalian pre-mRNAs have been shown to be situated near exon–intron or intron–exon borders. Frequently, intron sequence is also used to create a double-stranded RNA structure complementary to the edited site (Burns et al. 1997; Seeburg et al. 1998; Rueter et al. 1999; Reenan et al. 2000). However, because the ECS for single edited sites almost exclusively is situated in introns, editing has to happen before splicing; therefore, it is likely that these two RNA processing events interact with each other. One possible explanation for having editing complementary sequences situated within introns is the option to create a double-stranded structure without affecting the sequence of the open reading frame. However, this arrangement requires that editing and splicing are sequential, with editing occurring before splicing of the pre-mRNA.
A simple way to coordinate editing and splicing would be to prevent splicing to initiate before editing by sequestering the 5′ splice site in the double-stranded structure required for editing. After the editing reaction has occurred, the structure is disrupted to allow splicing. Pre-mRNA secondary structures have previously been shown to regulate splicing. The stem-loop structure at the 5′ splice site of exon 10 in the tau pre-mRNA has been shown to influence alternative splicing (Grover et al. 1999). Likewise, a dsRNA structure plays an important role promoting exon skipping of exon 7B of the hnRNP A1 pre-mRNA (Blanchette and Chabot 1997). In GluR-B pre-mRNA, the R/G editing site is situated only one nucleotide from the exon–intron junction in a stable duplex structure, which is an absolute requirement for A to I editing. To investigate if the splicing efficiency was affected by a stable stem structure in edited pre-mRNA, editing and splicing in the GluR-B pre-mRNA was analyzed. Our current data shows that the stem-loop, where the R/G editing site is situated, has a negative effect on splicing in vitro (Figs. 2B, 3A ▶ ▶). Deleting the sequence complementary to the 5′ splice site increased the rate of splicing substantially. Because splicing has been shown to occur cotranscriptionally (Bauren and Wieslander 1994; Beyer and Osheim 1988), editing must either occur as an earlier cotranscriptional event or by delaying splicing. Editing at the R/G site was analyzed in the context of efficient splicing in vitro. Addition of ADAR2 to the in vitro splicing reaction significantly reduced splicing (Fig. 3A ▶). One possible reason for this inhibition is that the ADAR2 interaction blocks binding of splicing factors and that the editing enzyme binds with a higher affinity than the splicing factors. ADAR2 binding has previously been shown to protect an area covering both the R/G editing site and the 5′ splice site (Ohman et al. 2000; Yi-Brunozzi et al. 2001). In the transcript where the R/G stem was disrupted by a 20 nucleotide deletion, ADAR2 had no effect on the rate of splicing (Fig. 3A ▶). These results indicate that splicing can be inhibited by ADAR2 binding, perhaps with the assistance of the stable stem-loop structure. The opposite effect was observed when interaction of the splicing machinery was allowed to occur before editing. Under these conditions no editing of the RNA was observed. Because this inhibition was not dependent on ATP, the binding of initial splice factors to the 5′ splice site rather than splicing as such, is sufficient to block editing (Fig. 3B ▶). Taken together, these observations imply that editing and splicing factors compete for the same binding site(s), and that their activities interfere when acting on an in vitro transcribed substrate. Further, editing has a priority over splicing when simultaneous editing and splicing is possible.
RNA helicase A may prevent ADAR2 binding and thereby facilitate splicing
A link between editing and splicing has previously been suggested in Drosophila melanogaster (Reenan et al. 2000). Mle, an RNA helicase, was shown to regulate splicing of the sodium channel para pre-mRNA, which is edited by the Drosophila ADAR2 homolog, dADAR. It was proposed that Mle disrupts the dsRNA structure required for editing to achieve removal of the downstream intron. Mutations in Mle led to altered splicing and editing of the para pre-mRNA. However, so far, no direct connections between an RNA helicase and the editing enzyme have been shown. In the present study, the mammalian homolog of Mle, RNA helicase A (RHA), was shown to facilitate in vitro splicing of a pre-mRNA containing the R/G stem-loop. Addition of RHA to the splicing reaction gave nearly a twofold increase in the efficiency of splicing (Fig. 3C ▶). It has generally been assumed that dsRNA helicases’ sole function is to unwind double-stranded RNA when acting on RNA. However, NPH-II a dsRNA helicase that belongs to the DExH RNA helicase family was recently shown to disrupt RNA–protein interactions in the splicing machinery (Jankowsky et al. 2001). Our initial results suggest that RHA can disrupt the R/G stem-loop and thereby prevent ADAR2 binding. It is possible that a splicing interference caused by ADAR2 can be overcome by RHA, by disrupting the interaction between ADAR2 and its substrate. Interestingly, transcription activators such as CBP and BRCA1 have been shown to be linked to the RNA pol II through an interaction with RHA, which is a part of the holoenzyme complex (Nakajima et al. 1997; Anderson et al. 1998; Aratani et al. 2001). It has still to be proven if RHA acts as a link between the transcription machinery and ADAR2, allowing editing to occur before splicing.
Editing and splicing are coordinated in vivo
Increasing evidence shows that posttranscriptional RNA processing events are tightly coupled to the transcription machinery of RNA polymerase II. The C-terminal domain, CTD, of the large subunit of RNA pol II has been shown to support capping, splicing, and 3′ end formation and function as a platform for RNA processing factors (reviewed in Bentley 1999; Hirose and Manley 2000; Maniatis and Reed 2002). So far, no direct interaction between RNA editing and the CTD and/or other RNA processing factors have been reported. However, A to I editing activity has been shown to be associated with large nuclear ribonucleoprotein (lnRNP) particles, previously demonstrated to contain splicing factors (Raitskin et al. 2001). To analyze editing and splicing in the context of ongoing transcription, the R/G editing/splicing construct was analyzed in transfected cells with or without coexpressed ADAR2. In vivo, R/G editing was not affected by the efficient splicing of the major late fusion construct (Fig. 4A ▶, right). Further, coexpressing ADAR2 did not inhibit splicing of the R/G reporter construct (Fig. 4B ▶).
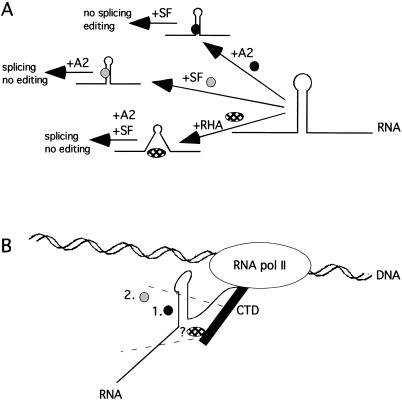
This suggests that editing and splicing are coordinated to happen sequentially during RNA pol II transcription. One possible reason why this coordination does not occur in vitro is that cotranscriptional RNA processing is not possible, and therefore editing and splicing compete for binding. Another possibility is that RNA pol II transcription is required for the coordination, mediated by the CTD. Preliminary data suggest that this coordination is lost during transcription with a RNA polymerase II lacking the CTD region. Initial data suggest that the efficient splicing prevents editing in this system (E. Bratt, G. Geiger, D. Bentley, and M. Öhman, in prep). We propose a model for how the editing and splicing linkage of the R/G substrate is different in vitro and in vivo (Fig. 5 ▶). In vitro, there is a competition between splicing factors and ADAR2 for binding to the presynthesized precursor RNA. Destabilization of the R/G stem either by deletion or by addition of the RNA helicase A (RHA) favor(s) splicing of intron 13, whereas binding of ADAR2 interferes with splicing (Fig. 5A ▶). These processes seem to be coordinated rather than competing in vivo (Fig. 5B ▶). Editing at the R/G site is not reduced in transcripts that are efficiently spliced, indicating that editing has priority over splicing. Other RNA processing events have been shown to be coordinated by the large subunit of RNA pol II. It is, therefore, possible that the CTD of RNA pol II acts as a platform for editing and splicing factors. One candidate enzyme for mediating the coordination of the two RNA processing events is RHA, which has previously been shown to mediate the interaction of transcriptional activators and the CTD. However, further experiments to fully elucidated the relationship of the splicing machinery with ADAR2, RHA, and the CTD of RNA pol II needs to be performed.
FIGURE 5.
Model of editing and splicing of GluR-B pre-mRNA in vitro and in vivo. (A) ADAR2 (black circle) competes with splicing factors (SF, gray circle) for binding to the R/G stem-loop in vitro. Disruption of the stem-loop by RNA helicase A (RHA, checked oval) facilitates the binding of splicing factors and thereby enhances splicing. (B) In vivo, editing and splicing is coordinated, possibly by the CTD (black rectangle) of RNA pol II. RHA might be the link between RNA processing factors and the polymerase. Editing (1.) and splicing (2.) are sequential events occurring in vivo.
MATERIALS AND METHODS
Plasmids and deoxyoligonucleotides
Exon 13, intron 13, and exon 14 (flop) of rat glutamate receptor subunit B (pGRG 988) were inserted into pGEM-TA cloning vector (Promega). The intron was shortened using PCR, facilitating in vitro transcription and in vitro splicing (pGRG SI). In the pGRG SS construct the 5′ splice site of GRG SI and 80 bp of intronic sequence containing the ECS were fused to a modified adeno major late pre-mRNA 3′ splice site including a branch point adenosine, polypyrimidine tract, 3′ splice site, and a small exon. The pTA10 vector containing the adeno major late pre-mRNA was a generous gift from Professor M. Garcia-Blanco, Duke University. The 20 nucleotide deletion (Δ20) of the editing complementary sequence of the R/G site was introduced by site-directed mutagenesis using the QuikChange™ kit (Stratagene). In the Glu5′SS construct the 5′ exon, splice site, and 5′ part of the intron of adenoviral pre-mRNA was replacing the GluR-B exon 13, 5′ splice site, and 80 bp of the intron 13 including the ECS. For in vivo expression of the editing and/or splicing competent constructs, a mammalian expression vector pcDNA3 was used, modified to contain a FLAG tag. For expression of ADAR2 in mammalian cells, a sequence containing rat ADAR2a was cloned into the pcDNA3 expression vector (Ohman et al. 2000).
Deoxyoligonucleotides used in this study were: GluR/G RT (5′-attgttatactattccaccc-3′); GluR/G exon 14 (5′-tttagtactgcgaggttaac cgc-3′); pTA10 exon 2 (5′-gagaccgcgaagagtttgtc-3′); GluR/G A (5′-ccttgtgacaccatgaaagtggg-3′); SP6 (5′-atttaggtgacactatag-3′); T7 (5′-taatacgactcactataggg-3′). All deoxyoligonucleotides were synthesized and purified by HPLC by Thermo Hybaid Interactiva Division, Germany.
In vitro splicing
In vitro transcripts were made using 20 units T7 RNA polymerase, 1 μg linearized plasmid, T7 transcription buffer supplemented with DTT, and 40 units RNaseOUT (Life Technologies), ribonucleotide mixture, α32P-CTP 800 Ci/mmole and G(5′)ppp(5′)G cap analog to generate capped and uniformly labeled pre-mRNA. After 2 h of transcription, the plasmid was digested with DNase and unincorporated nucleotides were removed using a G25 spin column (Amersham Bioscience). The RNA was gel purified on a 6% polyacrylamide/7 M urea/1×TBE gel and eluted from gel slice overnight in 500 μL crush and soak (0.5 M ammonium acetate, 1 mM EDTA, 0.1% SDS), followed by acid phenol (Ambion) extraction, chloroform extraction, and ethanol and sodium acetate precipitation. The RNA pellet was resuspended in water.
In vitro splicing reactions (20 μL) contained 20 fmole pre-mRNA, 1.2 mM MgCl2, 1 μg yeast tRNA, 4 mM DTT, 40 units RNaseOUT™ (Invitrogen), 1.5 mM ATP, 5 mM phosphocreatine, and 8 μL HeLa nuclear extract (4C Computer Cell Culture Center, Belgium, prepared according to Dignam et al. 1983). When indicated, 50 ng of recombinant rat ADAR2a (Ohman et al. 2000) or 40 ng of recombinant RNA helicase A, RHA, generously provided by Professor C.-G. Lee, New Jersey Medical School, was added. The splicing reactions were incubated at 30°C for 2 h or as indicated. After treatment with proteinase K, the samples were ethanol precipitated and resuspended in formamide gel loading dye or water. Splicing products were analyzed either by denaturing gel electrophoresis (6% polyacrylamide/7 M urea/1×TBE gel) or RT-PCR (see in vivo editing and splicing). Splicing efficiency was defined as (mRNA/[mRNA+precursor] × 100). For RT-PCR analysis of in vitro splicing, a total of 50 fmole of RNA was used and the splicing reactions were increased proportionally.
In vitro editing
GRG SI (500 fmole) or GRG SS pre-mRNA was edited by 125 ng recombinant rat ADAR2a as previously described (Ohman et al. 2000). Editing was detected using a limited primer extension; 1 pmole of 5′ end-labeled RT deoxyoligonucleotide and 500 fmole of edited GRG SI or GRG SS RNA in AMV-RT buffer (Boehringer Mannheim) was denatured for 10 min at 70°C and thereafter allowed to anneal for 3 h at 55°C. After the annealing, 1 μL dNTP/ddNTP mix (100 μM dATP, 100 μM dCTP, 100 μM dGTP, and 2.5 mM ddTTP) and 4.5 units AMV-RT (Boehringer Mannheim) were added and incubated for 45 min at 42°C. Reaction was stopped by addition of formamide gel loading dye (88% deionized formamide, 10 mM EDTA, 0.1% bromphenol blue, and 0.1% xylene cyanol) and extended products resolved on 20% polyacrylamide/7 M urea/1×TBE gel. The amount of editing was calculated by dividing the radioactivity associated with edited bands by the sum of the radioactivity in the nonedited and edited bands, using a FLA-3000, Fujifilm.
In vivo editing and splicing
Cell line 293, generously provided by Professor Akusjarvi, Uppsala University, was used for transient cotransfections of editing reporter vector with either rADAR2a expression vector or control vector. Transfections were performed using the DEAE-dextran method as described (Ausubel et al. 1987). A subconfluent 60-mm Petri dish was transfected with totally 2 μg DNA in 2 mL transfection media (2.5% newborn calf serum in DMEM, 100 μM chloroquine, and 100 μg/mL DEAE-dextran) and incubated for 3 h at 37°C, 5% CO2. The transfection media was removed and the cells were permeabilized for 5 min with 10% DMSO in PBS. The cells were rinsed with PBS and normal media (10% newborn calf serum in DMEM supplemented with L-glutamine and penicillin/streptomycin) was added. RNA isolation was performed after approximately 40 h using GenElute™ mammalian total RNA isolation kit (Sigma).
RT-PCR
RNA isolated from one dish/construct or 50 fmole RNA from in vitro splicing reactions were used for first-strand synthesis (cDNA) using 15 pmole SP6 deoxyoligonucleotide and MMLV-RT (Life Technologies) according to manufacturer’s instructions. Clontech advantage cDNA polymerase mix was used for PCR amplification, using SP6 and GluR/G A deoxyoligonucleotides, according to the manufacturer’s instructions. A quantitative PCR investigating the relation between spliced and nonspliced transcripts revealed that the amount of spliced material and the ratio between pre-mRNA and mRNA was constant from 15 to 35 cycles of PCR. For amplification of products after reverse transcription 25 to 35 cycles of PCR was used. PCR products corresponding to pre-mRNA and mRNA were isolated from a 2% agarose gel using av QIAQuick gel extraction kit (Qiagen). The PCR products were precipitated and sequenced using a DYEnamic™ ET terminator cycle sequencing kit (Amersham Biosciences). Splicing efficiency was defined as (mRNA/[mRNA+precursor] × 100), by measurements of intensities of spliced and nonspliced PCR products using BioRad Geldoc equipment.
Acknowledgments
We are grateful to professors C.-G. Lee, New Jersey Medical School, for the gift of recombinant RNA helicase A, G. Akusjarvi, Uppsala University, for supplying 293 cells, and M. Garcia-Blanco, Duke University Medical Center, for the pTA10 vector. This work was supported by grants from Swedish Natural Science Research Council, Carl Tryggers Stiftelse, Magnus Bergvalls Stiftelse, and Åke Wibergs Stiftelse.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2750803.
REFERENCES
- Anderson, S.F., Schlegel, B.P., Nakajima, T., Wolpin, E.S., and Parvin, J.D. 1998. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 19: 254–256. [DOI] [PubMed] [Google Scholar]
- Aratani, S., Fujii, R., Oishi, T., Fujita, H., Amano, T., Ohshima, T., Hagiwara, M., Fukamizu, A., and Nakajima, T. 2001. Dual roles of RNA helicase A in CREB-dependent transcription. Mol. Cell Biol. 21: 4460–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K., eds. 1987. Current protocols in molecular biology. John Wiley and Sons, New York.
- Bauren, G. and Wieslander, L. 1994. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192. [DOI] [PubMed] [Google Scholar]
- Bentley, D. 1999. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11: 347–351. [DOI] [PubMed] [Google Scholar]
- Beyer, A.L. and Osheim, Y.N. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev. 2: 754–765. [DOI] [PubMed] [Google Scholar]
- Blanchette, M. and Chabot, B. 1997. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA 3: 405–419. [PMC free article] [PubMed] [Google Scholar]
- Burnashev, N., Monyer, H., Seeburg, P.H., and Sakmann, B. 1992. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron 8: 189–198. [DOI] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., and Emeson, R.B. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308. [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, A.P. and Keller, W. 2001. RNA editing by base deamination: More enzymes, more targets, new mysteries. Trends Biochem. Sci. 26: 376–384. [DOI] [PubMed] [Google Scholar]
- Ghosh, S. and Garcia-Blanco, M.A. 2000. Coupled in vitro synthesis and splicing of RNA polymerase II transcripts. RNA 6: 1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover, A., Houlden, H., Baker, M., Adamson, J., Lewis, J., Prihar, G., Pickering-Brown, S., Duff, K., and Hutton, M. 1999. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J. Biol. Chem. 274: 15134–15143. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Maas, S., Single, F.N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R., and Seeburg, P.H. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Single, F.N., Kohler, M., Sommer, B., Sprengel, R., and Seeburg, P.H. 1993. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron–exon structure determines position and efficiency. Cell 75: 1361–1370. [DOI] [PubMed] [Google Scholar]
- Hirose, Y. and Manley, J.L. 2000. RNA polymerase II and the integration of nuclear events. Genes & Dev. 14: 1415–1429. [PubMed] [Google Scholar]
- Hume, R.I., Dingledine, R., and Heinemann, S.F. 1991. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science 253: 1028–1031. [DOI] [PubMed] [Google Scholar]
- Jankowsky, E., Gross, C.H., Shuman, S., and Pyle, A.M. 2001. Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science 291: 121–125. [DOI] [PubMed] [Google Scholar]
- Keegan, L.P., Gallo, A., O’Connell, M.A. 2001. The many roles of an RNA editor. Nat. Rev. Genet. 2: 869–878. [DOI] [PubMed] [Google Scholar]
- Kim, U., Wang, Y., Sanford, T., Zeng, Y., and Nishikura, K. 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. 91: 11457–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.G. and Hurwitz, J. 1992. A new RNA helicase isolated from HeLa cells that catalytically translocates in the 3′ to 5′ direction. J. Biol. Chem. 267: 4398–4407. [PubMed] [Google Scholar]
- Linder, P., Tanner, N.K., and Banroques, J. 2001. From RNA helicases to RNPases. Trends Biochem. Sci. 26: 339–341. [DOI] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J.R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., and Seeburg, P.H. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266: 1709–1713. [DOI] [PubMed] [Google Scholar]
- Maas, S. and Rich, A. 2000. Changing genetic information through RNA editing. Bioessays 22: 790–802. [DOI] [PubMed] [Google Scholar]
- Maas, S., Melcher, T., Herb, A., Seeburg, P.H., Keller, W., Krause, S., Higuchi, M., and O’Connell, M.A. 1996. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J. Biol. Chem. 271: 12221–12226. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Melcher, T., Maas, S., Herb, A., Sprengel, R., Seeburg, P.H., and Higuchi, M. 1996. A mammalian RNA editing enzyme. Nature 379: 460–464. [DOI] [PubMed] [Google Scholar]
- Nakajima, T., Uchida, C., Anderson, S.F., Lee, C.G., Hurwitz, J., Parvin, J.D., and Montminy, M. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90: 1107–1112. [DOI] [PubMed] [Google Scholar]
- Ohman, M., Kallman, A.M., and Bass, B.L. 2000. In vitro analysis of the binding of ADAR2 to the pre-mRNA encoding the GluR-B R/G site. RNA 6: 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitskin, O., Cho, D.S., Sperling, J., Nishikura, K., and Sperling, R. 2001. RNA editing activity is associated with splicing factors in lnRNP particles: The nuclear pre-mRNA processing machinery. Proc. Natl. Acad. Sci. 98: 6571–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan, R.A., Hanrahan, C.J., and Barry, G. 2000. The mle(napts) RNA helicase mutation in Drosophila results in a splicing catastrophe of the para Na+ channel transcript in a region of RNA editing. Neuron 25: 139–149. [DOI] [PubMed] [Google Scholar]
- Rueter, S.M., Dawson, T.R., and Emeson, R.B. 1999. Regulation of alternative splicing by RNA editing. Nature 399: 75–80. [DOI] [PubMed] [Google Scholar]
- Schwer, B. 2001. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 8: 113–116. [DOI] [PubMed] [Google Scholar]
- Seeburg, P.H., Higuchi, M., and Sprengel, R. 1998. RNA editing of brain glutamate receptor channels: Mechanism and physiology. Brain Res. Brain Res. Rev. 26: 217–229. [DOI] [PubMed] [Google Scholar]
- Sommer, B., Kohler, M., Sprengel, R., and Seeburg, P.H. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19. [DOI] [PubMed] [Google Scholar]
- Yi-Brunozzi, H.Y., Stephens, O.M., and Beal, P.A. 2001. Conformational changes that occur during an RNA-editing adenosine deamination reaction. J. Biol. Chem. 276: 37827–37833. [DOI] [PubMed] [Google Scholar]