Abstract
Archaeal RNA splicing involves at least two protein enzymes, a specific endonuclease and a specific ligase. The endonuclease recognizes and cleaves within a characteristic bulge-helix-bulge (BHB) structure formed by pairing of the regions near the two exon–intron junctions, producing 2‘,3′-cyclic phosphate and 5′-hydroxyl termini. The ligase joins the exons and converts the cyclic phosphate into junction phosphate. The ligated product contains a seven-base hairpin loop, in which the splice junction is in between the two 3′ terminal residues of the loop. Archaeal splicing endonucleases are also involved in rRNA processing, cutting within the BHB structures formed by pairing of the 5′ and 3′ flanking regions of the rRNAs. Large free introns derived from pre-rRNAs have been observed as stable and abundant circular RNAs in certain Crenarchaeota, a kingdom in the domain Archaea. In the present study, we show that the cells of Haloferax volcanii, a Euryarchaeote, contain circular RNAs formed by 3′,5′-phosphodiester linkage between the two termini of the introns derived from their pre-tRNAs. H. volcanii ligase, in vitro, can also circularize both endonuclease-cleaved introns, and non-endonuclease-produced substrates. Exon joining and intron circularization are mechanistically similar ligation reactions that can occur independently. The size of the ligated hairpin loop and position of the splice junction within this loop can be changed in in vitro ligation reactions. Overall, archaeal RNA splicing seems to involve two sets of two symmetric transesterification reactions each.
Keywords: Archaea, bulge-helix-bulge structure, C/D box small RNA, circular RNA, cyclic phosphate, RNA ligase, rRNA processing, splicing endonuclease
INTRODUCTION
Intron-containing genes have been observed in Archaea, Bacteria, and Eukarya, the three domains of life (Woese et al. 1990). There are two major kingdoms in the domain Archaea: Euryarchaeota and Crenarchaeota (Woese et al. 1990). Introns have been found in the tRNA genes of both kingdoms (Kaine et al. 1983; Daniels et al. 1985, 1986; Kaine 1987; Wich et al. 1987; Datta et al. 1989; Kjems et al. 1989a,b). Indeed, introns have been observed in the tRNA genes of all published archaeal genomes. Introns have also been found in the 16S and 23S rRNA genes of some Crenarchaeota (Kjems and Garrett 1985, 1991; Dalgaard and Garrett 1992; Burggraf et al. 1993; Itoh et al. 1998; Nomura et al. 1998). Some of these rRNA gene introns are large and contain open reading frames (ORF) that can encode LAGLIDADG amino acid motifs containing homing endonucleases. Recently, mRNA splicing has also been reported in the Crenarchaeota (Watanabe et al. 2002).
At least two separate protein enzymes, an endonuclease and a ligase, are known to be involved in RNA splicing in Archaea. All archaeal introns contain a bulge-helix-bulge (BHB) structure (Figs. 1 A,D, ▶ 5A ▶), formed by pairing of the regions near the two intron–exon junctions. The splicing endonucleases recognize these BHB structures and cleave each of the two strands between the middle and 3′ residues of the three-nucleotide bulges that are separated by four base pairs (Kjems et al. 1989a; Thompson et al. 1989; Thompson and Daniels 1990; Lykke-Andersen and Garrett 1994, 1997). Archaeal splicing endonucleases are also involved in rRNA processing, cleaving within the BHB structures present in the processing helices formed by pairing of the 5′ and 3′ flanking regions of the rRNAs (Durovic and Dennis 1994; Dennis et al. 1998; Ciammaruconi and Londei 2001; Tang et al. 2002).
FIGURE 1.
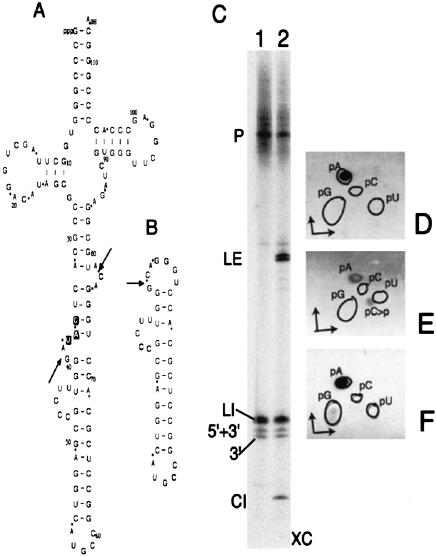
Sequences of pre-tRNATrp (A), elongator pre-tRNAMet (D), and their spliced products: ligated exons (B and E) and circularized introns (C and F). Arrowheads in A and D denote the splice sites within the bulge-helix-bulge (BHB) motifs. Anticodons are highlighted. Asterisks in B, C, E, and F denote the splice junctions. Positions corresponding to the oligonucleotides used for RT-PCR (Figs. 2B, 3A ▶ ▶) and Northern hybridizations (Fig. 4 ▶) are labeled and marked by the lines ending in half arrows. Bases within parentheses in C indicate the changes in various synthetic intron-like transcripts (Fig. 8 ▶).
FIGURE 5.
Splicing of truncated elongator pre-tRNAMet. Sequences of the precursor (A) and its circularized intron (B). The anticodon is highlighted in A. Arrows indicate the splice sites within the BHB motif in A and the junction in B. Phosphates derived from [α-32P]ATP labeling of the precursor are denoted by asterisks in A and B. Products of endonuclease (C, lane 1) followed by ligase (C, lane 2) using [α-32P]ATP-labeled precursor are separated on a 50-cm long, denaturing 15% gel. (P) Precursor; (LI) linear intron; (5′+3′) two exons together; (3′) only 3′ exon; (LE) ligated exons; (CI) circular intron; (XC) xylene cyanol. The gel-eluted precursor and its intron products (shown in C) were digested with nuclease P1 and resolved by TLC (D–F) using solvent a in the first dimension and solvent b in the second dimension. (D) Precursor (P). (E) Linear intron (LI). (F) Circular intron (CI). Outlines indicate the positions of nonradioactive markers in D–F.
Archaeal RNA splicing endonucleases and eukaryal tRNA splicing endonucleases are homologous, although the mechanisms of their substrate recognition are somewhat different (Di Nicola Negri et al. 1997; Kleman-Leyer et al. 1997; Lykke-Andersen and Garrett 1997; Trotta et al. 1997; Abelson et al. 1998; Li et al. 1998; Trotta and Abelson 1999). Eukaryal endonuclease can even recognize and cleave an archaeal BHB motif containing synthetic RNA substrate (Fabbri et al. 1998; Fruscoloni et al. 2001). It is suggested that eukaryal enzymes cleave their substrates after forming a structure resembling BHB (Fruscoloni et al. 2001).
Cleavage by archaeal endonuclease within the BHB structure followed by ligation of the exons forms a hairpin loop of seven residues, with the splice junction being between the 3′ ultimate and penultimate residues of the loop (Fig. 1B,E ▶). Potentially, two such hairpin loops of seven residues each can be formed after two cleavages within the BHB structure followed by two ligations, one joining the two exons and the other circularizing the intron (Fig. 1B,C,E,F ▶). This can occur because archaeal endonucleases cut symmetrically within the bulges of the BHB structure and produce 2‘,3′-cyclic phosphate and 5′-hydroxyl termini of the exons as well as the introns (Thompson and Daniels 1988; Thompson et al. 1989; Zofallova et al. 2000). The ligase converts the terminal cyclic phosphate into linking phosphate at the splice junction (Zofallova et al. 2000), which is similar to the action of tRNA splicing ligase present in animal cells (Nishikura and De Robertis 1981; Filipowicz and Shatkin 1983; Filipowicz et al. 1983; Laski et al. 1983; Perkins et al. 1985). Normally, intron products of the cleavage are quickly degraded and are not present in the cell in significant amounts (Kjems and Garrett 1991; Dalgaard and Garrett 1992). However, large ORF-containing introns derived from the rRNA genes of some Crenarchaeota are maintained as stable and abundant circles (Kjems and Garrett 1988; Dalgaard and Garrett 1992; Burggraf et al. 1993). In these cases, RNA splicing produces ligated exons (spliced rRNAs) as well as circularized introns. Therefore, the questions are whether other archaeal introns are also converted into circles after splicing, and whether archaeal RNA splicing in general produces two different ligated products.
The genes for tRNATrp and elongator tRNAMet of Haloferax volcanii, a Euryarchaeote, contain introns (Daniels et al. 1985; Datta et al. 1989). Pre-tRNATrp of H. volcanii also contains C/D box small RNA features in its intron and corresponding residues for ribose methylation in its exon regions (Omer et al. 2000; Dennis et al. 2001; d’Orval et al. 2001). We report here that circular introns are formed, both in vivo and in vitro, during tRNA splicing in H. volcanii. The mechanism of circularization of introns is the same as that reported for the ligation of exons (Gomes and Gupta 1997; Zofallova et al. 2000). The ligations of introns and exons are independent of one another. The ligase can circularize both endonuclease-cleaved introns and non-endonuclease-produced substrates. We also show that, in vitro, the size of the loop formed after ligation can be varied from the natural size of seven residues, and the splice junction need not be between the last two 3′ residues of the ligated loop.
RESULTS
Splicing reactions in vitro produce two different products: Ligated exons and circularized introns
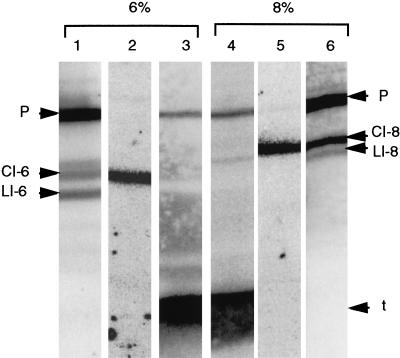
Sequences of the intron-containing precursors of tRNATrp and elongator tRNAMet, and their spliced products are shown in Figure 1 ▶. Splicing ligation reactions of pre-tRNATrp showed two new products beyond those observed for the endonuclease reactions (Fig. 2A ▶). One of these, as expected, is ligated exons or mature tRNA (Fig. 2A ▶, LE). The other ligated product has distinctly different mobility during polyacrylamide gel electrophoresis (PAGE) < 6% and 8% acrylamide concentrations (Fig. 2A ▶, CI-6 and CI-8). This anomalous mobility indicated that the product might be a circular RNA. The size of the RT-PCR product (Fig. 2B ▶) using this ligated product as the template with appropriate tRNATrp intron-specific primers was 95 bp, the predicted size of the derivative of the circularized intron. The circular nature of this intron was further confirmed by sequencing the RT-PCR product (Fig. 2C ▶, compare with sequence in Fig. 1C ▶). The sequence indicates that, during circularization of the intron, its two endonuclease-derived termini are joined precisely, with no addition, deletion, or misincorporation of any residue. A normal reverse transcription through the splice junction during RT-PCR further indicates that the two ends of the intron are ligated via a 3′,5′-phosphodiester linkage. Reverse transcriptase reaction can occur through a 2‘,5′-phosphodiester linkage (Lorsch et al. 1995), but it causes misincorporation of residues opposite this linkage (Vogel et al. 1997).
FIGURE 2.
Analyses of the ligase reaction products. (A) [α-32P]CTP labeled pre-tRNATrp transcripts (lanes 1 and 4) are used for endonuclease reactions (lanes 2 and 5) followed by ligase reactions (lanes 3 and 6). The products are resolved by denaturing PAGE. Lanes 1–3, 6% gel, and lanes 4–6, 8% gel. (P) Precursor; (LI) linear intron; (E) exons; (LE) ligated exons; (CI-6) circular intron in 6% gel; (CI-8) circular intron in 8% gel; (XC) xylene cyanol; (BB) bromophenol blue. (B) RT-PCR products using tRNATrp intron-specific primers H and I (Fig. 1C ▶), and gel-eluted circular introns (A, CI-6 and CI-8) as template are resolved by native 6% PAGE (lane 1). Lane 2 contains a 25-bp DNA ladder. (C) The 95-bp RT-PCR product (B, lane 1) is sequenced using primer I. Arrow denotes the position of the splice junction (asterisk in Fig. 1C ▶) in the sequence of the circularized intron.
Two termini of the introns can also join in vivo, producing circular introns
RT-PCR with H. volcanii total RNA and corresponding primers were done to determine the in vivo presence of the intron-containing precursors of tRNATrp and elongator tRNAMet, and their circularized intron products. Sizes of the RT-PCR products (Fig. 3A ▶) were as predicted. The sequencing (not shown) confirmed that the products in Figure 3A ▶, lanes 1 and 2, were derived from pre-tRNATrp and, in lanes 6 and 7, from elongator pre-tRNAMet. The sequence of the product in Figure 3A ▶, lane 5, indicated that it was derived from an RNA formed by end-to-end joining of the two termini of the intron of elongator tRNAMet (Fig. 3B ▶, compare with sequence in Fig. 1F ▶). This indicates that these introns are present in the cells as circles or as linear dimers/multimers. Similarly, the sequence of the product in Figure 3A ▶, lane 3 (not shown), was identical to that in Figure 2C ▶, which indicated that it was derived from the in vivo ligated (circularized or dimerized) intron of tRNATrp. As mentioned for the in vitro reactions, the junctions in these in vivo ligations of the introns are also formed by 3′,5′-phosphodiester linkages, with no addition or deletion of any residue.
FIGURE 3.
Determination of the in vivo presence of pre-tRNAs and circular introns. (A) RT-PCR products using H. volcanii total RNA and various primers are separated on a native 6% polyacrylamide gel. The primers (Fig. 1 ▶) used, expected sizes of the products, and the template regions corresponding to these products (thick lines in the schematic diagrams) are indicated below the lanes. Lane 4 contains a 25-bp DNA ladder. (B) The 69-bp RT-PCR product in A, lane 5, is sequenced using primer L. The arrow denotes the position of the splice junction (Fig. 1F ▶, asterisk) in the sequence of the circularized intron derived from elongator pre-tRNAMet.
Determination of whether the in vivo ligated products were circular monomers and not dimers was done by Northern analysis of total RNA derived from early log phase H. volcanii cells. Figure 4 ▶ shows the hybridization of tRNATrp intron- and exon-specific probes to the total RNA separated on 6% and 8% gels. The gels were intentionally overloaded with the RNA to detect the introns. Intron-specific probe hybridizes to the precursors and several intron-containing bands (lanes 1 and 6). Only one of these bands contained circular introns (lanes 2 and 5). The linear introns appear as two bands (LI-6 and a light band above CI-6 in lane 1), one of which comigrates with the circular intron band in 8% gel. (There are only two intron bands in lane 6 and CI-8 band in this lane is darker than CI-6 in lane 1.) However, the positions of circular and linear introns (103 b) between the precursor (about 180 b) and the tRNA (77 b) indicated that the cells contain both forms of introns as monomers. A linear or circular dimer of the intron would run slower than the precursor RNA. Although ligated intron derivatives of elongator pre-tRNAMet were detected by RT-PCR (Fig. 3 ▶, lane 5), Northern analysis for determining their presence in the cells was not very successful. The introns were barely detectable even after prolonged exposure to the phosphorimager screen, and linear and circular forms were not distinguishable from each other (data not shown). This indicates that free introns derived from elongator pre-tRNAMet are rapidly degraded.
FIGURE 4.
Northern analysis to detect in vivo presence of circular introns. Northern blots of H. volcanii total RNA separated by denaturing 6% (lanes 1–3) and 8% (lanes 4–6) PAGE are hybridized to 5′ 32P-labeled tRNATrp-specific oligonucleotides (marked in Figs. 1A,C). The oligonucleotides used are as follows: H (intron-specific) in lanes 1 and 6, N (circular intron-specific, overlapping splice junction region) in lanes 2 and 5, and B (exon-specific) in lanes 3 and 4. (P) Precursor; CI-6 and CI-8, circular introns in 6% and 8% gels, respectively; LI-6 and LI-8, linear introns in 6% and 8% gels, respectively; t, tRNA.
Circularization of introns and joining of exons are similar ligation reactions that can occur independent of each other
The junction phosphate during splicing of tRNA exons in H. volcanii is derived from the 2‘,3′-cyclic phosphate at the 3′ end of the 5′ exon (Zofallova et al. 2000). Nearest neighbor analysis using nuclease P1 was done to determine whether the junction phosphate in the circular intron was also derived from the 2‘,3′-cyclic phosphate at the 3′ end of the intron. A substrate used previously (Gomes and Gupta 1997; Zofallova et al. 2000) was again used for these splicing reactions. It is a truncated elongator pre-tRNAMet (Fig. 5A ▶) that contains a 36-base deletion (Fig. 1D ▶, positions 45–80) within its intron. Pre-tRNAMet and pre-tRNATrp containing full-sized introns could not be used in these reactions. In vitro-produced full-sized elongator pre-tRNAMet is not cleaved by the endonuclease (Gomes and Gupta 1997), a reaction necessary to produce introns needed for the ligation reaction, and the specific sequences at the intron–exon junctions of pre-tRNATrp are not suitable for the nearest neighbor analyses (Zofallova et al. 2000).
Ligation reactions of [α-32P]ATP-labeled truncated elongator pre-tRNAMet (Fig. 5C ▶) show a product, apparently a circular intron (CI in lane 2, Fig. 5C ▶) that migrates faster than the linear introns and exons. All products of splicing reactions (Fig. 5C ▶) were analyzed by thin layer chromatography (TLC) after nuclease P1 digestion (some not shown). As expected, the precursor produced only pA as the labeled product (Fig. 5D ▶), which was derived from the [α-32P]ATP in the in vitro transcription reaction. The digests of linear intron showed small amounts of labeled pC>p (>p indicates 2‘,3′-cyclic phosphate) along with the labeled pA (Fig. 5E ▶). This pC>p is derived from the 3′ end of the intron produced by cleavage of the precursor (Fig. 5A ▶). The >p in this pC>p is labeled, which is derived from the labeled phosphate preceding the A at the 3′ splice site of the precursor. Joining together of the two ends of the intron would convert labeled cyclic phosphate of the pC>p into labeled junction phosphate between C and G residues at the two termini of the intron (Fig. 5B ▶), if the mechanism of circularization of the intron is the same as that for the ligation of exons (Zofallova et al. 2000). Nuclease P1 (Fig. 5F ▶) digest of the apparently circular intron (Fig. 5C ▶, CI) does indeed show some labeled pG in addition to the labeled pA. This transfer of labeled phosphate to the 5′ side of a G indicates that the ligation reactions for joining exons and circularizing introns are similar.
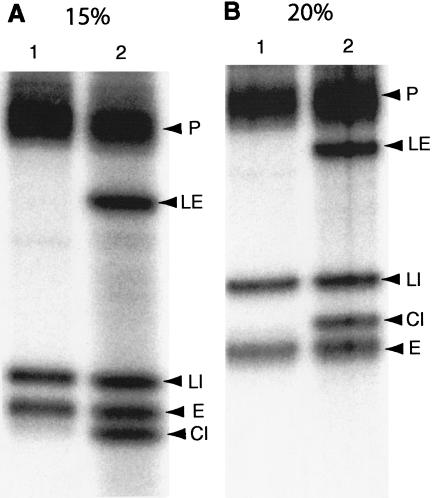
The faster migration of the ligated (apparently circular) intron product than its corresponding linear intron (Fig. 5C ▶) is the reverse of what is seen for the 103-b intron of pre-tRNATrp (Fig. 2A ▶). This may be due to its small size and compact structure (Fig. 5B ▶); 11–13 base pairs can be formed within the 39-b circle. Alternatively, it could be a linear product of <39 bases derived from the 39-b circular intron by specific action of some component present in our ligase preparations. To distinguish between these two possibilities, we resolved the reactions similar to those shown in Figure 5C ▶ by denaturing 15%, 20%, and 25% PAGE. Again, the band corresponding to CI in Figure 5C ▶ migrates faster than both linear introns and exons in 15% gel (Fig. 6A ▶). However, it runs in between the linear introns and exons in 20% gel (Fig. 6B ▶). The alteration in mobility of this RNA by the acrylamide concentration indicates that this RNA indeed is a 39-b circular intron. Even in 25% gels (not shown), this circular intron migrated between the linear intron and exons, although it was much closer to the linear intron than that observed in 20% gel.
FIGURE 6.
Analyses of splicing products of truncated elongator pre-tRNAMet by different concentration gels. Reactions similar to those shown in Figure 5C ▶ using [α-32P]CTP-labeled transcripts were resolved on 20-cm long, denaturing 15% (A) and 20% (B) gels. Lane 1, endonuclease products; lane 2, ligase products. (P) Precursor; (LE) ligated exons; (LI) linear intron; (CI) circular intron; (E) exons.
Two bands for the 3′ exons of this truncated elongator pre-tRNAMet substrate (Fig. 5C ▶) in the long 15% gels have also been observed previously (Zofallova et al. 2000). Sometimes in vitro-generated T7 RNA polymerase transcripts contain an extra non-template-coded residue at their 3′ ends. This may explain the presence of two bands for the 3′ exons. These 3′ exons, when ligated to the 5′ exons, do appear as a doublet (Fig. 5C ▶, LE, lane 2).
Normally, joining of exons and circularization of introns occur together. However, gel-purified exons and introns can also be ligated in separate reactions (Fig. 7 ▶). This indicates that each of the two ligation reactions, at least in vitro, can occur independently.
FIGURE 7.
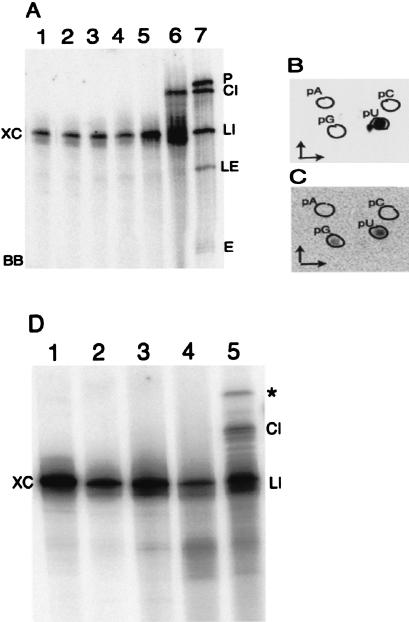
Ligase reactions using purified substrates. Gel-eluted endonuclease products of [α-32P]CTP labeled pre-tRNATrp are used in ligase reactions. The products are separated by denaturing 6% PAGE. Lane 1, Pre-tRNA; lane 2, standard endonuclease reaction; lane 3, standard ligase reaction; lanes 4 and 6, gel-eluted linear introns and exons, respectively; lanes 5 and 7, ligase reactions using gel-eluted linear introns and exons, respectively. (P) Precursor; (LI) linear intron; (E) exons; (LE) ligated exons; (CI) circularized intron; (XC) xylene cyanol; (BB) bromophenol blue.
Ligase in vitro can join non-endonuclease-produced termini
Independent ligation of the introns and exons indicated the possibility of conducting ligation reactions using substrates that have the proper termini but are produced by endonuclease-independent methods. A [α-32P]UTP-labeled 103-b transcript comparable to the intron of pre-tRNATrp (except for the presence of G instead of A at position 1) was used in these reactions. A combination of enzymatic and chemical procedures (see Materials and Methods) were used to produce a hydroxyl at its 5′ end and a 2‘,3′-cyclic phosphate at its 3′ end. This modified transcript was circularized by the ligase, just as the endonuclease-produced introns were (Fig. 8A ▶, cf. CI in lanes 6 and 7). A 2‘,3′-cyclic phosphate at the 3′ end was essential; a noncyclic 3′ phosphate was not a substrate for the ligase (Fig. 8A ▶, lane 4).
FIGURE 8.
Ligation of synthetic intron-like transcripts. (A): [α-32P]UTP-labeled synthetic tRNATrp intron transcripts are modified to create proper termini and then used as substrate for the ligase reactions. The products are analyzed on a denaturing 8% polyacrylamide gel. Lane 1, transcript; lane 2, dephosphorylated transcript; lane 3, dephosphorylated transcript after periodate oxidation and amine cleavage; lane 4, ligase reaction using substrate shown in lane 3; lane 5, substrate shown in lane 3 treated with RNA 3′ terminal phosphate cyclase; lane 6, ligase reaction using substrates shown in lane 5; lane 7, standard ligase reaction using endonuclease treated pre-tRNATrp as substrate. (LI) Linear intron and intron-like substrate; (CI) circular RNA; (P) precursor; (E) exons; (LE) ligated exons; (XC) xylene cyanol; (BB) bromophenol blue. Gel-eluted products from A are digested with nuclease P1 and separated by TLC in B (LI, lane 1) and C (CI, lane 6) using solvent a in the first dimension and solvent c in the second dimension. Outlines indicate the positions of nonradioactive markers in B and C. (The labeled spot near pU in B is derived from an unidentified contaminant present in [α-32P]UTP; see Materials and Methods.) (D) Synthetic intron transcripts similar to those in A with a specific CC-to-AA change (Fig. 1C ▶) were used for ligation reactions like those shown in A. Lane 1, transcript; lane 2, dephosphorylated transcript; lane 3, dephosphorylated transcript after periodate oxidation and amine cleavage; lane 4, substrate shown in lane 3 treated with RNA 3′ terminal phosphate cyclase; lane 5, ligase reaction using substrates shown in lane 4. Ligated dimer of the substrate is indicated by an asterisk in D.
Ligation of these synthetic substrates is similar to the ligation of endonuclease-cleaved substrates. The terminal U (Fig. 1A ▶, position 141) in these intron-like transcripts is deleted after the chemical procedures, and the newly produced 2‘,3′-cyclic phosphate is radioactive when [α-32P]UTPlabeled transcripts are used. During ligation, this cyclic phosphate becomes junction phosphate and is linked to the 5′ side of the G present at position 1. Nearest neighbor analysis confirms this. Nuclease P1 digestion of the transcript produces labeled pU (Fig. 8B ▶), whereas that of circular intron has an additional labeled pG (Fig. 8C ▶).
As mentioned earlier, the normal products of archaeal splicing ligation are hairpin loops of seven residues each because the normal substrates are endonuclease products and endonucleases cleave within a very specific BHB structure. However, the earlier-mentioned endonuclease-independent ligation of synthetic substrates produced a hairpin loop of six residues. A loop of seven residues retaining 3′ terminal U (Fig. 1A ▶, position 141) of the transcript would not produce labeled pG in Figure 8C ▶.
Similar ligation experiments (data not shown), after modifications of the ends of [α-32P]CTP-labeled 75-b transcripts comparable to the full-size introns of elongator pre-tRNAMet, also produced circular RNAs. (As mentioned earlier, these full-sized introns could not be produced by the endonuclease in vitro.) This 75-b circular intron migrates more slowly than the corresponding linear intron during PAGE. Nearest neighbor analysis of these circles (data not shown) also confirmed the transfer of labeled phosphate to the 5′ - end G residue of the transcript.
In order to determine the possibility of increasing the size of the ligated hairpin loop, similar endonuclease-independent ligations were done using [α-32P]UTP-labeled pre-tRNATrp intron-like transcripts that had specific CC residues changed to AA (Fig. 1C ▶). These transcripts had the potential to form 12 base loops (again, the 3′ terminal U is deleted) after proper modifications and ligation. (It could be a 15-base loop if an alternate folding to that in Fig. 1A ▶, and correspondingly in Fig. 1C ▶, is considered, where U44–A133 and G45–C132 pairs are replaced by one G45–C131 pair.) As seen in Figure 8D ▶ (CI in lane 5), these were also circularized. Nearest neighbor analyses (not shown) confirmed proper ligation of these substrates. The splice junction in the 12-base loop was not between the two 3′ terminal residues of the loop, but was between the seventh and eighth residue from the 5′ side. In these reactions, a new product (Fig. 8D ▶, asterisk in lane 5) appeared after ligation. It seems to be a linear dimer of the transcript because RT-PCR with this product and primers H and I (not shown) provided only a single-sized product of 94 bp. The length of this product is similar to the RT-PCR product of circular monomer derived from endonuclease-cleaved intron and the same primers (Fig. 2B ▶), except for it being shorter by 1 bp. (The substrate for the ligase reaction here is 102 b; see Materials and Methods.) RT-PCR of circular dimers under these conditions would have shown two products: one of 94 bp and the other of 196 bp.
DISCUSSION
As mentioned earlier, archaeal splicing endonucleases recognize and cleave the BHB structures present at the intron–exon junctions in all archaea. This creates a potential for forming two hairpin loops of seven residues each after two independent ligation reactions, one joining the two exons, the other circularizing introns. Large ORF-containing circular introns derived from rRNA genes have been observed in some Crenarchaeota (Kjems and Garrett 1988; Dalgaard and Garrett 1992; Burggraf et al. 1993). In some cases, these large introns are stably retained as linear monomers; but, there, too, trace amounts of their topoisomers (probably circles) with different mobility in two-dimensional denaturing agarose gels were observed (Nomura et al. 1998). In vitro circularization of a large intron derived from pre-23S rRNA of Desulfurococcus mobilis, a Crenarchaeote, using crude cell extract has also been reported (Kjems and Garrett 1988).
The two introns studied in the present work are comparatively small, do not seem to contain ORFs, and are derived from tRNA genes of a Euryarchaeote. Circular derivatives of these introns can be produced by in vitro splicing reactions (Figs. 2, 5, 6 ▶ ▶ ▶). Others have also observed circular intron-like products after treating pre-tRNATrp with H. volcanii S100 extracts (d’Orval et al. 2001). Our work further indicates that circular forms of the same two introns are also present in vivo (Figs. 3, 4 ▶ ▶).
Archaeal splicing endonuclease and ligase may also be involved in rRNA processing (Tang et al. 2002). BHB motifs are found in the processing stems of pre-16S and pre-23S rRNAs of several archaea. There is some evidence for the in vivo presence of two specific products of the processing of these pre-rRNAs in Archaeoglobus fulgidus, a Euryarchaeote, and Sulfolobus solfataricus, a Crenarchaeote; one product is linear and contains flanking regions of the rRNAs, whereas the other is circular and contains rRNAs and some flanking regions (Tang et al. 2002). We also have some evidence for the in vivo presence of circular intermediates of the pre-rRNA processing reactions in H. volcanii (F. Elazzouzi and R. Gupta, unpubl.).
Although joining of exons and circularization of introns may occur concurrently, they are separate and independent ligation reactions. This has been demonstrated by conducting ligation reactions using exons and introns separately (Fig. 7 ▶). However, the endonuclease cleavage simultaneously produces exons and introns. This is supported by the symmetric dimeric structure of the archaeal splicing endonucleases, and the structure of the BHB motif, where both bulges are on the same minor groove face of the central 4-bp helix (Diener and Moore 1998; Li et al. 1998; Li and Abelson 2000). These features indicate that both bulges could be cleaved after a single encounter with a splicing endonuclease (Diener and Moore 1998).
Normally, the endonuclease products, which have 5′-hydroxyl and 2‘,3′-cyclic phosphate termini, are the substrates for the ligase reactions. However, the ligase can also use the substrates that are produced independent of endonuclease cleavage (Fig. 8 ▶). Ligation of these synthetically produced substrates confirms that the termini required for the ligation (i.e., those produced by the endonuclease) indeed are 5′-hydroxyl and 2‘,3′-cyclic phosphate. The mechanisms of ligation of endonuclease-cleaved and synthetic substrates are the same.
The size of the seven-residue hairpin loop formed after ligation, and the position of the splice junction within this loop as between the last two 3′ residues, are normally fixed. These are not specified by the ligase, but are due to the specificity of the endonuclease cleavage within the BHB structure, which provides the substrates for the ligase reactions. In endonuclease-independent reactions, the size of the ligated hairpin loop can be changed from a normal of 7 bases to 6 bases, and to 12 to 15 bases (depending on the pairing within the intron) (Fig. 8 ▶). Furthermore, the position of the splice junction in the large loop can be changed from near its one end to about the middle. Dimers of introns have only been observed in the endonuclease-independent reactions when large hairpin loops are formed (Fig. 8D ▶). These dimers are not observed in in vivo (Fig. 4 ▶) and in vitro (Figs. 2A, 5C, 6, 7 ▶ ▶ ▶ ▶) reactions, when normal-sized hairpin loops are formed. Present experiments do not determine whether pairing between two strands (to form a hairpin) is required for the ligation, or ligase can join two completely unpaired strands.
It seems that overall archaeal RNA splicing involves two sets of two symmetric transesterification reactions each (Fig. 9 ▶). The endonuclease causes two symmetric cleavages of the 3′,5′-phosphodiester linkages between the middle and 3′ residues of the three nucleotides bulges in the two strands of the BHB structure, producing two 2‘,3′-cyclic phosphodiesters; one at the 3′ end of the 5′ exon and the other at the 3′ terminus of the intron. This is followed by two ligation reactions converting 2‘,3′-cyclic phosphodiesters to 3′,5′-phosphodiester linkages, one joining the two exons and other joining (circularizing) the two ends of the intron. The ligation reactions are also symmetric, creating the new junctions between the last two 3′ residues of newly formed seven-base hairpin loops. No external source of energy would be required in these reactions, which is the case for the H. volcanii in vitro ligation system (Gomes and Gupta 1997; Zofallova et al. 2000).
FIGURE 9.
A model for the reactions occurring during RNA splicing in Archaea. (A) Symmetric nature of the BHB-containing endonuclease substrate and two seven-base hairpin loops in ligase products. (B) Reactions involving specific phosphodiester linkages during RNA splicing. 1–9 and 1*–9* are residues involved in the formation of BHB in the substrates and hairpins in the products. Arrows in A indicate the splice sites in the substrate and products. The two phosphates (p1 and p2) and 2‘, 3′, and 5′ positions involved in the reactions are indicated. (E1 and E2) Two exons; (I) intron.
The earlier-mentioned model (Fig. 9 ▶) indicates that archaeal RNA splicing normally creates two products: one linear and one circular. One or both products that are useful for the cellular metabolism would be retained and the one that is not used would be degraded. The retained product may be processed further to convert it into a functional species. Mostly the linear products (spliced exons) are retained. Sometimes both circular and linear products are retained. One such case is of circular introns that contain ORFs for homing endonucleases and linear rRNAs derived from pre-rRNA splicing in certain Crenarchaeota (Kjems and Garrett 1988; Dalgaard and Garrett 1992; Burggraf et al. 1993). Another case is of the earlier-mentioned pre-rRNA processing at the BHB structure in the flanking regions of rRNAs of A. fulgidus and S. solfataricus (Tang et al. 2002); the circular products are processed further to produce mature rRNAs and the linear products appear to function as C/D box sRNAs (archaeal homologs of eukaryal snoRNAs). A third case is shown in this work, where the circular intron and mature tRNA derived from pre-tRNATrp are retained (Fig. 4 ▶). Therefore, although the production of circular intron by itself may not have biological significance, its retention in the cell may.
The free introns derived from the pre-tRNATrp are retained (Fig. 4 ▶), whereas those from the elongator pre-tRNAMet are degraded (data not shown) in the cell. This may be due to the presence of C/D box sRNA features in the tRNATrp introns. Ribose-methylation of two H. volcanii (and some other Euryarchaeota) tRNATrp residues appears to be C/D box RNA mediated (Omer et al. 2000; Dennis et al. 2001; d’Orval et al. 2001). These residues are C34 and U39 (tRNA numbering) corresponding to C35 and U143, respectively, in Figure 1A ▶. Normally, C/D box RNA-mediated modifications involve separate guide and target RNAs. However, C/D box sequences (consensus sequences RUGAUGA/CUGA) corresponding to Cm34 and Um39 modifications in H. volcanii tRNATrp are present in cis, within the intron of the pre-tRNATrp, and these boxes are retained within the intron after splicing. The C/D boxes for Cm34 are at positions 52–58/122–125, and C‘/D‘ boxes for Um39 are at positions 103–109/75–78 in Figure 1A ▶ (d’Orval et al. 2001). The folding of pre-tRNATrp shown in Figure 1A ▶ is splicing competent and has C/D and C‘/D‘ boxes in proper alignment (d’Orval et al. 2001). However, the structures required for splicing and 2‘-O-methylation of the two residues are mutually exclusive. In vivo, methylation reactions precede splicing because the target sequences that pair with the guide sequences overlap exon–intron junctions. It is not known whether the methylation reactions are intramolecular or intermolecular. It is possible that tRNATrp introns, after splicing, act in trans, as C/D box RNAs for the modification of one or the other or both residues in the pre-tRNA. This intron acting as sRNA may be somewhat similar to the Eukarya, in which most of the snoRNAs are derived from the introns.
Eukarya have two different types of tRNA splicing ligases: yeast type and animal type (Phizicky and Greer 1993; Arn and Abelson 1998; Trotta and Abelson 1999). Archaeal ligase appears to be somewhat similar to the animal type, because, in both cases, the junction phosphate is derived from the precursor (Zofallova et al. 2000). Neither archaeal nor animal enzymes have been sufficiently purified for detailed characterization. It has been reported that mammalian cells contain a default pathway that can join 2‘,3′-cyclic phosphate and 5′-hydroxyl termini and can circularize ribozyme-processed RNAs (Reid and Lazinski 2000). This ligation activity can join RNA in both bimolecular and intramolecular reactions in a sequence-independent manner (Reid and Lazinski 2000). It is possible that this default pathway ligase and animal-type tRNA splicing ligase is the same enzyme that is capable of joining exons (bimolecular) and circularizing RNAs (intramolecular). Both join the 5′-hydroxyl and the 2‘,3′-cyclic phosphate termini. In such cases, there may be a possible homology between the archaeal and animal-type ligases, because, as shown in the present work, the archaeal ligase can do both bimolecular and intramolecular ligations. Archaeal and eukaryal endonucleases have already been shown to be homologous. This raises a question. Are the archaeal and eukaryal (animal) protein enzyme-based RNA splicing systems derived from a common ancestor?
MATERIALS AND METHODS
Standard procedures as described by Sambrook and Russell (2001) or as provided by the manufacturers of the reagents were followed in most of the work.
Oligonucleotides
Sequences of the oligonucleotides used for PCR, RT-PCR, Northern hybridizations, and sequencing are as follows:
A: TAATACGACTCACTATAGGGGCTGTGGCCAAGC,
B: TGGGGCCGGAGGGATTTGAAC,
C: TAATACGACTCACTATAGCCCGGGTGGCTTAGC,
D: TGCCCGGGGTGGGCTCCGAAC,
E: TAATACGACTCACTATAGGGCTTGGCGCCCGGGA,
F: ATCTCCGGTGGGCACCT,
G: ATCTCCTTTGGGCACCT,
H: TCAGTATATCAGCTGGAGTGTC,
I: CTGATCATCGGTCGTGTTGACG,
J: GGGGCTGTGGCCAAGC,
K: AGGCTTCCAAGGCGTTC,
L: CCCGCGAGGTCATGCC,
M: GCCCGGGTGGCTTAGCT,
N: CGCCAAGCCTATCACCGG.
Positions corresponding to some of these are marked in Figure 1 ▶.
RNA substrates for splicing reactions
Internally labeled RNA substrates were prepared and purified as described previously (Zofallova et al. 2000) with minor modifications. A typical 100-μL transcription reaction performed at 37°C for 2–3 h contained 40 mM Tris-Cl (pH 8.0), 13 mM MgCl2, 20 mM DTT (Dithiothreitol), 2 mM spermidine, 50 μg/mL bovine serum albumin, 4% PEG (polyethylene glycol) 8000, 0.001% Triton X-100, 100 μCi of one [α-32P]NTP (ICN Biomedicals, sp. act. 3000 Ci/mmol), unlabeled NTPs (0.3 mM of the NTP used for the label and 0.75 mM of each of the rest), varying amounts of PCR-amplified DNA, and 0.1–0.15 μg T7 RNA polymerase. Transcripts were purified by denaturing PAGE. Approximate amounts of the labeled transcripts were estimated by Cerenkov counting. The templates and primers to produce PCR-amplified DNAs for various transcripts are pVT9P11 (see following) with primers A and B for pre-tRNATrp; pHVMΔi36 (Gomes and Gupta 1997) with primers C and D for truncated elongator pre-tRNAMet; pVT9P11 with primers E and F for synthetic tRNATrp intron; pVT9P11 with primers E and G for synthetic tRNATrp intron having a CC-to-AA change. Plasmid pVT9P11 has about a 1-kb PstI fragment of pVT9 (Daniels et al. 1985) cloned into pBluescript KS+ (D. Shao and R. Gupta, unpubl.); the fragment contains an H. volcanii tRNATrp-coding region. Synthetic tRNATrp intron transcripts had an A-to-G change at the first position (indicated in Fig. 1C ▶) for the initiation of T7 RNA polymerase transcription.
Termini of the synthetic tRNATrp introns were converted to 5′-hydroxls and 2‘,3′-cyclic phosphates as follows. Gel-eluted transcripts were treated with calf intestinal alkaline phosphatase, phenol extracted, and ethanol precipitated. Then periodate oxidation and amine cleavage of the dephosphorylated RNA was done by a combination of published procedures (Neu and Heppel 1964; Gupta 1984). A volume of 10 μL freshly prepared 0.5 M NaIO4 in 0.5 M NaOAc (pH 5.2) was added to 40 μL of RNA in water and incubated in the dark at 25°C for 30 min, followed by the addition of 50 μL 1 M lysine (pH 9.0), and further incubation in the dark at 45°C for 90 min. Then 8 μL 1 M ethylene glycol and 20 μL 1 M Tris-Cl (pH 8.5) were added and incubated in the dark at 25°C for 15 min, followed by ethanol precipitation. The 3′-phosphate of the RNA thus produced was then converted to the 2‘,3′-cyclic phosphate. RNA (10,000–25,000 cpm) was incubated for 20 min at 25°C, in a 10-μL reaction containing 20 mM K-HEPES (potassium-N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) (pH 7.6), 150 mM NaCl, 2 mM MgCl2, 0.15 mM EDTA (ethylenediamine tetra acetate), 0.1 mM spermidine, 1.25 mM DTT, 0.005% Triton X-100, 4.5% glycerol, 0.02 mM ATP, and 2–20 ng/mL recombinant Escherichia coli RNA 3′-terminal phosphate cyclase (Genschik et al. 1997). During this whole process, one nucleoside residue from the 3′ side of the transcript is deleted. Therefore, normal 103-b tRNATrp intron transcripts (corresponding to positions 39–141 in Fig. 1A ▶) are converted to 102 b.
Endonuclease and ligase reactions
Endonuclease reactions were done by incubating 5000 to 20,000 cpm of gel-eluted transcripts for 30 min at 37°C in a 100-μL reaction containing 40 mM Tris-Cl (pH 7.5), 20 mM MgCl2, 10% glycerol, and 2 μg recombinant H. volcanii endonuclease (Kleman-Leyer et al. 1997). The ligase preparations and reactions were as described previously (Zofallova et al. 2000). Partially purified ligase preparations (L. Zofallova and R. Gupta, unpubl.) were used in later work. The substrates for the ligase reactions were either endonuclease-treated precursors or synthetic introns after modification of their termini as described earlier. Endonuclease and 3′-terminal phosphate cyclase products were not purified before the ligase reaction. Recovery of small RNAs after phenol extraction and ethanol precipitation, especially from the high-salt-containing ligase reactions, was not complete. Ligase reactions of truncated elongator tRNAMet were desalted through Sephadex G-25 columns before ethanol precipitation. This enhanced the recovery of exons and truncated introns, both linear and circular.
Reverse transcriptase-polymerase chain reactions and DNA sequencing
The templates for the RT-PCR were either ligase reaction products eluted from the gels or H. volcanii total RNA from early log phase cells. The initial RT reaction was done with the primer complementary to the RNA and M-MLV (Moloney murine leukemia virus) reverse transcriptase (GIBCO BRL). Then the PCR was done after adding the other primer. RNase A treatment was mostly done after the PCR. The products were resolved by nondenaturing PAGE. Appropriate RT-PCR products were eluted from the native gels and sequenced with the same primers used for the RT-PCR. Cycle sequencing with 32P-end-labeled primers was done by SequiTherm EXCEL II kit (Epicentre).
Northern hybridizations
Total RNA was extracted from H. volcanii cells grown to an A550 of 0.5, with TRIzol reagent (Invitrogen). The RNA was then treated with DNase I, phenol–chloroform extracted, and precipitated with ethanol. The RNA was separated by denaturing PAGE either in 6% or in 8% gel. For each of these separations, ∼1 mg of total RNA was loaded into a 17-cm-wide well of a 20 × 20 × 0.08 cm gel. Northern blots were prepared as described previously (Datta et al. 1989). The blots were cut longitudinally into several strips. The strips were hybridized to 5′ 32P-labeled oligonucleotides.
Thin layer chromatography
Gel-purified RNA samples were digested with nuclease P1 (Sigma) and resolved by two-dimensional TLC on cellulose plates (EM Science or Machery-Nagel, Polygram Cel 300). The solvents for the TLC were (a) isobutyric acid/0.5 N NH4OH, 5:3, v/v; (b) isopropanol/HCl/H2O, 70:15:15, v/v/v; and (c) 0.1 M sodium phosphate, pH 6.8/ammonium sulfate/n-propanol, 100:60:2, v/w/v (Gupta 1984). Nuclease P1 digests of yeast total RNA were used as the source of the unlabeled markers. Some batches of [α-32P]UTP (from ICN Biomedical) contained an unidentified contaminant, which was incorporated in the transcript by T7 RNA polymerase (data not shown). Digestion of these transcripts by nuclease P1 produced an extra (unidentified) labeled product in addition to the normal labeled pU (see Fig. 8B ▶).
Acknowledgments
We thank W. Filipowicz and C.J. Daniels for providing the clones for the recombinant E. coli RNA 3′-terminal phosphate cyclase and H. volcanii splicing endonuclease, respectively, and Jack Parker for comments on the manuscript. This work was supported by NIH grant (GM55945) to R.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Abelson, J., Trotta, C.R., and Li, H. 1998. tRNA splicing. J. Biol. Chem. 273: 12685–12688. [DOI] [PubMed] [Google Scholar]
- Arn, E.A. and Abelson, J. 1998. RNA ligases: Function, mechanism, and sequence conservation. In RNA structure and function (eds. R.W. Simons and M. Grunberg-Manago), pp. 695–726. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Burggraf, S., Larsen, N., Woese, C.R., and Stetter, K.O. 1993. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. 90: 2547–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciammaruconi, A. and Londei, P. 2001. In vitro processing of the 16S rRNA of the thermophilic archaeon Sulfolobus solfataricus. J. Bacteriol. 183: 3866–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard, J.Z. and Garrett, R.A. 1992. Protein-coding introns from the 23S rRNA-encoding gene from stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene 121: 103–110. [DOI] [PubMed] [Google Scholar]
- Daniels, C.J., Gupta, R., and Doolittle, W.F. 1985. Transcription and excision of a large intron in the tRNATrp gene of an archaebacterium, Halobacterium volcanii. J. Biol. Chem. 260: 3132–3134. [PubMed] [Google Scholar]
- Daniels, C.J., Douglas, S.E., and Doolittle, W.F. 1986. Genes for transfer RNAs in Halobacterium volcanii. Syst. Appl. Microbiol. 7: 26–29. [Google Scholar]
- Datta, P.K., Hawkins, L.K., and Gupta, R. 1989. Presence of an intron in elongator methionine-tRNA of Halobacterium volcanii. Can. J. Microbiol. 35: 189–194. [DOI] [PubMed] [Google Scholar]
- Dennis, P.P., Ziesche, S., and Mylvaganam, S. 1998. Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 180: 4804–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, P.P., Omer, A., and Lowe, T. 2001. A guided tour: Small RNA function in Archaea. Mol. Microbiol. 40: 509–519. [DOI] [PubMed] [Google Scholar]
- Diener, J.L. and Moore, P.B. 1998. Solution structure of a substrate for the archaeal pre-tRNA splicing endonucleases: The bulge-helix-bulge motif. Mol. Cell 1: 883–894. [PubMed] [Google Scholar]
- Di Nicola Negri, E., Fabbri, S., Bufardeci, E., Baldi, M.I., Gandini Attardi, D., Mattoccia, E., and Tocchini-Valentini, G.P. 1997. The eucaryal tRNA splicing endonuclease recognizes a tripartite set of RNA elements. Cell 89: 859–866. [DOI] [PubMed] [Google Scholar]
- d’Orval, B.C., Bortolin, M.-L., Gaspin, C., and Bachellerie, J.-P. 2001. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res. 29: 4518–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durovic, P. and Dennis, P.P. 1994. Separate pathways for excision and processing of 16S and 23S rRNA from the primary rRNA operon transcript from the hyperthermophilic archaebacterium Sulfolobus acidocaldarius: Similarities to eukaryotic rRNA processing. Mol. Microbiol. 13: 229–242. [DOI] [PubMed] [Google Scholar]
- Fabbri, S., Fruscoloni, P., Bufardeci, E., Negri, E.D.N., Baldi, M.I., Attardi, D.G., Mattoccia, E., and Tocchini-Valentini, G.P. 1998. Conservation of substrate recognition mechanisms by tRNA splicing endonucleases. Science 280: 284–286. [DOI] [PubMed] [Google Scholar]
- Filipowicz, W. and Shatkin, A.J. 1983. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell 32: 547–557. [DOI] [PubMed] [Google Scholar]
- Filipowicz, W., Konarska, M., Gross, H.J., and Shatkin, A.J. 1983. RNA 3′-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res. 11: 1405–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruscoloni, P., Baldi, M.I., and Tocchini-Valentini, G.P. 2001. Cleavage of non-tRNA substrates by eukaryal tRNA splicing endonucleases. EMBO Rep. 2: 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik, P., Billy, E., Swianiewicz, M., and Filipowicz, W. 1997. The human RNA 3′-terminal phosphate cyclase is a member of a new family of proteins conserved in Eucarya, Bacteria and Archaea. EMBO J. 16: 2955–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, I. and Gupta, R. 1997. RNA splicing ligase activity in the archaeon Haloferax volcanii. Biochem. Biophys. Res. Commun. 237: 588–594. [DOI] [PubMed] [Google Scholar]
- Gupta, R. 1984. Halobacterium volcanii tRNAs: Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 259: 9461–9471. [PubMed] [Google Scholar]
- Itoh, T., Suzuki, K., and Nakase, T. 1998. Occurrence of introns in the 16S rRNA genes of members of the genus Thermoproteus. Arch. Microbiol. 170: 155–161. [DOI] [PubMed] [Google Scholar]
- Kaine, B.P. 1987. Intron-containing tRNA genes of Sulfolobus solfataricus. J. Mol. Evol. 25: 248–254. [Google Scholar]
- Kaine, B.P., Gupta, R., and Woese, C.R. 1983. Putative introns in tRNA genes of prokaryotes. Proc. Natl. Acad. Sci. 80: 3309–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems, J. and Garrett, RA. 1985. An intron in the 23S ribosomal RNA gene of the archaebacterium Desulfurococcus mobilis. Nature 318: 675–677. [DOI] [PubMed] [Google Scholar]
- ———. 1988. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium Desulfurococcus mobilis. Cell 54: 693–703. [DOI] [PubMed] [Google Scholar]
- ———. 1991. Ribosomal RNA introns in archaea and evidence for RNA conformational changes associated with splicing. Proc. Natl. Acad. Sci. 88: 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems, J., Jensen, J., Olesen, T., and Garrett, R.A. 1989a. Comparison of transfer RNA and ribosomal RNA intron splicing in the extreme thermophile and archaebacterium Desulfurococcus mobilis. Can. J. Microbiol. 35: 210–214. [DOI] [PubMed] [Google Scholar]
- Kjems, J., Leffers, H., Olesen, T., and Garrett, R.A. 1989b. A unique tRNA intron in the variable loop of the extreme thermophile Thermofilum pendens and its possible evolutionary implications. J. Biol. Chem. 264: 17834–17837. [PubMed] [Google Scholar]
- Kleman-Leyer, K., Armbruster, D.W., and Daniels, C.J. 1997. Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell 89: 839–847. [DOI] [PubMed] [Google Scholar]
- Laski, F.A., Fire, A.Z., RajBhandary, U.L., and Sharp, P.A. 1983. Characterization of tRNA precursor splicing in mammalian extracts. J. Biol. Chem. 258: 11974–11980. [PubMed] [Google Scholar]
- Li, H. and Abelson, J. 2000. Crystal structure of a dimeric archaeal splicing endonuclease. J. Mol. Biol. 302: 639–648. [DOI] [PubMed] [Google Scholar]
- Li, H., Trotta, C.R., and Abelson, J. 1998. Crystal structure and evolution of a transfer RNA splicing enzyme. Science 280: 279–284. [DOI] [PubMed] [Google Scholar]
- Lorsch, J.R., Bartel, D.P., and Szostak, J.W. 1995. Reverse transcriptase reads through a 2‘–5′ linkage and a 2‘-thiophosphate in a template. Nucleic Acids Res. 23: 2811–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J. and Garrett, R.A. 1994. Structural characteristics of the stable RNA introns of archaeal hyperthermophiles and their splicing junctions. J. Mol. Biol. 243: 846–855. [DOI] [PubMed] [Google Scholar]
- ———. 1997. RNA-protein interactions of an archaeal homotetrameric splicing endoribonuclease with an exceptional evolutionary history. EMBO J. 16: 6290–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu, H.C. and Heppel L.A. 1964. Nucleotide sequence analysis of polyribonucleotides by means of periodate oxidation followed by cleavage with an amine. J. Biol. Chem. 239: 2927–2934. [PubMed] [Google Scholar]
- Nishikura, K. and De Robertis, E.M. 1981. RNA processing in microinjected Xenopus oocytes. Sequential addition of base modifications in the spliced transfer RNA. J. Mol. Biol. 145: 405–420. [DOI] [PubMed] [Google Scholar]
- Nomura, N., Sako, Y., and Uchida, A. 1998. Molecular characterization and postsplicing fate of three introns within the single rRNA operon of the hyperthermophilic archaeon Aeropyrum pernix K1. J. Bacteriol. 180: 3635–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer, A.D., Lowe, T.M., Russell, A.G., Ebhardt, H., Eddy, S.R., and Dennis, P.P. 2000. Homologs of small nucleolar RNAs in archaea. Science 288: 517–522. [DOI] [PubMed] [Google Scholar]
- Perkins, K.K., Furneaux, H., and Hurwitz, J. 1985. Isolation and characterization of an RNA ligase from HeLa cells. Proc. Natl. Acad. Sci. 82: 684–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky, E.M. and Greer, C.L. 1993. Pre-tRNA splicing: Variation on a theme or exception to the rule? Trends Biochem. Sci. 18: 31–34. [DOI] [PubMed] [Google Scholar]
- Reid, C.E. and Lazinski, D.W. 2000. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. 97: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Tang, T.H., Rozhdestvensky, T.S., d’Orval, B.C., Bortolin, M.-L., Huber, H., Charpentier, B., Branlant, C., Bachellerie, J.-P., Brosius, J, and Hüttenhofer, A. 2002. RNomics in archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 30: 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, L.D. and Daniels, C.J. 1988. A tRNATrp intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties. J. Biol. Chem. 263: 17951–17959. [PubMed] [Google Scholar]
- ———. Recognition of exon–intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J. Biol. Chem. 265: 18104– 18111. [PubMed]
- Thompson, L.D., Brandon, L.D., Nieuwlandt, D., and Daniels, C.J. 1989. Transfer RNA intron processing in the halophilic archaebacteria. Can. J. Microbiol. 35: 36–42. [DOI] [PubMed] [Google Scholar]
- Trotta, C.R. and Abelson, J. 1999. tRNA splicing: An RNA world add-on or an ancient reaction? In: The RNA World, 2nd ed. (eds. R.F. Gesteland, T.R. Cech, and J.F. Atkins), pp. 561–584. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Trotta, C.R., Miao, F., Arn, E.A., Stevens, S.W., Ho, C.K., Rauhut, R., and Abelson, J.N. 1997. The yeast tRNA splicing endonuclease: A tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 89: 849–858. [DOI] [PubMed] [Google Scholar]
- Vogel, J., Hess, W.R., and Börner, T. 1997. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 25: 2030–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y.-i., Yokobori, S.-i., Inaba, T., Yamagishi, A., Oshima, T., Kawarabayasi, Y., Kikuchi, H., and Kita, K. 2002. Introns in protein-coding genes in archaea. FEBS Lett. 510: 27–30. [DOI] [PubMed] [Google Scholar]
- Wich, G., Leinfelder, W., and Bock, A. 1987. Genes for stable RNA in the extreme thermophile Thermoproteus tenax: Introns and transcription signal. EMBO J. 6: 523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R., Kandler, O., and Wheelis, M.L. 1990. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. 87: 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofallova, L., Guo, Y., and Gupta, R. 2000. Junction phosphate is derived from the precursor in the tRNA spliced by the archaeon Haloferax volcanii cell extract. RNA 6: 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]