Abstract
GNRA tetraloops (N is A, C, G, or U; R is A or G) are basic building blocks of RNA structure that often interact with proteins or other RNA structural elements. Understanding sequence-dependent structural variation among different GNRA tetraloops is an important step toward elucidating the molecular basis of specific GNRA tetraloop recognition by proteins and RNAs. Details of the geometry and hydration of this motif have been based on high-resolution crystallographic structures of the GRRA subset of tetraloops; less is known about the GYRA subset (Y is C or U). We report here the structure of a GUAA tetraloop determined to 1.4 Å resolution to better define these details and any distinctive features of GYRA tetraloops. The tetraloop is part of a 27-nt structure that mimics the universal sarcin/ricin loop from Escherichia coli 23S ribosomal RNA in which a GUAA tetraloop replaces the conserved GAGA tetraloop. The adenosines of the GUAA tetraloop form an intermolecular contact that is a commonplace RNA tertiary interaction called an A-minor motif. This is the first structure to reveal in great detail the geometry and hydration of a GUAA tetraloop and an A-minor motif. Comparison of tetraloop structures shows a common backbone geometry for each of the eight possible tetraloop sequences and suggests a common hydration. After backbone atom superposition, equivalent bases from different tetraloops unexpectedly depart from coplanarity by as much as 48°. This variation displaces the functional groups of tetraloops implicated in protein and RNA binding, providing a recognition feature.
Keywords: A-minor motif, conformational change, hydration, RNA motifs, RNA-protein recognition, RNA structure, RNA tertiary contacts, sarcin/ricin loop
INTRODUCTION
RNA hairpins are ubiquitous elements of RNA secondary structure that are frequently capped by tetraloops with a GNRA consensus sequence (N is A, C, G, or U; R is A or G). Located next to Watson–Crick (WC) helical regions or non-WC motifs, these GNRA tetraloops are closed off by at least one WC base pair. GNRA tetraloops help fold RNA: They allow single-stranded RNA to fold back on itself by reversing the direction of the phosphodiester backbone, and they bring together distant regions of the ribonucleotide sequence by providing sites for RNA tertiary contacts. By providing sites for protein binding, they also facilitate assembly of ribonucleoprotein particles. These interactions can involve specific recognition of GNRA tetraloops by proteins (Wool et al. 1992) and by RNAs (Doherty et al. 2001; Nissen et al. 2001). The structural basis of this specificity is still emerging.
Each tetraloop sequence is thought to share a common fold based on structures of a subset of the eight possible GNRA sequences that were determined first by NMR spectroscopy (Heus and Pardi 1991) and later by X-ray crystallography (Pley et al. 1994b; Scott et al. 1995; Cate et al. 1996; Correll et al. 1999). The fold presents distinctive functional groups to the accessible minor groove, including those at the Watson–Crick edges of the last three bases of the tetraloop. Complementary contacts to these functional groups by proteins and RNAs are thought to enable specific recognition. Specific recognition may also occur through shape complementarity to the RNA backbone because structural comparisons have suggested conformational differences between GRRA tetraloops and GYRA (Y is C or U) tetraloops (Correll et al. 1998). These comparisons have been limited by the small number of GNRA tetraloop structures that were available (Correll et al. 1998) and by the relatively low resolution (3 Å) of the only GYRA tetraloop structure used in the comparison—a GUAA tetraloop from a hammerhead ribozyme (Scott et al. 1995). Similar limitations apply to structural comparisons that indicated that all GNRA tetraloops share a common fold. Two structures determined to better than 1.6 Å resolution have defined the details of the geometry and hydration for the GRRA subclass of tetraloops (Correll et al. 1999; Batey et al. 2000, 2001); less is known about the GYRA subclass of tetraloops. It is therefore unknown if there is any sequence-dependent structural variation among the different GNRA tetraloops that would provide additional recognition opportunities.
To help address these questions, we have studied a GUAA tetraloop that is part of a 27-nt RNA that mimics a universal stem-loop structure of Escherichia coli 23S ribosomal RNA. The GUAA tetraloop is a mutant of the conserved GAGA tetraloop. The toxins sarcin and ricin are site-specific enzymes that target this GAGA tetraloop: Sarcin cleaves the backbone on the 3′-side of G2661 (E. coli 23S RNA numbering is used throughout) and ricin depurinates A2660. This stem-loop structure is therefore referred to as the sarcin/ricin loop (SRL). Action by either toxin disrupts binding of elongation factors (EF-Tu and EF-G) to the ribosome, thereby inhibiting protein synthesis. Cell death then results from a caspase III apoptotic pathway that is triggered by an unknown mechanism (Olmo et al. 2001). The SRL RNA is required for protein synthesis because it is a critical component of the binding site for EFs and possibly for other GTPase protein factors, such as initiation factor IF2 (La Teana et al. 2001; Cameron et al. 2002). Synthetic oligonucleotides (∼30-mers) with the SRL nucleotide sequence mimic the form and function of the SRL in the ribosome (Ban et al. 2000); they have therefore served as a minimal substrate for EF-G binding (Munishkin and Wool 1997), for sarcin and ricin activity (Wool 1997), and for structural studies (Szewczak et al. 1993; Szewczak and Moore 1995; Correll et al. 1998, 1999; Seggerson and Moore 1998; Rife et al. 1999).
We report here the crystal structure of a GUAA tetraloop mutant of the SRL RNA determined to 1.4 Å resolution. Structural comparisons show that the eight GNRA tetraloop sequences share a common backbone geometry and have an unexpectedly large variation in the orientation of the last three bases of the tetraloop. Interestingly, these bases are implicated in RNA tertiary contacts and protein binding. The lattice of the GUAA mutant crystals is stabilized by an intermolecular contact that is an A-minor motif. This motif is a common and stabilizing RNA tertiary contact (Doherty et al. 2001; Nissen et al. 2001) that has been implicated in the decoding of messenger RNA during translation (Ogle et al. 2001). Analysis of the GUAA structure suggests a structural basis for the observed sequence preference of A-minor interactions.
RESULTS
Structure determination and overview
Of the seven possible GNRA mutants of the SRL GAGA tetraloop that were investigated, only the GUAA mutant produced data-quality crystals. Previously, the GYRA tetraloops in the 50S subunit structure determined to 2.4 Å resolution provided the highest resolution view of this subclass of tetraloops. We report here the structure of the GUAA mutant of the SRL RNA determined by molecular replacement (see Materials and Methods; Table 1 ▶); the crystal data, which extended to 1.4 Å resolution, were refined to a final Rfree of 0.205, with a conventional R of 0.162. As expected, the structure consists of a GUAA tetraloop, a bulged-G motif, and an A-form stem region (Fig. 1 ▶). The stem is connected to the motifs by two water-mediated non-WC base pairs (Correll et al. 1999). The overall features of the tetraloop are typical: The first and fourth bases form a G•A pair, the backbone reverses direction between the first and the second nucleotide, and the last three bases stack on one another.
TABLE 1.
Data and refinement statistics
| GUAAa | |
| Resolution range (Å) | 40–1.4 |
| Space group | C2 |
| Unit cell | |
| a, b, c (Å) | 57.97, 22.53, 58.47 |
| β (°) | 101.72 |
| Unique reflections (fold redundancy) | 14,372 (6.3) |
| Completeness (%) | 97.9 (80.0) |
| I/σI | 21.8 (2.3) |
| Rmergeb | 0.072 (0.323) |
| R-factorc | 0.164 (0.278) |
| Rfreec | 0.207 (0.458) |
| % of data used to calculate Rfree | 10 |
| RNA atoms | 582 |
| Solvent molecules | 109 |
| R.m.s deviation from ideality | |
| bonds (Å) | 0.01 |
| angles (Å) | 0.028 |
| Average B-factors (Å2) | |
| RNA | 11.8 |
| solvent | 27.9 |
aThe numbers in parentheses are statistics for data between 1.42 Å and 1.40 Å, except for fold redundancy.
bRmerge = ∑hklΣ|Ii − 〈I〉|/∑hkl∑/i for all data with I/σI > −3.
cR-factor = ∑hkl | |Fobs| − k|Fcalc| | /∑hkl |Fobs|
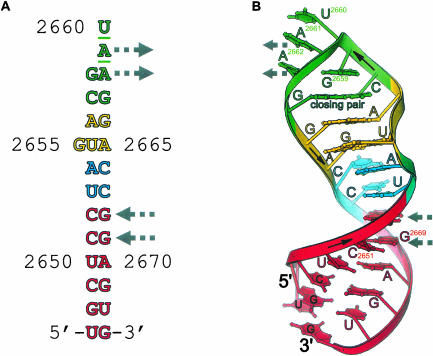
FIGURE 1.
Structural overview and lattice contacts. (A) Schematic of the RNA sequence in the crystal structure showing the tetraloop (green), bulged-G motif (yellow), the flexible region (blue), and the stem region (red). Gray arrows indicate the two adenosines and WC base pairs involved in lattice contacts that mimic A-minor like interactions (Fig. 3 ▶). The sites of mutation are underlined. (B) A ribbon drawing of the GUAA mutant structure.
Common and distinctive features of GNRA tetraloops
Structural comparisons of the GUAA tetraloop and the other GNRA tetraloops show a common backbone geometry and a variable base orientation (Fig. 2 ▶). The GAGA tetraloop and the GUAA tetraloop of the SRL RNA share a common backbone geometry; backbone atoms superimpose with a pairwise root mean square (rms) deviation of 0.6 Å. To better characterize any sequence-dependent structural variations among different GNRA tetraloop sequences, we compared the structures of 20 tetraloops determined by X-ray crystallography to at least 2.4-Å resolution (see Materials and Methods). The resolution cutoff is a compromise between having sufficient crystallographic data to define the tetraloop geometry and including at least one structure for each of the eight different tetraloop sequences. GNRA tetraloops in this set share a common backbone geometry; backbone atoms superimpose with a pairwise rms deviation that varies between 0.6 Å and 0.9 Å.
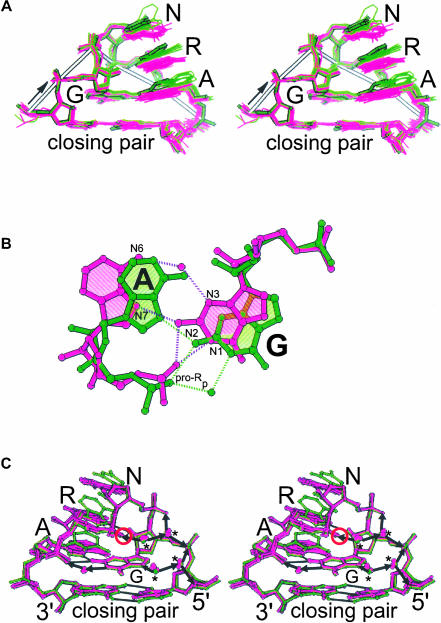
FIGURE 2.
Comparison of the GUAA structure (thick lines) with other GNRA tetraloop structures. (A) Stereodiagram of the backbone atom superposition of 20 tetraloop structures determined to at least 2.4 Å resolution (see Materials and Methods); the standard orientation of the tetraloop bases (purple) is contrasted with the altered orientation (green). (B) Two extremes in the conformation of the base pair between the first and the fourth nucleotide of the tetraloop. (C) Stereodiagram of the superposition of the GUAA tetraloop reported here (green) and the other tetraloop structures determined to at least 2.0 Å resolution (see Materials and Methods). The GUAA structure illustrates the altered orientation of the last three bases of the tetraloop whereas the other structures illustrate the standard orientation. In the tetraloop, the pro-Rp oxygen of the third nucleotide (circled) is within 3.2 Å of the first guanine base. Arrows marking solvent-mediated hydrogen bonds are pointing toward proton acceptors. Most of the conserved solvent sites are obligate proton acceptors (asterisks).
Unlike the similarities seen in the tetraloop backbone geometries, equivalent bases in different GNRA tetraloops depart from coplanarity by as much as 48° after backbone atom superposition (Fig. 2A ▶). As a result, equivalent atoms in the last three bases of the tetraloops are displaced by as much as 3 Å. The last three bases of each tetraloop remain stacked on one another regardless of orientation. If these bases are nearly parallel to the bases of the closing WC pair, we refer to them as being in the “standard orientation”; otherwise they are referred to as being in the “altered orientation.” The GUAA structure illustrates the altered orientation and the GAGA of the wild-type GAGA SRL RNA structure illustrates the standard orientation (Correll et al. 1999). Base flipping and unstacking of these last three bases, as observed in NMR analyses (Jucker et al. 1996) and dynamic simulations (Sorin et al. 2002) of GNRA tetraloops, are not observed in crystal structures. Three of the seven GYRA tetraloop structures adopt the standard orientation and the others adopt the altered orientation. In contrast, only 2 out of 13 of the GRRA tetraloop structures adopt the altered orientation. Possibly, GYRA tetraloops are more likely to vary from the standard orientation than GRRA tetraloops; however, additional tetraloop structures are needed to substantiate this trend.
The first and fourth nucleotides of the tetraloop form an A•G base pair that is a variant of a regular sheared pair (Fig. 2B ▶). Several structures determined by NMR spectroscopy have reported that the first and fourth nucleotides of the tetraloop form a sheared G•A base pair, where N3 of the G contacts N6 of the A, and N2 of the G contacts N7 of the A. In accord with analysis of the NMR data, the exocyclic amine (N2) of the G forms a bifurcated hydrogen bond with the base (N7) and the backbone (O1P) of the A in each of the 20 crystal structures of GNRA tetraloops. This interaction is believed to contribute about 0.7 kcal/mole of stabilization to the tetraloop based on comparison of thermal denaturation data for unmodified tetraloops and site-modified ones in which this contact is absent (SantaLucia et al. 1992). In contrast, N3 of G is too far from N6 of A to form a hydrogen bond in each of the 20 structures surveyed (4.4 ± 0.7 Å). In some structures this distance is larger than 5 Å, creating a cavity; in several structures a solvent molecule is observed filling this cavity (Fig. 2B ▶). Interaction between the pro-Rp oxygen of the third nucleotide and the π orbitals of the first G nucleotide may help stabilize GNRA tetraloop structures (Fig. 2C ▶). Sulfur substitution of this pro-Rp oxygen increases tetraloop stability by ∼2.9 kcal/mole compared to unmodified tetraloops; modifications at other nonbridging oxygen atoms in the tetraloop did not significantly change stability (Horton et al. 2000).
Consensus geometry and hydration of the GNRA tetraloop
For tetraloops to create unique surface features for RNA and protein recognition, one or more of the six backbone torsion angles per nucleotide of these tetraloops must deviate from standard values. Torsion angles are often underdetermined because X-ray diffraction of RNA usually extends only to ∼3 Å resolution, and NMR spectroscopy determines relatively few constraints on the backbone geometry (Rife et al. 1999). The definition of torsion angles is, hence, critical for establishing restraints in the refinement of RNA structures and for building accurate models. To define the consensus geometry and hydration, we compared the six structures that were determined to better than 2 Å resolution; these include GAGA, GAAA, and GUAA tetraloops (see Materials and Methods). The resolution cutoff is necessary to adequately determine the torsion angles and hydration of the GNRA tetraloops.
The torsion angles in GNRA tetraloops adopt predominantly A-form values (Table 2 ▶). Of the 42 possible backbone and glycosidic torsion angles in the tetraloop and the closing WC pair, only 5 deviate greater than 36° from A-form geometry. Each sugar in the tetraloop adopts a 3′-endo pucker. Reversal of the loop chain direction is the result of two torsion angle deviations in the backbone of the second nucleotide of the tetraloop. Without additional deviation from A-form geometry, the WC edges of the bases of the closing pair lie ∼6 Å apart, precluding base-pair formation. Deviations in three torsion angles of the 3′-closing nucleotide increase the overall twist in the backbone and unstack the fourth base of the tetraloop from the 3′-adjacent closing base (Figs. 1B, 3A ▶ ▶). As a consequence, the bases of the closing pair are brought together, allowing base-pair formation.
TABLE 2.
Torsion angles
| Nucleotide | χa | α | β | γ | δ | ɛ | ζ |
| 5′ closing pair | 200 ± 5 (Δ7)b | 299 ± 8 (Δ14) | 171 ± 4 (−Δ13) | 47 ± 4 (−Δ7) | 81 ± 1 (Δ0) | 213 ± 4 (−Δ1) | 293 ± 3 (Δ4) |
| G | 195 ± 7 (Δ2) | 291 ± 5 (Δ6) | 177 ± 3 (−Δ7) | 49 ± 4 (−Δ4) | 80 ± 2(−Δ2) | 224 ± 4 (Δ10) | 296 ± 4 (Δ7) |
| N | 196 ± 8 (Δ3) | 168 ± 5 (Δ117)c | 140 ± 8 (−Δ44) | 54 ± 5 (Δ1) | 87 ± 4 (Δ6) | 231 ± 6 (Δ17) | 282 ± 9 (−Δ11) |
| R | 210 ± 12 (Δ17) | 298 ± 6 (Δ13) | 152 ± 10 (−Δ32) | 64 ± 6 (Δ11) | 87 ± 3 (Δ6) | 206 ± 4 (−Δ8) | 291 ± 12 (−Δ6) |
| A | 219 ± 8 (Δ26) | 289 ± 9 (Δ4) | 173 ± 8 (−Δ11) | 52 ± 4 (−Δ1) | 87 ± 3 (Δ6) | 218 ± 3 (Δ4) | 296 ± 8 (−Δ1) |
| 3′ closing pair | 200 ± 5 (Δ7) | 139 ± 8 (Δ146) | 227 ± 11 (Δ43) | 188 ± 9 (Δ135) | 86 ± 3(Δ5) | 221 ± 6 (Δ7) | 298 ± 8 (Δ1) |
aBackbone torsion angles: 5′→O3′-P−αO5′−βC5′−γC4′−δC3′−ɛO3′−ζP−O5′→3′.
bThe delta is the amount the values differs from A-form values (Gelbin et al. 1996).
cUnderlined values differ from A-form values by more than 36°.
FIGURE 3.
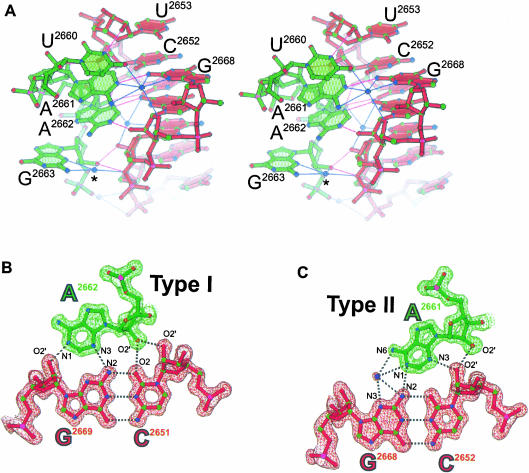
An interaction that mimics an A-minor motif. (A) Stereoview of the GUAA tetraloop (green) docking into the minor groove of a symmetry related stem region (red). The asterisk marks a solvent site that is conserved in tetraloop structures. The refined models of type I (B) and type II (C) A-minor contacts are superimposed on a 1.4 Å SIGMAA-weighted 2Fo–Fc electron density map. Weak C-H…O hydrogen bonds (as reviewed in Desiraju 1996) between C2 of A2662 and N3 of G2669 (3.2 Å) and between C2 of A2661 and O2 of C2652 (3.2 Å) may contribute to the stability of this A-minor interaction.
Comparison of tetraloop structures suggests common sites of hydration (Fig. 2C ▶). Solvent molecules, which coat the surface of RNA molecules, sometimes act as extensions of the RNA molecule, and can therefore play a role in recognition. These solvent molecules are considered to be structural if their location relative to an atom of the RNA molecule is conserved and independent of buffer conditions and lattice environment. By this criterion 7 of the 50 solvent molecules are structural to the tetraloop: Five line the inside of the phosphodiester backbone turn on the major groove side, and the remaining two contact the minor groove side of the closing pair (see Materials and Methods). Four of the major groove solvent molecules are obligate proton acceptors and adopt an unusual geometry by lying on the vertices of a rectangle. These distinctive features may provide recognition opportunities. Both of the minor groove solvent sites form bridges between the 2′-hydroxyl groups of the closing WC pair and the acceptor groups on the bases. These minor groove sites fail to provide a distinctive recognition opportunity because equivalent hydration is observed in WC base pairs.
RNA recognition of GNRA tetraloops: A-minor motif
A lattice contact provides the first high-resolution view (1.4 Å) of a tetraloop participating in what is believed to be the most common type of RNA tertiary interaction: the A-minor motif (Doherty et al. 2001; Nissen et al. 2001). This type of A-minor interaction was first observed as an intermolecular contact between neighboring molecules in the crystal structure of the hammerhead ribozyme (Pley et al. 1994a); the structure of the P4-P6 domain of group I intron was the first to illustrate this motif as a tertiary contact (Cate et al. 1996). A-minor contacts involve tandem adenosines docking into the minor groove of an RNA helix. Ribose zippers as well as interactions between tetraloops and tetraloop receptors are types of A-minor interactions (Doherty et al. 2001; Nissen et al. 2001). In the crystal lattice of the GUAA structure reported here, both adenosines from the GUAA tetraloop of one SRL RNA molecule form an A-minor motif by docking into the minor groove of the WC stem region of a second, symmetry-related SRL RNA molecule (Figs. 1, 3 ▶ ▶). This contact is the most common type of A-minor motif in which type I and type II base triples form tertiary interactions between two adenosines and two receptor C•G base pairs (Nissen et al. 2001). Nucleotides that flank either side of the adjacent adenosines also make direct and solvent-mediated contacts to the backbone of the receptor, which presumably provide additional stabilization. In the type I interaction, the adenosine makes four favorable hydrogen bonds to the receptor C•G base pair, spanning across the minor groove. Contacts to the 2′-hydroxyl groups are energetically significant (Abramovitz et al. 1996; Silverman and Cech 1999; Doherty et al. 2001) and presumably guide the docking of the adenosine into the minor groove. In the type II interaction, the adenosine makes direct and solvent-mediated contacts to the receptor C•G base pair that span only part of the way across the minor groove. This solvent interaction was predicted but previously unobserved due primarily to insufficient resolution; previously, the P4-P6 structure determined to 2.25 Å resolution provided the highest resolution view of these motifs. In the type I interaction, the adenine base and the base pair of the receptor deviate from coplanarity by 32°; in the type II interaction, this deviation is 22°. These are typical deviations that are less than the largest deviation (45°) observed in other type I and type II base triples (Nissen et al. 2001).
Binding studies of mutant RNAs and phylogenetic covariation analyses indicate that type I interactions favor a receptor C•G base pair over the other three WC base pairs (Doherty et al. 2001; Nissen et al. 2001). Analysis of the A-minor motif found in the GUAA mutant structure suggests that the molecular basis of sequence preference of type I interactions hinges on one hydrogen bond (Pley et al. 1994a). The most striking difference among the functional groups presented to the minor groove by the four WC base pairs is the presence of the exocyclic amine (N2) of guanine in G•C and C•G base pairs and its absence in U•A and A•U base pairs. The type I interaction has a hydrogen bond between the N3 of the adenine base and the N2 of the guanine base in the receptor C•G base pair (Fig. 3B ▶). Loss of this hydrogen bond involving N2 may explain why receptors with U•A and A•U base pairs are less common than G•C and C•G base pairs. Based on sequence-specific contacts between synthetic polyamide polymers and the minor groove of DNA (Kielkopf et al. 1998; Dervan and Burli 1999), type I discrimination between G•C and C•G base pairs is believed to rely on recognition of a subtler difference: the orientation of the proton donor group of the exocyclic amine found in these 2 base pairs (see below). The resolution of the GUAA structure is insufficient to identify the position of hydrogen atoms; however, it is possible to calculate their positions according to well-established geometric criteria using the program X-PLOR (Brunger 1992). In the type I interaction shown, the angle between the N3 and the N2-proton bond is ∼170°, whereas the equivalent angle for a receptor G•C base pair, assuming an isosteric substitution of a G•C base pair for a C•G base pair, is ∼124°. In a survey of equivalent hydrogen bonds, the most probable angles lie near 180° and angles less than 141° are not observed (Llamas Saiz and Foces-Foces 1990), in accord with the underlying energetic preference for a linear geometry. An energetic preference for a linear hydrogen bond with the exocyclic amine (N2) of the receptor base pair may, therefore, discriminate C•G base pairs from G•C base pairs. Type II interactions also have a phylogenetic preference for receptors with C•G base pairs. Unlike type I interactions, there appears to be little energetic (Doherty et al. 2001) or structural rationale to explain the sequence preference of type II interactions.
DISCUSSION
An important step toward deciphering the molecular basis by which proteins and RNA receptors recognize specific GNRA tetraloops is to define the distinctive and common structural features of this basic building block of RNA structure. To our knowledge, this is the first study to verify by structural comparisons that all eight tetraloop sequences fold into a common backbone geometry. These comparisons have also been used to define the consensus backbone geometry and hydration of the GNRA tetraloop. Common features of GNRA tetraloops include a shared backbone geometry, several structural solvent molecules, and two unstacked bases: the 5′-side of the second base and the 3′-side of the fourth base (Figs. 1B, 2 ▶ ▶; Table 2 ▶). Protein and RNA contacts to common features are important to discriminate GNRA tetraloops from other elements of RNA structure and to guide the docking of sequence specific contacts. Formation of complementary contacts to features in the minor groove of GNRA tetraloops provides the most direct route to sequence-specific recognition; the A-minor interaction is a relevant example. The tetraloop minor groove presents distinctive functional groups on Watson–Crick edges of the second, third, and fourth bases of the tetraloop, which permit direct sequence specific recognition by proteins or RNAs. Variation in the orientation of the last three bases of the tetraloop offers another option for specific recognition by displacing the distinctive functional groups presented to the minor groove. Differences in the docking geometry between the tetraloop and the receptor (Doherty et al. 2001; Nissen et al. 2001) and the variation in the orientation of the tetraloop bases presumably allows for a larger repertoire of RNA receptors.
Specific recognition between a GNRA tetraloop and an RNA receptor involves A-minor interactions and is accompanied by minimal conformational adaptation. Complementary minor groove contacts between the receptor and the distinctive functional groups of GNRA tetraloops are thought to mediate specific recognition (Pley et al. 1994a; Doherty et al. 2001; Nissen et al. 2001). These specific contacts are guided by tetraloop contacts to the backbone of the receptor RNA helix (Fig. 3A ▶). Recognition of the minor groove of A-form helices is challenging because the accessible functional groups at the edge of the base pairs present few features to discriminate between the four possible WC base pairs. Protein recognition of the DNA minor groove is believed to occur via indirect strategies, such as sequence-dependent distortability. The only well-documented case of sequence-specific direct recognition of the minor groove is between DNA and synthetic polyamide polymers (Dervan and Burli 1999). Like the type I interactions, G•C base pairs are thought to be distinguished from C•G base pairs by preference for a linear hydrogen bond to the exocyclic amine (N2) of the G base (Kielkopf et al. 1998). Polyamide polymers and adenosines are more rigid than protein side-chains. Possibly, these rigid keys—adenosines and polyamides—are better able to unlock the subtleties displayed in the minor groove than the more adaptable protein moiety.
Unlike A-minor interactions, conformational adaptation by the protein, the RNA, or both is a hallmark of protein–RNA complex formation (Williamson 2000). Sarcin and ricin (Wool et al. 1992) are different enzymes that typify protein recognition of a specific GNRA tetraloop. These enzymes are toxins that target a conserved GAGA tetraloop in ribosomes. Structural studies of complexes between the sarcin homolog restrictocin and substrate analogs indicate that this toxin binds to a base-flipped form of the tetraloop portion of the SRL RNA. This bound form of the substrate positions the nucleophile for in-line attack on the scissile bond and is believed to be a prerequisite for cleavage (Yang et al. 2001). Structural studies also suggest that in order for the active site of ricin to gain access to the scissile bond, the tetraloop unfolds (Weston et al. 1994; Yang et al. 2001); however, the structure of this unfolded form is unknown. It is also unclear if EF-G or EF-Tu binds to the canonical form or an altered form of the SRL RNA. These studies indicate that proteins may recognize unfolded tetraloop substrates.
There is a growing consensus that most GNRA tetraloop molecules (∼80%) fold into a canonical structure described by NMR and X-ray analyses (Fig. 2 ▶; Table 2 ▶). The remainder of these molecules (∼20%) adopt one or more different noncanonical conformations; molecular dynamic simulations suggest that tetraloop bases are flipped and restacked relative to those in the canonical tetraloop structure (Jucker et al. 1996; Sorin et al. 2002). It is unknown whether proteins—such as sarcin and ricin—induce the unfolding of GNRA tetraloops or merely bind to a preexisting unfolded form. NMR data indicate a mixed population of sugar puckers for positions two, three, and four of the tetraloop (Szewczak and Moore 1995; Jucker et al. 1996; Seggerson and Moore 1998). Each sugar in the canonical structure adopts a 3′-endo conformation, whereas noncanonical tetraloops appear to adopt 2′-endo puckers. Fluorescence studies indicate that the bases of the noncanonical tetraloops stack in a different manner than those of canonical tetraloops, but these noncanonical structures are inaccessible to NMR structure determination due to fast exchange (Menger et al. 2000).
The structural analyses reported here are a comprehensive reexamination of GNRA tetraloop geometry, hydration, and recognition. Structural comparisons have been used to define a consensus backbone geometry and hydration, which are common tetraloop features. We have also shown that the last three bases of GNRA tetraloops vary in orientation. This unexpectedly large variation results in significant displacement of distinctive functional groups in the minor groove recognition surface of tetraloops. We therefore expect that this variation may provide proteins and RNAs with an additional recognition strategy. A lattice contact forms the most common type of A-minor contact, consisting of a tandem type I and a type II base triple. The GUAA structure provides new details of the geometry and hydration of this motif. We show that sequence preference for type I interactions is likely to depend on the orientation of a single hydrogen bond.
MATERIALS AND METHODS
RNA synthesis, crystallization, and data collection
The GUAA oligoribonucleotide (27-mer) in the crystal was synthesized at the Yale Keck Microchemical Facility; it was deprotected with triethylamine trihydrofluoride and purified by gel electrophoresis. The 27-mer reproduced the nucleotides at positions 2647–2673 in E. coli 23S RNA r(UGCUCCUAGUACGUAAGGACCGGAGUG) with the A2660U and G2661A mutations underlined. Before crystallization trials, the concentration of the RNA was adjusted to ∼2.5 mg mL−1 in 1.0 mM Na•EDTA, 10 mM Tris (pH 8.0) and annealed by heating for 10 min at 55°C then slowly cooling to 24°C. Crystals of the annealed RNA grew in 1–3 days by vapor diffusion at 19°C in drops having 2 μL of the annealed RNA, 1 μL of well solution, and 0.3 μL of microseed, derived from crushed wild-type crystals (Correll et al. 1999). The well solution contained 3.0–3.2 M (NH4)2SO4 and buffer M (50 mM K•MOPS (pH 7.0), 10 mM MgCl2, 10 mM MnCl2). The crystals grew as clusters of triangular plates. To separate the clusters into single crystals, ∼0.3 mL of 2.0 M (NH4) 2SO4 and buffer M were added to the drop. After ∼2 min, when the clusters separated, the single crystals (150 × 100 × ∼5 μm3, approximate size) were stored in stabilizer solution, which had 3.5 M (NH4)2SO4 and buffer M. Microseeds were prepared by crushing a crystal in stabilizer solution.
The diffraction data were collected at the Structural Biology Center (SBC) beamline 19-ID at the Advanced Photon Source using a 3 × 3 charge-coupled device detector and 1° oscillations. Before collection of diffraction data at low temperature, the crystals were soaked for 3–5 min in a stabilizing solution that had 15% (w/v) xylitol and were then frozen in propane. Data were integrated and scaled with HKL-2000 (Otwinowski and Minor 1997).
Structure determination and refinement
The GUAA mutant structure was determined by molecular replacement and refined initially with CNS (Brunger et al. 1998) using an E. coli SRL RNA structure as a search model (Correll et al. 1999). After Patterson correlation refinement, a direct rotation search produced a 6.3 σ peak; a subsequent translation search produced a 2 σ peak. Although the height of the translation peaks only modestly exceeded that of the next highest peak, the solution led directly to the refined structure. Refinement was continued with SHELX-97 (Sheldrick and Schneider 1997) to fit individual anisotropic thermal parameters, which were tightly restrained to avoid overfitting the data. Models of the structure were built with the program O (Jones et al. 1991) and figures where generated with RIBBONS (Carson 1991).
Base-stacking interactions, which frequently stabilize RNA crystal lattices, are unusual in the GUAA mutant structure because the SRL RNAs stack stem to stem about a crystallographic twofold axis that passes through two atoms in the terminal G•U wobble base pair (C6 of U2697 and N1 of G2673). Each molecule about the twofold axis contributes 1 nt to the wobble pair and the nucleotide that could form the intramolecular closing pair is disordered. Therefore, the occupancy of the closing pair is 0.5.
Structures used for comparison
Twenty tetraloop structures were used for comparison: the 1.4 Å GUAA tetraloop structure; the 2.4 Å structure of the 50S subunit of the ribosome, containing at least one of each tetraloop, totaling 12 examples (Klein et al. 2001); a 1.52 Å GAAA structure (Batey et al. 2001); a 2.25 Å GAAA tetraloop (Juneau et al. 2001); a GAGA 1.11 Å tetraloop (Correll et al. 1999); a 2.3 Å GGAA tetraloop (Hainzl et al. 2002); a 1.75 Å GAGA tetraloop, three independent copies in the asymmetric unit cell (M. Lubbers, Y. Chan, I. Wool, and C. Correll, unpubl.). One of the tetraloops in the 50S subunit structure is in the 5S rRNA (89–94); the remaining 11 tetraloops are in the 23S rRNA (468–473, 576–581, 690–695, 804–809, 1326–1331, 1862–1867, 2248–2253, 2411–2416, 2629–2634, 2695–2700, 2876–2881). Tetraloops that contain a bulged nucleotide between the fourth and the 3′-adjacent nucleotide were not included. The following Protein Data Bank (PDB) entries were used: 1JJ2, 1HQ1, 1HR2, 483D, 1LNG. For analysis of hydration, we used five of the six structures that were determined to 2.0 Å or higher resolution. Only two of the three molecules in the asymmetric unit of the 1.75 Å GAGA structure were used because the third copy deviates significantly from the other five tetraloop structures shown in Figure 2C ▶. After superimposing the backbone atoms of the tetraloops, solvent sites were considered structural if a cluster of at least four solvent molecules existed in which no two atoms in the cluster were farther than 0.8 Å apart.
Coordinate accession codes
Coordinates and structure factors for the GUAA structure were deposited with the PDB as entry number 1MSY.
Acknowledgments
We are grateful to Ya-Ling Wu and Matthew Plantinga for help in model building, and the staff of SBC for help with data collection. Use of the Argonne National Laboratory Structural Biology Center beamline at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under Contract No. W-31-109-ENG-38. The work was supported by a grant from the National Institutes of Health to C.C.C. (GM59782).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2147803.
REFERENCES
- Abramovitz, D.L., Friedman, R.A., and Pyle, A.M. 1996. Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science 271: 1410–1413. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Rambo, R.P., Lucast, L., Rha, B., and Doudna, J.A. 2000. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 287: 1232–1239. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Sagar, M.B., and Doudna, J.A. 2001. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 307: 229–246. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T. 1992. X-PLOR Version 3.1: A system for X-ray crystallography and NMR. Yale University Press, New Haven, CT.
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Cameron, D.M., Thompson, J., March, P.E., and Dahlberg, A.E. 2002. Initiation factor IF2, thiostrepton and micrococcin prevent the binding of elongation factor G to the “Escherichia coli” ribosome. J. Mol. Biol. 319: 27–35. [DOI] [PubMed] [Google Scholar]
- Carson, M. 1991. Ribbons. Methods Enzymol. 277: 493–505. [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., and Doudna, J.A. 1996. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science 273: 1678–1685. [DOI] [PubMed] [Google Scholar]
- Correll, C.C., Munishkin, A., Chan, Y.L., Ren, Z., Wool, I.G., and Steitz, T.A. 1998. Crystal structure of the ribosomal RNA domain essential for binding elongation factors. Proc. Natl. Acad. Sci. 95: 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correll, C.C., Wool, I.G., and Munishkin, A. 1999. The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: The structure at 1.11 A resolution. J. Mol. Biol. 292: 275–287. [DOI] [PubMed] [Google Scholar]
- Dervan, P.B. and Burli, R.W. 1999. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 3: 688–693. [DOI] [PubMed] [Google Scholar]
- Desiraju, G.R. 1996. The C-H…O hydrogen bond: Structural implications and supramolecular design. Acc. Chem. Res. 29: 441–449. [DOI] [PubMed] [Google Scholar]
- Doherty, E.A., Batey, R.T., Masquida, B., and Doudna, J.A. 2001. A universal mode of helix packing in RNA. Nat. Struct. Biol. 8: 339–343. [DOI] [PubMed] [Google Scholar]
- Gelbin, A., Schneider, B., Clowney, L., Hsieh, S., Olson, W.K., and Berman, H.M. 1996. Geometric parameters in nucleic acids: Sugar and phosphate constraints. J. Am. Chem. Soc. 118: 519–529. [Google Scholar]
- Hainzl, T., Huang, S., Sauer-Eriksson, A.E. 2002. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature 417: 767–771. [DOI] [PubMed] [Google Scholar]
- Heus, H.A. and Pardi, A. 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253: 191–194. [DOI] [PubMed] [Google Scholar]
- Horton, T.E., Maderia, M., and DeRose, V.J. 2000. Impact of phosphorothioate substitutions on the thermodynamic stability of an RNA GAAA tetraloop: An unexpected stabilization. Biochemistry 39: 8201–8207. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Jucker, F.M., Heus, H.A., Yip, P.F., Moors, E.H., and Pardi, A. 1996. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 264: 968–980. [DOI] [PubMed] [Google Scholar]
- Juneau, K., Podell, E., Harrington, D.J., and Cech, T.R. 2001. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure (Camb.) 9: 221–231. [DOI] [PubMed] [Google Scholar]
- Kielkopf, C.L., Baird, E.E., Dervan, P.B., and Rees, D.C. 1998. Structural basis for G.C recognition in the DNA minor groove. Nat. Struct. Biol. 5: 104–109. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana, A., Gualerzi, C.O., and Dahlberg, A.E. 2001. Initiation factor IF 2 binds to the α-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA 7: 1173–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas Saiz, A.L. and Foces-Foces, C. 1990. N-H. . .Nsp2 hydrogen interactions in organic crystals. J. Mol. Struct. 238: 367–382. [Google Scholar]
- Menger, M., Eckstein, F., and Porschke, D. 2000. Dynamics of the RNA hairpin GNRA tetraloop. Biochemistry 39: 4500–4507. [DOI] [PubMed] [Google Scholar]
- Munishkin, A. and Wool, I.G. 1997. The ribosome-in-pieces: Binding of elongation factor EF-G to oligoribonucleotides that mimic the sarcin/ricin and thiostrepton domains of 23S ribosomal RNA. Proc. Natl. Acad. Sci. 94: 12280–12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B., and Steitz, T.A. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 98: 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons Jr., W.M., Tarry, M.J., Carter, A.P., and Ramakrishnan, V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902. [DOI] [PubMed] [Google Scholar]
- Olmo, N., Turnay, J., De Buitrago, G.G., De Silanes, I.L., Gavilanes, J.G., and Lizarbe, M.A. 2001. Cytotoxic mechanism of the ribotoxin α-sarcin induction of cell death via apoptosis. Eur. J. Biochem. 268: 2113–2123. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pley, H.W., Flaherty, K.M., and McKay, D.B. 1994a. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature 372: 111–113. [DOI] [PubMed] [Google Scholar]
- ———. 1994b. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74. [DOI] [PubMed] [Google Scholar]
- Rife, J.P., Stallings, S.C., Correll, C.C., Dallas, A., Steitz, T.A., and Moore, P.B. 1999. Comparison of the crystal and solution structures of two RNA oligonucleotides. Biophys. J. 76: 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia Jr., J., Kierzek, R., and Turner, D.H. 1992. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science 256: 217–219. [DOI] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., and Klug, A. 1995. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 81: 991–1002. [DOI] [PubMed] [Google Scholar]
- Seggerson, K. and Moore, P.B. 1998. Structure and stability of variants of the sarcin-ricin loop of 28S rRNA: NMR studies of the prokaryotic SRL and a functional mutant. RNA 4: 1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick, G.M. and Schneider, T.R. 1997. SHELXL: High-resolution refinement. Methods Enzymol. 277: 319–343. [PubMed] [Google Scholar]
- Silverman, S.K. and Cech, T.R. 1999. RNA tertiary folding monitored by fluorescence of covalently attached pyrene. Biochemistry 38: 14224–14237. [DOI] [PubMed] [Google Scholar]
- Sorin, E.J., Engelhardt, M.A., Herschlag, D., and Pande, V.S. 2002. RNA simulations: Probing hairpin unfolding and the dynamics of a GNRA tetraloop. J. Mol. Biol. 317: 493–506. [DOI] [PubMed] [Google Scholar]
- Szewczak, A.A. and Moore, P.B. 1995. The sarcin/ricin loop, a modular RNA. J. Mol. Biol. 247: 81–98. [DOI] [PubMed] [Google Scholar]
- Szewczak, A.A., Moore, P.B., Chan, Y.L., and Wool, I.G. 1993. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc. Natl. Acad. Sci. 90: 9581–9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston, S.A., Tucker, A.D., Thatcher, D.R., Derbyshire, D.J., and Pauptit, R.A. 1994. X-ray structure of recombinant ricin A-chain at 1.8 A resolution. J. Mol. Biol. 244: 410–422. [DOI] [PubMed] [Google Scholar]
- Williamson, J.R. 2000. Induced fit in RNA–protein recognition. Nat. Struct. Biol. 7: 834–837. [DOI] [PubMed] [Google Scholar]
- Wool, I.G. 1997. Structure and mechanism of action of the cytotoxic ribonuclease a-sarcin. In Ribonucleases structures and functions (eds. G. D’Alessio et al.), pp. 131–162. Academic Press, Inc., San Diego, CA.
- Wool, I.G., Gluck, A., and Endo, Y. 1992. Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends. Biochem. Sci. 17: 266–269. [DOI] [PubMed] [Google Scholar]
- Yang, X., Gerczei, T., Glover, L., and Correll, C.C. 2001. Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping. Nat. Struct. Biol. 8: 968–973. [DOI] [PubMed] [Google Scholar]