Abstract
Regulated expression of proteins involved in mammalian iron metabolism is achieved in part through the interaction of the iron regulatory proteins IRP1 and IRP2 with highly conserved RNA stem-loop structures, known as iron-responsive elements (IREs), that are located within the 5′ or 3′ untranslated regions of regulated transcripts. As part of an effort to determine the structures of the IRP–IRE complexes using crystallographic methods, we have developed an efficient process for obtaining functionally pure IRP1 and IRP2 that relies upon the improved overexpression (>10 mg of soluble IRP per liter of culture) of each human IRP in the yeast Pichia pastoris and large-scale purification using RNA affinity chromatography. Despite the utility of RNA affinity chromatography in the isolation of RNA-binding proteins, current methods for preparing RNA affinity matrices produce columns of low capacity and limited stability. To address these limitations, we have devised a simple method for preparing stable, reusable, high-capacity RNA affinity columns. This method utilizes a bifunctional linker to covalently join a 5′-amino tethered RNA with a thiol-modified Sepharose, and can be used to load 150 nmole or more of RNA per milliliter of solid support. We demonstrate here the use of an IRE affinity column in the large-scale purification of IRP1 and IRP2, and suggest that the convenience of this approach will prove attractive in the analysis of other RNA-binding proteins.
Keywords: 5′-aminoalkyl tether, affinity chromatography, iron regulatory proteins, iron-responsive element (IRE), protein purification, RNA-binding proteins, thiol-modified sepharose
INTRODUCTION
The isolation and characterization of many RNA-binding proteins has been greatly facilitated by the use of RNA affinity chromatography (Rouault et al. 1989; Neupert et al. 1990; Copeland and Driscoll 1999; Sela-Brown et al. 2000). Although numerous strategies have been employed in the preparation of RNA affinity matrices (for review, see Kaminski et al. 1998), these methods generally involve the covalent or noncovalent immobilization of the RNA on a solid support. Typically, noncovalent immobilization methods rely upon either the hybridization of poly-A sequences within the target RNA to anchored oligo-U or oligo-dT, or the association of biotinylated RNAs with streptavidin-bearing matrices. Two recent innovations have used RNA aptamers with affinities for streptavidin (Srisawat and Engelke 2001) or streptomycin (Bachler et al. 1999) to immobilize the target RNA.
Covalent attachment of RNA has most typically been achieved through either the reaction of unmodifed RNAs with cyanogen bromide activated Sepharose, or the coupling of periodate-oxidized RNA with adipic acid dihydrazide-agarose beads (Caputi et al. 1999). In the former, reaction between the RNA and the matrix happens at random locations, which presumably limits the number of accessible protein binding sites. The latter method requires the treatment of the RNA with an oxidant and limits attachment to the 5′ end of the RNA.
Despite the obvious advantages of RNA affinity chromatography, it is rarely used for the routine purification of overexpressed RNA-binding proteins. For large-scale purification, the benefits of RNA affinity chromatography are often discarded in favor of more traditional methodologies. Practical preparative RNA affinity chromatography would require an RNA affinity column that has both high capacity and long-term stability. To the best of our knowledge, none of the above methods, covalent or noncovalent, has ever succeeded in loading more than a few nanomoles of RNA per milliliter of solid support. Furthermore, the intrinsic reversibility of noncovalent associations limits the lifetime and reusability of columns prepared by those methods.
Two extensively studied mammalian mRNA-binding proteins are the iron regulatory proteins IRP1 and IRP2. These cytosolic proteins regulate the expression of proteins involved in iron metabolism through their interaction with conserved RNA stem-loop structures, referred to as iron-responsive elements (IREs), that are located within the 5′ or 3′ untranslated regions (UTR) of regulated transcripts (for review, see Hentze and Kuhn 1996; Harford and Rouault 1998). Under conditions of intracellular iron depletion, both IRP1 and IRP2 are capable of binding IREs with high affinity, resulting in either repression of translation or stabilization of the transcript. When iron levels rise, the two IRPs respond via distinct mechanisms, both of which result in the loss of IRE-binding activity. Whereas IRP2 is targeted for degradation by the proteasome (Guo et al. 1995; Iwai et al. 1995, 1998), IRP1 is loaded with a [4Fe-4S] cluster that impedes its ability to bind IREs while converting it into a functional cytosolic aconitase (for review, see Beinert et al. 1996; Hentze and Kühn 1996; Rouault and Klausner 1996; Schneider and Leibold 2000).
Efforts to understand the manner in which IRP1 and IRP2 bind to their IRE substrates have been hindered by both a lack of structural information and the absence of any recognizable RNA recognition motif within the IRP sequences. However, the similarity between IRP1 and other aconitases has provided a useful framework for analyzing IRP structure (Rouault et al. 1991, 1992). Crystal structures of mitochondrial aconitases reveal a common folding motif in which the aconitase active site lies in a cleft formed between the N-terminal globular domains 1–3 and the C-terminal domain 4 (Robbins and Stout 1989). Several lines of evidence suggest that both IRP1 and IRP2 adopt similar conformations, and that the IRE-binding site lies within the active site cleft (Basilion et al. 1994b; Hirling et al. 1994; Philpott et al. 1994; Swenson and Walden 1994; Butt et al. 1996). NMR studies (Laing and Hall 1996; Addess et al. 1997) and analysis of IRE mutations (Allerson et al. 1999; Meehan and Connell 2001) have led to the identification of nucleotides that are likely to be involved in specific contacts with the IRPs. A recent study of IRP mutants has further identified short stretches of amino acids that are likely to interact with these critical nucleotides (Kaldy et al. 1999). However, a complete understanding of the IRP–IRE complexes awaits their analysis by X-ray crystallographic methods.
As part of our effort to determine the structures of the IRP–IRE complexes, we set out to develop an optimized strategy for overexpressing and purifying both human IRPs. We have developed an overexpression system using the methylotrophic yeast Pichia pastoris, in which yields greatly exceed those of a previously developed baculovirus expression system (Basilion et al. 1994a; Samaniego et al. 1994). Use of the P. pastoris system provides the combined benefits of high-level expression, easy scale-up, and the ability to express larger eukaryotic proteins (Cregg et al. 2000).
The eventual crystallization of the IRP–IRE complexes will require functionally pure IRPs that are fully capable of being bound to IREs. However, the IRE-binding activities of expressed IRP1 and IRP2 have been shown to be sensitive to oxidation in vitro (Phillips et al. 1996), giving rise to the possibility of functional isomers. To eliminate the nonbinding isomers and ensure functional homogeneity, we have adopted a purification strategy that employs RNA affinity chromatography as a final step. To circumvent the limitations of existing methodologies, we have developed a novel procedure for covalently linking an amine-modified RNA to a Sepharose support that should provide the capacity and stability required of a truly reusable column. We report here the overexpression of human IRP1 and IRP2, the preparation of an RNA affinity column with an estimated capacity of ∼15 mg of IRP, and its use in the isolation of functionally enriched IRPs.
RESULTS AND DISCUSSION
Expression of human IRP1 and IRP2
To obtain the quantities of protein required for future crystallographic studies, we overexpressed human IRP1 and IRP2 in the yeast P. pastoris. Our laboratory had previously reported good expression of IRPs in insect cells using a baculovirus system (Basilion et al. 1994a; Samaniego et al. 1994), but the yields of this system were variable and insufficient for our purposes. Although the expression of rat IRPs in Saccharomyces cerevisiae has achieved modest levels (1–2 mg/L of culture; Phillips et al. 1996), the methylotrophic yeast P. pastoris has been known to occasionally reach grams per liter protein yields (Sreekrishna et al. 1988). This system, which takes advantage of the strong alcohol oxidase (AOX1) promoter that is activated when the optimal carbon source of glucose is replaced with methanol, seemed ideally suited for IRP expression.
Of the many vectors available for expression in P. pastoris, we used the intracellular expression vector pHIL-D2, which was included with the original expression kit from Invitrogen. We selected intracellular expression in an effort to minimize posttranslational modification of the IRPs and to protect the expressed protein (especially IRP2) from unchelated metals within the expression media. Based on a report that expression levels can be dramatically improved by placing the start codon immediately adjacent to the end of the AOX1 5′ UTR (Sreekrishna et al. 1997), we made several modifications to pHIL-D2. In that report, a modified expression plasmid was designed to take advantage of a BstBI (TTCGAA) site located in the last 9 bp of the AOX1 5′ UTR. By appending the coding region insert with the sequence TTCGAAACG ATG . . . (start codon in italics), the BstBI site can be used to obtain the optimal 5′ UTR sequence. To employ a similar strategy with pHIL-D2, we removed a second BstBI site from the 3′ AOX1 region of the plasmid, and introduced a simple multiple cloning site (containing unique SacII, BamHI, and XhoI restriction sites) to create the expression vector pCA10.3.
From plasmids containing the coding sequence of either human IRP1 (pGEM-hIRF; Hirling et al. 1992) or a myc epitope-tagged form of human IRP2 (Samaniego et al. 1994), coding regions were isolated and subcloned into pBluescript II SK(+) to facilitate the introduction of the sequence TTCGAAACG upstream of each start codon, the elimination of a preexisting BstBI site in IRP2, and the removal of the myc-epitope tag from IRP2. Subsequent subcloning into pCA10.3 created the IRP1 and IRP2 expression constructs pCA19.8 and pCA22.1, respectively.
The P. pastoris strain GS115 was transformed with pCA19.8, pCA22.1, or control plasmid pCA10.3. After screening of the transformants, small-scale expression tests led to the identification of the optimal IRP1-expressing clone CA1302 (Fig. 1A ▶, lane 5), IRP2-expressing clone CA1506 (Fig. 1B ▶, lane 5), and a control nonexpressing clone CA1103 (Fig. 1A,B ▶, lane 3). Each of the optimal expression clones appeared to contain multiple copies of integrated insert (data not shown). Both IRP1 and IRP2 were optimally expressed after 36 h of growth in methanol-containing media and were readily visualized in unpurified total lysates.
FIGURE 1.
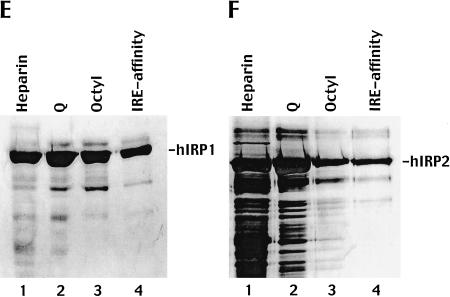
Unlabeled forms of both human IRP1 and human IRP2 were specifically and efficiently expressed in Pichia pastoris and purified to near homogeneity. (A) Comparison of lysates from yeast transformed with vector alone to those of yeast transformed with the IRP1-containing vector by 8% SDS-PAGE shows that despite their near identity in the absence of methanol induction (lanes 2,4), methanol induction of the IRP1-transformed yeast causes the appearance of a unique species (lane 5) with a molecular weight of the approximately correct mass for IRP1 (∼98 kD). (B) A similar comparison of vector-transformed and IRP2-vector transformed yeast also highlights the appearance of a unique species (lane 5) with an apparent molecular weight that closely matches the expected mass of IRP2 (∼104 kD). (C) The purification of IRP1 from P. pastoris lysates through a series of four chromatographic columns (Heparin Sepharose, lane 1; Q Sepharose, lane 2; Octyl Sepharose, lane 3; and preparative IRE-affinity Sepharose, lane 4) yielded a nearly homogeneous protein preparation as determined by Coomassie-stained SDS-PAGE. The band appearing at ∼75 kD may correspond to a nicked form of IRP1 that fragments upon denaturation onto the gel (data not shown). (D) The purification of IRP2 from lysates through an identical procedure also yields protein of very high purity, especially following preparative IRE-affinity chromatography. Each lane of C and D contained approximately 4 μg of total protein (as determined by BCA assay). (E,F) Analyses of IRP1 (E) and IRP2 (F) purification identical to those depicted in C and D, respectively, except that silver staining was used to visualize the protein and less protein was used per lane.
Initial purification of IRP1 and IRP2
Yeast lysates were subjected to one or more rounds of chromatographic purification, then analyzed by both Coomassie-stained and silver-stained SDS-PAGE. An initial pass through a Heparin-Sepharose column resulted in a substantial enrichment of both IRPs relative to the initial lysates (Fig. 1C,D ▶, lane 1). Subsequent fractionation through a Q-Sepharose column (Fig. 1C,D ▶, lane 2), followed by an octyl-Sepharose column (Fig. 1C,D, lane 3) yielded IRPs of very high purity. The efficiency of this stepwise purification was confirmed using the enhanced sensitivity of silver-staining methods (Fig. 1E,F, lanes 1–3). Typically, the lysis and purification of 0.5 L of yeast culture yielded 7–9 mg of purified IRP after these three chromatographic steps.
To determine the specific IRE-binding activity of the IRPs after these three chromatographic steps, each preparation was used in band-shift RNA-binding titration experiments, as illustrated for IRP1 in Figure 5A ▶ (see below). Despite their relative purity, the amount of IRE-binding activity of the IRPs varied considerably between batches of purified protein, and was always much less than expected based on determinations of protein concentration. In one batch of IRP1 (Fig. 5A ▶, see below), the specific activity was measured as approximately 30% of the total protein. Although some of the deviation from the expected maximum is likely to arise from the inaccuracies of determining both protein concentration (calibrated against bovine serum albumin standards) and RNA concentration (calculated from absorbance at 260 nm), the batch-wise variation seemed to be a separate phenomenon linked to the IRPs themselves. In several instances, the specific activity was lower than 10% of maximum, and was consistently lower for IRP2 (data not shown) than for IRP1. Although not rigorously investigated, there also appeared to be a correlation between the elapsed time of purification and measured activity, where longer purification times reflected lower activities. These observations are in general agreement with previous reports of diminished IRP2 activity during purification (Phillips et al. 1996). Although some of the loss of IRE-binding activity can be reversed by the addition of reducing agents, there is no assurance that this reversion is complete.
FIGURE 5.
The specific activity of IRPs, both before and after purification through an IRE-affinity column, can be determined through the use of electrophoretic band-shift assays. (A) A constant concentration of 50 nM of 32P-labeled C-IRE was treated with increasing concentrations of nonaffinity-purified IRP1, then electrophoresed on a 10% nondenaturing polyacrylamide gel. Autoradiography revealed that the specific binding activity of IRP1 was approximately 30%. Although not shown, nonaffinity-purified IRP2 had even lower specific activity. (B) Affinity-purified IRP1 was analyzed in an experiment similar to that shown in A. Autoradiography revealed at least a twofold improvement in the specific activity in comparison to nonaffinity-purified protein. (C) Affinity-purified IRP2 was analyzed in a similar fashion to reveal an apparent specific activity of ∼50%, which was substantially greater than the activity of nonaffinity-purified protein (not shown).
Preparation of a high-capacity RNA affinity column
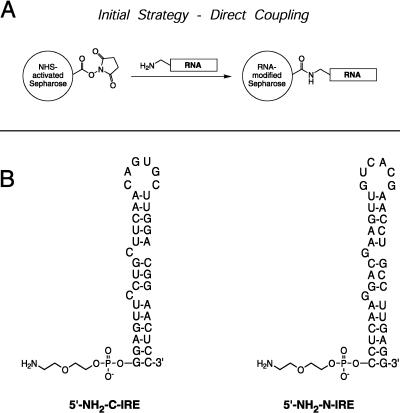
The variable specific activity and residual impurities in the purified IRP1 and IRP2 highlighted the need for RNA affinity purification. High-capacity DNA affinity columns have previously been prepared for use in purification of DNA-binding proteins through the covalent coupling of alkylamine-tethered DNA with NHS-activated Sepharose columns (Larson and Verdine 1992). This method was used to obtain a column with a loading of 250 nmole of DNA per milliliter of matrix and uses prepacked HiTrap columns (Amersham Pharmacia) that simplify column preparation and use. We reasoned that this strategy should work equally well with an alkylamine-modified RNA, as illustrated schematically in Figure 2A ▶.
FIGURE 2.
A conceptually simple method for the covalent attachment of an RNA to an NHS-modified support. (A) An RNA bearing an alkylamine tether, whether located at the 5′ end of the RNA or elsewhere, ought to react through a direct nucleophilic attack at the N-hydroxysuccinimidyl-ester activated carbonyl. This direct coupling proved too inefficient for the preparation of a large-capacity column. (B) Two sequences were used for the synthesis of RNA columns. The RNA 5′-NH2-C-IRE corresponds to a consensus iron-responsive element sequence, and has a 5-atom alkyl-ether amine tether appended to the 5′ terminal phosphate. The negative control, 5′-NH2-N-IRE is similarly modified with a 5′-alkylamine tether, but has a sequence that is the exact complement of the consensus IRE.
We designed both a consensus IRE sequence (C-IRE) and a non-IRE stem-loop (N-IRE) in which the sequence was inverted to generate the complementary stem-loop. Each of these RNAs was obtained bearing a 5′-alkylamine tether, yielding the modified RNAs 5′-NH2-C-IRE and 5′-NH2-N-IRE (Fig. 2B ▶). However, when we attempted to directly couple these RNAs to NHS-activated Sepharose HiTrap columns, we observed very low coupling efficiences, regardless of length of treatment or buffer conditions. In the best case, only a little over 3 nmole of RNA (of an initial 150 nmole) coupled to the resin for a coupling efficiency of 2%. This low efficiency might be the result of a number of factors. First, in the original report of this strategy for affinity column preparation (Larson and Verdine 1992), larger quantities of DNA were used (at least 500 nmole), which might exceed a concentration threshold for efficient coupling. Additionally, in one version, the DNA was ligated into a multimer that contained many alkylamines, in which case the reaction of a single amine would result in the loading of multiple copies of the DNA affinity site. Even when not ligated, the complementary overhanging ends of these DNAs might permit their association into noncovalent multimers that increase the apparent coupling efficiency. The RNA sequences used here are unable to associate through a similar mechanism, and thus must react with the activated support as true monomers. It is also possible that the reactivity of the alkylamine tether in 5′-NH2-C-IRE and 5′-NH2-N-IRE is somehow diminished, either sterically or electrostatically, by its local environment. Regardless, the direct coupling of the singly tethered IREs proved to be too inefficient for large-scale use.
Despite the low efficiency of direct coupling, we devised an alternative strategy in which a bridging linker would be used to connect the same alkylamine-modified RNA with the Sepharose matrix (illustrated schematically in Fig. 3A ▶). We realized that to obtain optimal coupling efficiency, the coupling of the RNA to the matrix ought to involve a process in which competing reactions are minimal or nonexistent. The chemistry involving the reaction of a thiol with the iodoacetamide moiety seemed to be ideally suited for our purposes.
FIGURE 3.
An alternative approach to covalent attachment of an alkylamine-modified RNA to an activated solid support. (A) This strategy utilizes a bifunctional linker (sulfo-SIAB) that bears an amine-reactive NHS-ester at one end and a thiol-reactive iodoacetamide group at the other. Starting with a thiol-modified solid matrix (Sepharose), a simple two-step procedure can be employed to covalently attach an alkylamine-modified RNA to the solid support. (B) One option for creating a thiol-modifed Sepharose is through the treatment of NHS-ester-activated Sepharose with the diamine cystamine. Subsequent reduction of the internal mixed disulfide with dithiothreitol produces the free-thiol. The yield of this reaction can be readily and reversibly measured through the treatment of the modified support with DTNB and measurement of the released chromophore at a wavelength of 412 nm. (C) The linker sulfo-SIAB reacts specifically and quantitatively with 5′-alkyamine modified RNAs (lanes 4,8). No reaction occurred between sulfo-SIAB and unmodified RNAs (lanes 2,6). Analysis is performed using 10% denaturing PAGE, 250 pmole of RNA per lane, and visualization by methylene blue.
The conversion of the hydrolytically unstable NHS-activated Sepharose HiTrap column into an alkyl-thiol column was achieved through the treatment of the column with the disulfide-containing diamine cystamine, followed by reduction with DTT (Fig. 3B ▶). The efficiency of this conversion was readily determined through the reversible reaction of the thiol group with 5,5′-dithiobis(2-nitrobenzoic acid) and is typically about 80% (8 μmole thiol per 1-mL column). In our experience, the column can be stored at 4°C in either the disulfide or DTNB-treated form for at least a month. Although several thiol matrices are commercially available (e.g., Thiol Sepharose 4B and Thiopropyl-activated agarose, from either Amersham Biotech or Sigma), the ease of performing the cystamine conversion and convenience of the HiTrap columns make this a simple and attractive alternative.
The 5′-amine tethered RNA is readily appended with a 5′-iodoacetamide group in a nearly quantitative reaction with the bifunctional linker sulfo-SIAB (Fig. 3C ▶), which reacts selectively with the 5′ amine tethers as demonstrated by the absence of any adduct with unmodified RNAs (Fig. 3C ▶, lanes 2,6). At this stage, the RNA can be stored in lyophilized form in the dark for at least several weeks. Upon regenerating the free thiol form of the column, the RNA is efficiently coupled to the column. Treatment of the column with 200 nmole of RNA typically resulted in the coupling of 150–170 nmole of RNA, for yields of 75%–85%. Any unreacted thiols on the column were then blocked by treatment with iodoacetamide. This method was used to prepare both the C-IRE and N-IRE columns, plus a control column in which an NHS-activated column had been treated with ethanolamine instead of cystamine.
Preparative RNA affinity purification of IRP1 and IRP2
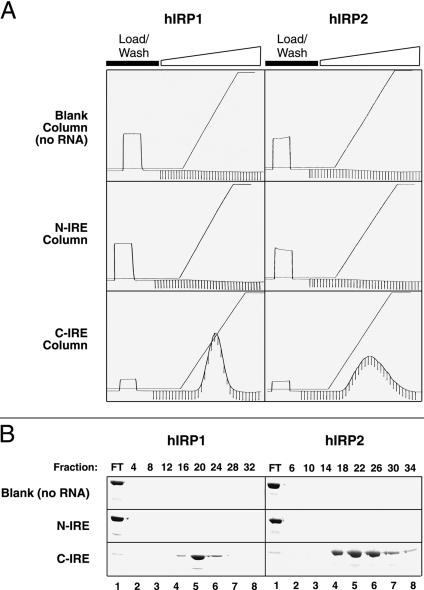
In turn, partially purified IRP1 and IRP2 were eluted through the blank (no RNA) column, the non-IRE (N-IRE) column, and the consensus IRE (C-IRE) column (Fig. 4A ▶). The binding and elution of both IRPs appeared to be highly specific. Elution of the IRPs from the C-IRE column using a salt gradient required approximately 1200 mM NaCl for IRP1 and 1500 mM NaCl for IRP2, reflecting extraordinarily tight binding. Proteins were bound at 250 mM NaCl. Fractions collected from each column were examined by Coomassie SDS-PAGE (Fig. 4B ▶) to determine purity and tested for IRE-binding activity (data not shown). Despite the appearance of residual Coomassie-detectable IRP in the C-IRE column flow-through (Fig. 4B ▶, lanes 1), this protein had negligible IRE-binding activity (data not shown). In comparison to the pre-affinity-purified protein, both IRP1 and IRP2 preparations appeared only marginally cleaner by both Coomassie-stain and silver-stain SDS-PAGE (Fig. 1C ▶‘F, cf lanes 3 and 4). Although there appear to be substantial amounts of contaminating proteins present in both IRP1 and IRP2 preparations, we suspect that many, if not all, of these species represent fragments of the respective parent IRP. In support of this suspicion, Western blot analysis confirms that at least some of the most prominent impurities are derived from the parent IRP sequence (data not shown). We speculate that the heat denaturation required for electrophoresis may cause the dissociation of backbone-nicked forms of the IRPs that, despite the accumulation of oxidative or chemical damage during expression and purification, have retained their proper high-affinity IRE-binding conformation. We are continuing to examine the nature of these contaminating species and have been investigating ways to minimize their abundance.
FIGURE 4.
The large-scale, high-affinity binding of hIRP1 and hIRP2 is specific for the consensus IRE (C-IRE) column. (A) Protein preparations (containing an estimated 7–10 mg of protein) of hIRP1 or hIRP2 collected at the completion of the first three chromatographic steps were equilibrated in 250 mM NaCl, then passed through a column containing no RNA, a column with a disrupted IRE (N-IRE), or a column with the consensus IRE (C-IRE). No binding of protein was detected with either the no-RNA or N-IRE columns, despite the large quantity of RNA on the latter. In contrast, both proteins bound in large quantity (7–10 mg), and with high affinity to the C-IRE column, requiring ∼1.2 M NaCl and ∼1.5 M NaCl, respectively, to elute. (B) SDS-PAGE analysis of fractions collected during chromatography indicated no Coomassie-detectable protein in either of the first two columns. Some IRP appears in the flow-through of the C-IRE column, and may arise from either slow binding kinetics during loading or may indicate the presence of inactive conformers.
The improvement in IRP quality afforded by affinity purification was more dramatically revealed by the measurement of specific activity. Following desalting, the affinity-purified IRPs were assayed for specific activity by RNA band-shift titration (Fig. 5B,C ▶). In comparison to activities measured prior to RNA affinity chromatography (IRP1, Fig. 5A ▶; IRP2, data not shown), both IRP1 and IRP2 had much greater specific activities. Even though the activities still appear to be less than 100% (∼70% for IRP1, ∼50% for IRP2), it seems more reasonable here to assume that much of this discrepancy reflects the inherent inaccuracies of protein and RNA concentration determination discussed above. Regardless, the specific activities of both proteins are significantly improved through the use of affinity chromatography.
Although we have prepared multiple C-IRE columns, a single column was sufficient for the purifications of both IRP1 and IRP2 shown above. One C-IRE column has been used repeatedly for up to a month without any noticeable deterioration in binding capacity (data not shown). For both IRPs, the column had sufficient capacity to bind all of the IRP from a single 0.5 L expression culture. Although not seen here, column saturation causes a change in the loading profile and is easily detected by visual inspection of the chromatographic detector output. The column used here, with an RNA loading of 150 nmole, should have theoretical capacities of 14.7 mg for IRP1 and 15.8 mg for IRP2, assuming all IRE sites are available.
The development of the high-capacity IRE column has allowed us to streamline the purification process, limiting the process to three chromatographic steps followed by affinity purification. Furthermore, the use of common elution buffers and the rapid flow rates allowed by the use of HiTrap columns make for a very simple, straightforward process.
One of the few drawbacks of this purification process is the elution of the IRPs into high salt. This necessitates a desalting step prior to the use of these preparations for crystallographic analysis. However, most protein samples require concentration before they can be used for crystallization, which makes ultrafiltration a convenient way to simultaneously desalt and concentrate the IRP samples. Another alternative to the final high-salt elution that we are considering is the use of RNA columns containing mutant IREs with specific yet slightly diminished affinities, which might allow elution with lower salt concentrations. Yet another possibility that we are examining is the use of a cleavable linker in the RNA-Sepharose attachment. This might permit the use of the column as a way of preassembling the IRP–IRE complexes for subsequent release in the bound state, which could then be used directly for structural studies.
Although in this example we used chemically synthesized 5′-aminoalkyl tethered RNAs, there are many other ways to incorporate alkylamino groups into RNA. Site-specific placement of reactive amines at internal sites can also be achieved through the use of convertible ribonucleosides (Allerson et al. 1997) in solid phase synthesis. The ability of T7 RNA polymerase to incorporate modified nucleosides and monophosphates into the 5′-most residue should also allow the preparation of amine-tethered RNAs that are much larger than those available through chemical synthesis. Together, these methods should permit the use of virtually any RNA for large-scale affinity chromatography.
This approach for preparing RNA affinity columns has the potential to be quite versatile. The entire process of column preparation is straightforward, requires no special equipment, and can be completed within a single day. The efficiency and yield of the coupling reaction between the linker-modified RNA and the thiol-modified column far exceeds what has been achieved by any other method. The reduced likelihood of side reactions during coupling permits the use of extended coupling times, thus assuring efficient coupling even with lower quantities of RNA. Even greater yields might be attained if the RNA solution is cycled through the column during coupling.
The use of large-scale RNA affinity column promises to greatly facilitate structural studies of IRP–IRE interactions, and may prove equally advantageous in the analysis of other RNA–protein complexes.
MATERIALS AND METHODS
General
Chemical reagents were obtained from Sigma-Aldrich, with the exception of the yeast nitrogen base (YNB), which was from BIO101, Inc., and sulfo-SIAB (Molecular Probes). Enzymes were from either Life Technologies or New England Biolabs. DNA oligonucleotides were custom synthesized by Life Technologies. The 5′-amino modified RNA oligomers were prepared by Dharmacon Research. Chromatographic purifications were performed at 4°C using a Pharmacia FPLC system. HiTrap Heparin-Sepharose, Q Sepharose, Phenyl-Sepharose, and NHS-activated Sepharose columns were from Amersham Biotech. Yeast breaking buffer (BB) is composed of 25 mM Tris-HCl (pH 8.3), 20 mM KCl, 1 mM EDTA, 0.1 mM desferrioximine, 1 mM 2-mercaptoethanol, 1 mM aminoethylbenzenesulfonyl fluoride (AEBSF), 5 μg/mL leupeptin, 10 μg/mL aprotinin. FPLC Chromatographic buffers are Buffer A: 25 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, 0.1 mM desferrioximine, 0.1 mM 2-mercaptoethanol; and Buffer B: 25 mM Tris-HCl (pH 8.0), 0.5 mM EDTA, 0.1 mM desferrioximine, 0.1 mM 2-mercaptoethanol, 3 M NaCl.
Construction of P. pastoris expression vector pCA10.3
The BstBI site at position 4832 in plasmid pHIL-D2 was changed to TTCGGA using two-step PCR. Briefly, primers Forward1 (5′-GCGCTTGTTTCGGCGTGGGTATGGTGGC-3′), which lies 39 bp upstream of the SphI site, and MutP (5′-GGCACCGAAAAA GTTCGGAACAAGAAGTTTGTTCC-3′) were used to generate an ∼430 bp product. This fragment was used in a second PCR reaction with primer Reverse1 (5′-GCATTTATCAGGGTTATTGTC TCATGAGCGG-3′) to yield a 1239-bp product containing the disrupted BstBI site. Following digestions with SphI and AatII, the resulting fragment was used to replace the corresponding region of pHIL-D2 to yield plasmid pCA7.3.
A 60-bp region of pCA7.3 between the remaining BstBI site and a downstream AgeI site of pCA7.3 was replaced with a 23-bp synthetic polylinker to create pCA10.3. This introduced unique SacII, BamHI, and XhoI sites, and was generated by the BstBI/AgeI double digestion of a duplex formed by two synthetic oligonucleotides (NL1, 5′-CCGCATACAATTCGAAACGACCTACCGCGGA TCCTCGAGACCGGTCTAGGCAAGG-3′, and NL2, 5′-CCTTG CCTAGACCGGTCTCGAGGATCCGCGGTAGGTCGTTTCGAA TTGTATGCGG-3′).
Construction of human IRP1 expression vector (pCA19.8)
A BamHI/HindIII fragment bearing all but the first 35 bp of the coding region of human IRP1, plus a portion of the 3′ UTR, was liberated from pGEM-hIRF (Hirling et al. 1992) and subcloned into the BamHI/HindIII site of pBluescript II SK(+) to generate plasmid pCA6.6. To replace the remainder of the coding region and add the BstBI site, a 72-bp duplex was made from two synthetic oligonucleotides (H1F1, 5′-CCATTGGCCCGCGGTTC GAAACGATGAGCAACCCATTCGCACACCTTGCTGAGCCAT TGGATCCCCGTACCG-3′; H1F2, 5′-cggtacggggatccaatggc tcagcaaggt gtgcgaatgg gttgctcatcgtttcgaaccgcgggccaat gg-3′). This duplex was double digested and inserted into the SacII/BamHI site of pCA6.6 to create plasmid pCA13.5. An ∼320-bp MscI/XhoI fragment was removed from pCA13.5 and replaced with a short synthetic duplex (top strand 5′-CCAAGTAGC-3′, bottom strand 5′-TCGAGCTACTTGG-3′) to generate plasmid pCA16.2. The BstBI/XhoI fragment was released from pCA16.2 and cloned into the BstBI/XhoI site of pCA10.3, creating the human IRP1 expression plasmid pCA19.8.
Construction of human IRP2 expression vector (pCA22.1)
A fragment encoding a myc-epitope tagged form of human IRP2, plus some vector-derived sequence (Samaniego et al. 1994), was subcloned into the XhoI/KpnI site of pBluescript II KS(+) to generate pCA12.1. A BstBI site upstream of the start codon was introduced by PCR amplification of the first ∼400 bp of the coding region using a mutagenic forward primer (H2F1, 5′-CCGCCTA ATCTAGATTCGAAACGATGGACGCCCAAAAGCAGGATACG CC-3′) and a reverse primer (H2R1, 5′-CCTCCAGGATTTGG TGCATTCTGTATTGC-3′), followed double digestion of the PCR product with XbaI/BglII and replacement of the existing region of pCA12.1 to yield pCA14.1.
A BstBI site within the IRP2 coding region was eliminated by introducing a silent amino acid codon change (F418, TTT to TTC). First, a 255-bp region was amplified using forward primer HIP2BF (5′-GCACCTCAGGCAAGTAGGAGTGGCTGG-3′) and the mutagenic reverse primer HIP2BM (5′-CCTGAAGAATTCT GGTCATTTCGGAACAATTTCACAGC-3′). The resulting PCR product was used in a second reaction using a new reverse primer (HIP2BR, 5′-CCTGGAGTGCAATTGGCTCTTTGGTAAG-3′) to generate an 1184-bp product, which was digested with BspEI and NcoI and substituted into pCA14.1 to create pCA17.7. The myc-epitope tag was removed and the wild-type stop codon reestablished by amplifying the terminal portion of the coding region with forward primer HIP2EF (5′-CGAGTAGAGGAAGAACAT GTTATACTATCC-3′) and a reverse primer HIP2ER (5′GGCGT GTATTGGATCCTATGAGAATTTTCGTGCCACAAAGTTTAAT AATCCTCC-3′) that adds a BamHI site adjacent to the new stop codon, digesting this product with NcoI/BamHI, and replacing the existing segment of pCA17.7 to make the new plasmid pCA21.2. The BstBI/BamHI fragment was subsequently released and cloned into the corresponding site of pCA10.3, creating the human IRP2 expression plasmid pCA22.1.
Transformation of P. pastoris and selection of optimally expressing clones
The IRP1 expression plasmid (pCA19.8) and the IRP2 expression plasmid (pCA22.1), along with empty pCA10.3 plasmid as a control, were linearized by digestion with SalI, then transformed into strain GS115 (his4, Mut+) of P. pastoris. Transformations were performed using the spheroplast method as described in the supplier’s handbook (Invitrogen). From these transformations, 20 (control), 30 (IRP1), and 12 (IRP2) colonies were selected for further screening. Genomic DNA for each was collected and examined by PCR for the presence of expression insert using the primer set NHA5 (5′-CAGAAGGAAGCTGCCCTGTCTTAAA CC-3′) and NHA3 (5′-GCGAGATAGGCTGATCAGGAGCAAG CTCGTACG-3′). These primers should generate products of ∼2200 bp, corresponding to the wild-type AOX1 gene, 338 bp for the CA11 (control) clones, 2994 bp for CA13 (IRP1) clones, and 3154 bp for CA15 (IRP2) clones. By this analysis, 10 of 20 CA11, 22 of 30 CA13, and 6 of 12 CA15 contained the appropriate insert.
To screen for optimal protein expression, single colonies were used to inoculate 10 mL of minimal glycerol media (1.34% YNB, 1% glycerol, 4 × 10−5% biotin) and were subsequently cultured at 30°C until they reached an O.D.600 of 2–3 (∼2 days), at which point they were gently pelleted and resuspended in minimal methanol media (1.34% YNB, 0.5% methanol, 4 × 10−5% biotin) at an O.D.600 of 1.0. These were incubated for an additional 2 days at 30°C (cultures were supplemented with methanol at 24 h to maintain 0.5% concentration). The yeast were then pelleted and resuspended in breaking buffer and lysed using glass beads (425–600 μm, acid washed, Sigma). Lysates were examined by 8% SDS-PAGE and stained with Coomassie brilliant blue. From this analysis, CA13-02 and CA13-17 showed the highest levels of IRP1 expression, whereas CA15-06 and CA15-10 showed the highest levels of IRP2 expression. Time course analysis found optimal expression after 36 h of induction with methanol.
Large scale expression of IRP1 and IRP2
Glycerol stocks of IRP1 expression clone CA13-02 and IRP2 expression clone CA15-06 were prepared and maintained at −80°C. Periodically, portions of these stocks were streaked onto minimal 2% dextrose plates, grown at 30°C for 2 days, then stored at 4°C. For optimal expression, colonies were picked within 2 wk of plating. Single colonies were used to inoculate 10 mL of minimal glycerol media, followed by incubation for 20 h at 30°C and 275 rpm. A 4-mL aliquot of this culture was used to inoculate 250 mL of minimal glycerol media, which was then incubated at 30°C and 275 rpm until the cell density had reached O.D.600 ∼6–8 (18–22 h). Yeast were pelleted by centrifugation (500g), then resuspended in mimimal methanol media to a density of O.D,600 = 1. Culture volumes of 250 mL were incubated in 1 L baffled flasks for 36 h. At 24 h, additional methanol was added to maintain 0.5% concentration. To harvest, yeast were collected at 6500 rpm (7500g) and stored at −80°C until ready for purification.
Lysis and initial purification of IRP1 and IRP2
Yeast pelleted from two 250-mL cultures were thawed on ice and resuspended in 60 mL of degassed ice-cold breaking buffer. Lysis was performed either with glass beads or using a French pressure cell, followed by centrifugation at 9500 rpm (12,500g), for 12 min at 4°C. The supernatant was collected, transferred to a fresh tube, and immediately subjected to chromatographic purification.
Chromatographic buffers were continually sparged with Argon to minimize the presence of oxygen. Elution of protein from each column was monitored by absorbance at 280 nm as 1-mL fractions were collected. Flow rates were typically 1 mL/min for loading, 0.5 mL/min for elution. Crude lysates were first loaded onto a 2 × 5 mL HiTrap Heparin Sepharose column (two 5-mL columns connected in series). The column was washed with 10 mM NaCl, then eluted with an gradient to 2400 mM NaCl. Typically, IRP1 eluted at ∼390 mM NaCl, IRP2 at ∼435 mM NaCl. The appropriate fractions (as determined by SDS-PAGE) were pooled, diluted to 40 mM NaCl, then loaded onto a 2 × 5 mL HiTrap Q Sepharose column. After washing with 40 mM NaCl, protein was eluted with a gradient up to 1400 mM NaCl, with IRP1 eluting at ∼225 mM NaCl and IRP2 eluting at ∼255 mM NaCl. Again, the appropriate fractions were pooled, brought to 3 M NaCl, then loaded onto a 4 × 1 mL HiTrap Octyl Sepharose column and eluted with a decreasing NaCl gradient. IRP1 eluted at ∼0 mM NaCl, whereas IRP2 eluted at ∼500 mM NaCl. Protein concentrations were determined using the Pierce/Endogen BCA assay kit.
RNA affinity column preparation
The 36-mer modified RNAs (5′-NH2-C-IRE, 5′-aminotetherGGAGUUCCUGCUUCAACAGUGCUUGGACGGAACUCC-3′; 5′NH2-N-IRE, 5′-aminotether-GGAGUUCCUGAUUCAAUACACCUU GGACGGAACUCC-3′), were received as lyophilized pellets and deprotected as recommended by the supplier. For direct coupling to the HiTrap NHS-activated Sepharose column (Amersham Biotech), 150 nmole of RNA were dissolved in 1.2 mL of 0.2 M NaHCO3, 0.5 M NaCl (pH 8.3) and loaded onto the column as recommended by the manufacturer. After elution through a sizing column to remove release N-hydroxysuccinimide, RNA was quantified by measuring the absorbance at 260 nm.
To prepare a thiol-modified column for the linker approach, a 1-mL HiTrap-NHS Sepharose column was washed with 5 mL of ice-cold 1-mM HCl (slowly, to avoid irreversible compression of the column matrix), then with 100 mM cystamine (pH 8.0) in three 2-mL portions. An additional column was treated instead with 100 mM ethanolamine as a control. All columns were subsequently washed with 50 mM Tris-HCl.
The amount of cystamine on the column was determined by measuring the release of 4-thio-2-nitrobenzoic acid upon treatment of the reduced columns with 2,2′-dithiobisnitrobenzoic acid (DTNB). Each column was reduced with 50 mM DTT then washed with 40 mM Tris-HCl (pH 7.5), 20 mM NaCl. Treatment with 5 mL of 30 mM DTNB in 200 mM Tris-HCl (pH 7.5) was achieved by passing 2 mL of the solution through the column, waiting 4 min, then repeating twice more with 1.5-mL portions. The combined DTNB eluents were pooled, diluted, and assayed for absorbance at 412 nm.
Both RNAs were modified with sulfo-SIAB by treating with 4.5 mM sulfo-SIAB in 200 mM sodium phosphate (pH 8) for 6 h at room temperature in the dark. The RNA was then ethanol precipitated, dried, and redissolved in 1.1 mL 180 mM sodium phosphate (pH 8), 5 mM NaCl. Folding of the IRE structure was promoted by heating to 85°C for 3 min, followed by rapid cooling on ice. After washing the thiol column first with 20 mM solution of DTT in 200 mM Tris-HCl (pH 7.5)/20 mM NaCl, then with 200 mM Tris-HCl (pH 7.5)/20 mM NaCl, the RNA solution was carefully loaded into the column. The column was sealed and incubated in the dark at room temperature for 12–16 h, after which it was washed with 4 mL of 200 mM Tris-HCl (pH 7.5). The RNA loading of the column was determined by comparing the A260 of the RNA solution from before and after the coupling reaction. Unreacted thiol groups were blocked in a subsequent treatment of the column with 10 mM iodoacetamide in 200 mM Tris-HCl (pH 7.5).
Elution of IRP1 and IRP2 through RNA columns
Solutions of IRP1 or IRP2 were loaded through the control, N-IRE, and C-IRE columns at 250 mM NaCl and 0.5 mL/min. Elution from the C-IRE column was achieved with a gradient of 250 mM to 3 M NaCl. Protein-containing fractions were desalted by ultrafiltration (Amicon).
Band-shift titrations
A 32P-labeled RNA corresponding to the C-IRE sequence (5′-GG AGUUCCUGCUUCAACAGUGCUUGGACGGAACUCC-3′) was generated using methods previously described (Allerson et al. 1999). For each mixture, 50 nM of 32P-labeled C-IRE was mixed with increasing concentrations of either IRP1 or IRP2 (from 0 to 225 nM) in the presence of 5% glycerol, 0.025 units/μL RNase Inhibitor (5 Prime 3 Prime, Inc.), 0.15 mg/mL yeast tRNA, 2 mM DTT, 25 mM Tris-HCl (pH 7.5), and 40 mM KCl. These reactions were incubated for 20 minutes at room temperature, then eletrophoresed on 10% nondenaturing polyacrylamide gels at 130 V for 4 h. After drying, the gels were exposed to either Kodak BioMax MR film, or a Molecular Dynamics Phosphorimaging screen. Quantitation was performed using the ImageQuant software packaged (Molecular Dynamics).
Acknowledgments
We thank Lukas Kühn for providing the plasmid pGEM-hIRF.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2143303
REFERENCES
- Addess, K.J., Basilion, J.P., Klausner, R.D., Rouault, T.A., and Pardi. A. 1997. Structure and dynamics of the iron responsive element RNA: Implications for binding of the RNA by iron regulatory binding proteins. J. Mol. Biol. 274: 72–83. [DOI] [PubMed] [Google Scholar]
- Allerson, C.R., Chen, S.L., and Verdine, G.L. 1997. A chemical method for site-specific modification of RNA: The convertible nucleoside approach. J. Am. Chem. Soc. 119: 7423–7433. [Google Scholar]
- Allerson, C.R., Cazzola, M., and Rouault, T.A. 1999. Clinical severity and thermodynamic effects of iron-responsive element mutations in hereditary hyperferritinemia-cataract syndrome. J. Biol. Chem. 274: 26439–26447. [DOI] [PubMed] [Google Scholar]
- Bachler, M., Schroeder, R., and Ahsen, U.V. 1999. StreptoTag: A novel method for the isolation of RNA-binding proteins. RNA 5: 1509–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basilion, J.P., Kennedy, M.C., Beinert, H., Massinople, C.M., Klausner, R.D., and Rouault, T.A. 1994a. Overexpression of iron-responsive element-binding protein and its analytical characterization as the RNA-binding form, devoid of an iron-sulfur cluster. Arch. Biochem. Biophys. 311: 517–522. [DOI] [PubMed] [Google Scholar]
- Basilion, J.P., Rouault, T.A., Massinople, C.M., Klausner, R.D., and Burgess, W.H. 1994b. The iron-responsive element-binding protein: Localization of the RNA-binding site to the aconitase active-site cleft. Proc. Natl. Acad. Sci. 91: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinert, H., Kennedy, M.C., and Stout, D.C. 1996. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 96: 2335–2373. [DOI] [PubMed] [Google Scholar]
- Butt, J., Kim, H.-Y., Basilion, J.P., Cohen, S., Iwai, K., Philpott, C.C., Altschul, S., Klausner, R.D., and Rouault, T.A. 1996. Differences in the RNA binding sites of iron regulatory proteins and potential target diversity. Proc. Natl. Acad. Sci. 93: 4345–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi, M., Mayeda, A., Krainer, A.R., and Zahler, A.M. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18: 4060–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland, P.R. and Driscoll, D.M. 1999. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J. Biol. Chem. 274: 25447–25454. [DOI] [PubMed] [Google Scholar]
- Cregg, J.M., Cereghino, J.L., Shi, J., and Higgins, D.R. 2000. Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 6: 23–52. [DOI] [PubMed] [Google Scholar]
- Guo, B., Phillips, J.D, Yu, Y., and Leibold, E.A. 1995. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 270: 21645–21651. [DOI] [PubMed] [Google Scholar]
- Harford, J.B. and Rouault, T.A. 1998. RNA structure and function in cellular iron homeostasis. In RNA structure and function (eds. R.W. Simons and M. Grunberg-Manago), pp. 575–602. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Hentze, M.W. and Kühn, L.C. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. 93: 8175–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirling, H., Emery-Goodman, A., Thompson, N., Neupert, B., Seiser, C., and Kuhn, L.C. 1992. Expression of active iron regulatory factor from a full-length human cDNA by in vitro transcription/translation. Nuc. Acids Res. 20: 33–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirling, H., Henderson, B.R., and Kühn, L.C. 1994. Mutational analysis of the [4Fe-4S]-cluster converting iron regulatory factor from its RNA-binding form to cytoplasmic aconitase. EMBO J. 13: 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai, K., Klausner, R.D., and Rouault, T.A. 1995. Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J. 14: 5350–5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai, K., Drake, S.K., Wehr, N.B., Weissman, A.M., LaVaute, T., Minato, N., Klausner, R.D., Levine, R.L., and Rouault, T.A. 1998. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: Implications for degradation of oxidized proteins. Proc. Natl. Acad. Sci. 95: 4924–4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldy, P., Menotti, E., Moret, R., and Kühn, L.C. 1999. Identification of RNA-binding surfaces in iron regulatory protein-1. EMBO J. 18: 6073–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski, A., Ostareck, D.H., Standart, N.M., and Jackson, R.J. 1998. Affinity methods for isolating RNA binding proteins. In RNA:protein interactions, a practical approach (ed. C.W.J. Smith), pp. 137–160. Oxford University Press, New York.
- Laing, L.G. and Hall, K.B. 1996. A model of the iron responsive element RNA hairpin loop structure determined from NMR and thermodynamic data. Biochemistry 35: 13586–13596. [DOI] [PubMed] [Google Scholar]
- Larson, C.J. and Verdine, G.L. 1992. A high-capacity column for affinity purification of sequence-specific DNA-binding proteins. Nucleic Acids Res. 20: 3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan, H.A. and Connell, G.J. 2001. The hairpin loop but not the bulged C of the iron responsive element is essential for high affinity binding to iron regulatory protein-1. J. Biol. Chem. 276: 14791–14796. [DOI] [PubMed] [Google Scholar]
- Neupert, B., Thompson, N.A., Meyer, C., and Kühn, L.C. 1990. A high yield affinity purification method for specific RNA-binding proteins: Isolation of the iron regulatory factor from human placenta. Nucleic Acids Res. 18: 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, J.D., Guo, B., Yu, Y., Brown, F.M., and Leibold, E.A. 1996. Expression and biochemical characterization of iron regulatory proteins 1 and 2 in Saccharomyces cerevisiae. Biochemistry 35: 15704–15714. [DOI] [PubMed] [Google Scholar]
- Philpott, C.C., Klausner, R.D., and Rouault, T.A. 1994. The bifunctional iron-responsive element binding protein/cytosolic aconitase: The role of active-site residues in ligand binding and regulation. Proc. Natl. Acad. Sci. 91: 7321–7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, A.H. and Stout, C.D. 1989. The structure of aconitase. Proteins 5: 289–312. [DOI] [PubMed] [Google Scholar]
- Rouault, T.A. and Klausner, R.D. 1996. Post-transcriptional regulation of genes of iron metabolism in mammalian cells. J. Bioinorg. Chem. 1: 494–499. [Google Scholar]
- Rouault, T.A., Hentze, M.W., Haile, D.J., Harford, J.B., and Klausner, R.D. 1989. The iron-responsive element binding protein: A method for the affinity purification of a regulatory RNA-binding protein. Proc. Natl. Acad. Sci. 86: 5768–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault, T.A., Stout, C.D., Kaptain, S., Harford, J.B., and Klausner, R.D. 1991. Structural relationship between an iron-regulated RNA-binding protein (IRE-BP) and aconitase: Functional implications. Cell 64: 881–883. [DOI] [PubMed] [Google Scholar]
- Rouault, T.A., Haile, D.J., Downey, W.E., Philpott, C.C., Tang, C., Samaniego, F., Chin, J., Paul, I., Orloff. D., Harford, J.B., et al. 1992. An iron-sulfur cluster plays a novel regulatory role in the iron-responsive element binding protein. BioMetals 5: 131–140. [DOI] [PubMed] [Google Scholar]
- Samaniego, F., Chin, J., Iwai, K., Rouault, T.A., and Klausner, R.D. 1994. Molecular characterization of a second iron-responsive element binding protein, iron regulatory protein 2. J. Biol. Chem. 269: 30904–30910. [PubMed] [Google Scholar]
- Schneider, B.D. and Leibold, E.A. 2000. Regulation of mammalian iron homeostasis. Curr. Opin. Clin. Nutr. Metab. Care 3: 267–273. [DOI] [PubMed] [Google Scholar]
- Sela-Brown, A., Silver, J., Brewer, G., and Naveh-Many, T. 2000. Identification of AUF1 as a parathyroid hormone mRNA 3′-untranslated region-binding protein that determines parathyroid hormone mRNA stability. J. Biol. Chem. 275: 7424–7429. [DOI] [PubMed] [Google Scholar]
- Sreekrishna, K., Potenz, R.H.B., Cruze, J.A., McCombie, W.R., Parker, K.A., Nelles, L., Mazzaferro, P.K., Holden, K.A., Harrison, R.G., Wood, P.J., et al. 1988. High level expression of heterologous proteins in methylotrophic yeast Pichia pastoris. J. Basic Microbiol. 28: 265–278. [DOI] [PubMed] [Google Scholar]
- Sreekrishna, K., Brankamp, R.G., Kropp, K.E., Blankenship, D.T., Tsay, J.-T., Smith, P.L., Wierschke, J.D., Subramaniam, A., and Birkenberger, L.A. 1997. Strategies for optimal synthesis and secretion of heterologous proteins in the methylotrophic yeast Pichia pastoris. Gene 190: 55–62. [DOI] [PubMed] [Google Scholar]
- Srisawat, C. and Engelke, D.R. 2001. Streptavidin aptamers: Affinity tags for the study of RNAs and ribonucleoproteins. RNA 7: 632–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson, G.R. and Walden, W.E. 1994. Localization of an RNA binding element of the iron responsive element binding protein within a proteolytic fragment containing iron coordination ligands. Nucleic Acids Res. 22: 2627–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]