Abstract
Switches and sensors play important roles in our everyday lives. The chemical properties of RNA make it amenable for use as a switch or sensor, both artificially and in nature. This review focuses on recent advances in artificial RNA switches and sensors. Researchers have been applying classical biochemical principles such as allostery in elegant ways that are influencing the development of biosensors and other applications. Particular attention is given here to allosteric ribozymes (aptazymes) that are regulated by small organic molecules, by proteins, or by oligonucleotides. Also discussed are ribozymes whose activities are controlled by various nonallosteric strategies.
Keywords: Switch, sensor, allosteric, ribozyme, aptazyme
INTRODUCTION
What is the first thing that most people do in the morning? A likely answer is “turn on a switch in response to a sensor”—a switch that controls a room’s light, in response to a sensor that detects some time on an alarm clock. Switches and sensors play a central role in our everyday lives, from the moment we wake up and throughout the day. Not surprisingly, molecular-scale scientists are seeking to impose the properties of switches and sensors on molecules. This is for the inherent intellectual pleasure derived from controlling the molecular domain and also for the practical goal of creating useful devices such as sensors for biological compounds. As is appropriate for the journal RNA, this review focuses on switches and sensors made from RNA. Because several excellent summaries of progress in the general area have appeared (Soukup and Breaker 1999c, 2000; Breaker 2002; Rajendran and Ellington 2002), this review describes selected recent advances. The primary focus is on artificial RNA switches and sensors, rather than the naturally occurring RNA switches and sensors that nature uses to control metabolite biosynthesis and presumably for other reasons as well.
The distinction between a “switch” and a “sensor” is rather blurry. In a trivial sense, every RNA molecule that changes conformation upon interacting with other molecules or ions is a chemical switch that acts as a sensor for the binding species. In the everyday world, at first glance most switches appear to be two-state: the light is switched either on or off. But what about dimmer switches that continuously modulate light output? Similarly, sensors can be either all or nothing (e.g., fire alarms, automobile engine warning lights, and circuit breakers) or continuous (Geiger counters, bathroom scales, computer mice, and automobile pedals). In the RNA context, switches and sensors are treated here as related facets of the same general phenomenon.
THE SIGNALING-APTAMER APPROACH TO BIOSENSORS
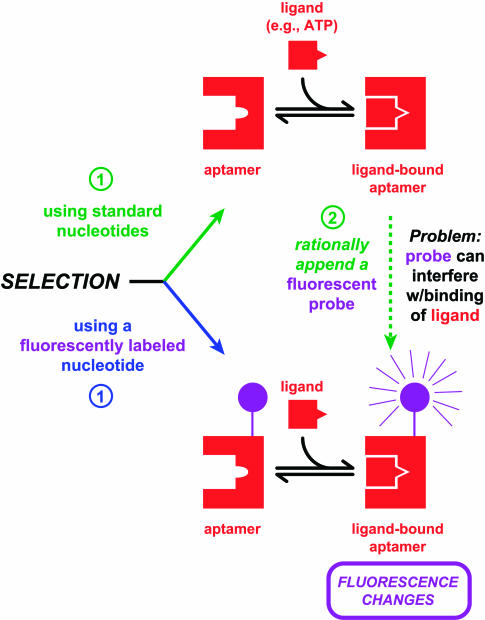
Aptamers are nucleic acids that bind small molecules (Jayasena 1999; Famulok et al. 2000; Hermann and Patel 2000). Coupling an aptamer to an appropriate detection system offers the possibility of sensing analytes in solution. Ellington and coworkers developed RNA and DNA aptamers for this purpose by rational incorporation of fluorescein into known ATP-binding aptamers (Fig. 1 ▶, green pathway; Jhaveri et al. 2000b). However, the fluorophore interfered with ligand binding, which prompted direct selection of ATP-sensitive fluorescent aptamers using mononucleotides that were covalently modified with fluorescein (Fig. 1 ▶, blue pathway; Jhaveri et al. 2000a). This general theme of rational design falling short and selection methods coming to the rescue is a common one that will be mentioned again. Whether developed by design or selection, signaling aptamers have a significant advantage over other sensors because of their ability to monitor analyte concentration continuously in solution without binding or washing steps. In addition, aptamers may, in principle, be selected to match any desired analyte (Wilson and Szostak 1999), offering a potentially general route to concentration-dependent sensors that rival antibodies for this purpose (Jayasena 1999; Hesselberth et al. 2000; Hoffman et al. 2001; Rajendran and Ellington 2002).
FIGURE 1.
In vitro selection of ligand-binding aptamers with fluorescence detection. The green pathway shows a relatively conventional two-step strategy of (1) selecting an aptamer responsive to a given ligand such as ATP, followed by (2) appending a fluorescent chromophore that renders the aptamer useful for signaling (Jhaveri et al. 2000b). The blue pathway shows an alternate one-step strategy in which fluorescent aptamers are selected directly by incorporation of one or more fluorescently labeled mononucleotides during the selection process (Jhaveri et al. 2000a).
ALLOSTERIC RIBOZYME SENSORS (APTAZYMES) AND GENERAL “COMMUNICATION MODULES”
Nature uses allosteric interactions to modulate protein enzyme activity (Perutz 1994), and it was logical for RNA scientists to ask whether allostery can be harnessed in the form of interactions between RNA and small organic molecules or other ligands (effectors). Although no allosterically regulated ribozymes have yet been identified in nature (but see the section below on oligonucleotide-regulated ribozymes), many have been developed artificially. This has been accomplished by the often complementary approaches of rational design (Tang and Breaker 1997, 1998; Kuwabara et al. 1998; Vaish et al. 2002; Wang and Sen 2002) and in vitro selection (Koizumi et al. 1999; Robertson and Ellington 1999; Soukup and Breaker 1999b). Allosteric regulation has been imposed onto the hammerhead (Kertsburg and Soukup 2002), HDV (Kertsburg and Soukup 2002), hairpin (Hartig et al. 2002), Tetrahymena group I intron (Kertsburg and Soukup 2002), and X-motif ribozymes (Kertsburg and Soukup 2002). The activity-modulating ligands include ATP (Tang and Breaker 1997; Robertson and Ellington 2000), theophylline (Soukup and Breaker 1999a; Robertson and Ellington 2000; Soukup et al. 2000), flavin mononucleotide (FMN; Araki et al. 1998, 2001; Soukup and Breaker 1999a, 1999b; Robertson and Ellington 2000), cyclic nucleotide monophosphates (cNMPs; Koizumi et al. 1999), doxycycline (Piganeau et al. 2000), 3-methylxanthine (Soukup et al. 2000), and pefloxacin (Piganeau et al. 2001), as well as various metal ions (Seetharaman et al. 2001), oligonucleotides (Porta and Lizardi 1995; Kuwabara et al. 1998; Robertson and Ellington 1999; Komatsu et al. 2000), and several proteins (Hartig and Famulok 2002; Hartig et al. 2002; Vaish et al. 2002). Allosteric regulation has also been extended to DNA enzymes that are activated by ATP (Levy and Ellington 2002). By coupling of ribozyme activation to a fluorescent read-out, such as that based on cleavage of a doubly labeled FRET probe, convenient biosensor assays have been established (Frauendorf and Jaschke 2001; Stojanovic et al. 2001).
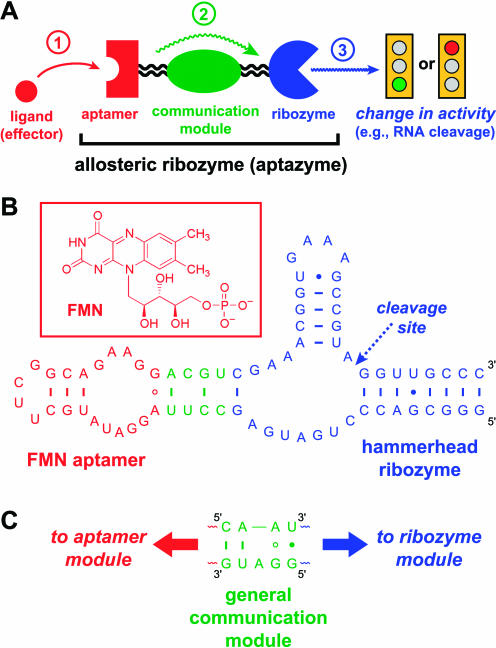
The basic principle of an allosteric nucleic acid sensor is shown in Figure 2A ▶, and a specific example of a FMN-sensitive ribozyme is shown in Figure 2B ▶. To create an allosteric ribozyme, a ribozyme is connected via a “communication module” to an aptamer that is specific for a certain ligand. The communication module couples the aptamer–ligand interaction to ribozyme activity (Robertson and Ellington 2000; Kertsburg and Soukup 2002), usually via up-regulation but sometimes via inhibition (Tang and Breaker 1997, 1998). The resulting RNA combines an aptamer with a ribozyme and has thus been termed an “aptazyme” (Robertson and Ellington 1999). The activity enhancement upon ligand activation of an aptazyme is typically between 10 and 103; the higher end of this range is more than sufficient for practical biosensor assays. In the most favorable case yet reported, that of a ribozyme obtained by Breaker and coworkers that is allosterically up-regulated by cGMP, the enhancement is 5000-fold (Koizumi et al. 1999). The mechanism by which ligand binding is coupled to ribozyme activation is usually not known in detail, but this is generally thought to involve structural stabilization of the catalytically active ribozyme conformation upon ligand binding (Soukup and Breaker 1999a). For one reported ATP-down-regulated ribozyme, simple steric clashes between the ligand-bound aptamer and the ribozyme are sufficient to explain the loss of activity (Tang and Breaker 1998).
FIGURE 2.
(A) General strategy for creation of an allosteric ribozyme (aptazyme), depicting the aptamer, communication, and ribozyme modules. The order of events starting with ligand binding is numbered. (B) Specific example of an allosteric ribozyme that is sensitive to flavin mononucleotide (FMN; Soukup and Breaker 1999b). (C) The general communication module of Kertsburg and Soukup (2002).
Ideally, the aptazyme approach is completely modular in that the aptamer, communication, and ribozyme modules are each fully interchangable among various aptazymes. Recent advances have brought this universal design nearer to reality. In one instance, a ribozyme dependent on the simultaneous presence of two effectors, theophylline and FMN, was created by appending the appropriate aptamer domains in series to the hammerhead ribozyme (Jose et al. 2001). However, in other cases, initial attempts to induce ligand activation of ribozyme activity were unsuccessful, and by examining these cases we might see where future challenges lie. Robertson and Ellington found that simply appending a FMN or chloramphenicol aptamer to their L1 ligase ribozyme (Robertson and Ellington 1999; Robertson et al. 2001) did not allow ligand activation (Robertson and Ellington 2000), contrary to the results with ATP or theophylline (Robertson and Ellington 1999). This instigated use of in vitro selection to identify communication modules that would successfully couple effector binding to ribozyme activation. The selection approach worked with FMN but failed with chloramphenicol (Robertson and Ellington 2000). Although one should be wary of overinterpreting a small number of experiments, these results suggest that there is a significant chance of both success and failure in using rational design to create a ligand-activated ribozyme from a known aptamer and ribozyme. In certain cases, selection methods may be imperative to identify aptazymes, but even then success is not assured. Recently, Kertsburg and Soukup (2002) reported a simple yet highly versatile 9-nt communication module that couples a variety of ribozymes and aptamers (Fig. 2C ▶). It will be interesting to see whether their new communication module proves completely general, particularly for those difficult cases that have not been amenable to rational design or even selection.
RIBOZYME ACTIVATION UPON BINDING OF PROTEINS
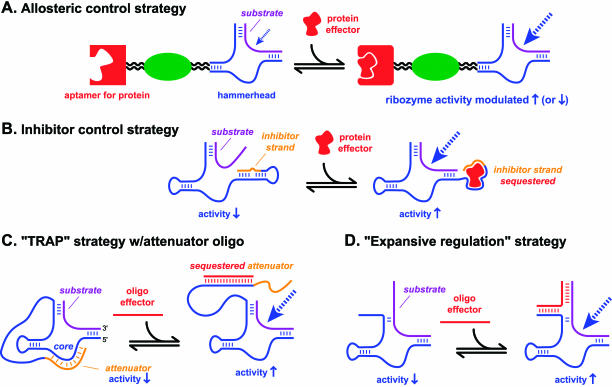
Ligand-activated ribozymes have been developed that use proteins instead of small organic molecules as their effectors. In an early study, Burke and coworkers rendered the hairpin ribozyme dependent upon binding of the R17 coat protein by fusing the appropriate protein-binding domain to one arm of the hairpin secondary structure (Sargueil et al. 1995). More recently, Famulok and coworkers converted a FRET-based hammerhead ribozyme assay (Jenne et al. 1998) into a protein-dependent method (Hartig et al. 2002). This was accomplished in two distinct ways that are here termed “allosteric” and “inhibitor” control strategies (Fig. 3 ▶). First, allosteric inactivation of the hammerhead ribozyme was achieved upon addition of the HIV Rev protein to a hammerhead that was fused with a Rev-binding element (RBE) domain via a 2-bp communication module. The mechanism by which Rev binding disrupts hammerhead activity is not obvious from their study. The Famulok group has also used rational design to obtain a hammerhead ribozyme allosterically down-regulated by HIV-1 reverse transcriptase, showing half-maximal inhibition at ∼10 nM protein (Hartig and Famulok 2002). Alternatively, ribozyme activity was modulated by appending an “inhibitor” oligonucleotide strand to the hammerhead ribozyme (Hartig et al. 2002). Upon Rev binding to nucleotides that included the inhibitor sequence, the inhibitor strand was displaced from the hammerhead. This allowed the substrate RNA to bind, and ribozyme activity was thereby induced. The role of the inhibitor strand in this process is analogous to the classical biochemical role of a competitive inhibitor, because the inhibitor strand directly prevents the substrate from binding.
FIGURE 3.
Allosteric (A) and inhibitor (B) control strategies for protein-mediated modulation of ribozyme activity, illustrated with the hammerhead ribozyme. In the classification proposed here, the inhibitor control strategies are not truly “allosteric” in that they do not involve a communication module. Instead, the inhibitor oligonucleotide strand binds in direct competition with the ribozyme’s substrate. In principle, small molecules could also function as the inhibitor (see text). (C) The TRAP strategy of Burke et al. (2002). (D) The expansive regulation strategy of Sen and coworkers (Wang and Sen 2001; Wang et al. 2002a).
Using both allosteric and inhibitor control approaches with various protein-responsive ribozymes, these studies allowed identification of both protein–antibiotic and protein–protein interactions. For example, antibiotics that disrupt the Rev–RBE allosteric interaction restored hammerhead activity by dissociating the Rev–RBE complex (Hartig et al. 2002). In principle, a small-molecule effector could also be used to up-regulate ribozyme activity in the framework of the inhibitor control strategy, if the small molecule binds and sequesters the inhibitor strand. Experimental realization of this approach has not yet been reported.
In another example of allosterically protein-activated ribozymes, Robertson and Ellington (2001) developed ribozymes for which RNA ligase activity was up-regulated 103-fold by either the tRNA synthetase protein CYT-18 or by lysozyme. However, simply appending an appropriate protein-binding module to the L1 ligase ribozyme did not generate a protein-dependent RNA enzyme. It was instead necessary to select directly for protein-dependent ribozyme function. This is in contrast to the study described above, in which merely fusing the RBE domain to the hammerhead led to a Rev-responsive ribozyme (Hartig et al. 2002). Failure of the rational approach in the former instance is reminiscent of the previously mentioned poor outcome in creating certain small-molecule-dependent aptazymes. In both instances, it was necessary to turn to selection after rational design was unsuccessful.
Despite examples of failure, sometimes rational design does work quite well. Vaish, Seiwert, and colleagues rationally appended a protein-binding RNA domain to the hammerhead ribozyme and achieved 50-fold activation upon addition of the corresponding protein, the unphosphorylated protein kinase ERK2 (Vaish et al. 2002). Inactivity of the ribozyme in the absence of the protein was enforced by Watson–Crick base pairs between one inhibitor strand of the protein-binding RNA domain and one strand of an essential hammerhead duplex arm. Protein binding sequestered the inhibitor RNA strand and allowed the hammerhead to adopt its native secondary and tertiary structure. This overall approach is categorized in Figure 3B ▶ as an inhibitor control strategy rather than “allosteric” control, and it is analogous to the inhibitor control strategy of Famulok and coworkers that was described above. Intriguingly, Vaish et al. also created hammerheads that were specifically activated by phosphorylated ERK2, by swapping in a protein-binding RNA domain specific to the phosphorylated form of the protein. The ERK2-activated ribozymes were competent both in vitro and in cell lysates, offering significant potential for detection of posttranslationally modified proteins.
OLIGONUCLEOTIDE-ACTIVATED RIBOZYMES
Natural RNA enzymes can adopt alternative base-paired structures that regulate their activity (Miller and Silver 1991; Pan and Woodson 1998); these could perhaps be considered as “self-regulated” ribozymes. Closer to the main topic of this review, several research groups have used intermolecularly associating oligonucleotides as effectors to modulate ribozyme activity (Porta and Lizardi 1995; Kuwabara et al. 1998; Robertson and Ellington 1999; Komatsu et al. 2000). The overall themes of these studies are similar to those using small-molecule or protein effectors. An interesting advance was recently reported by Burke et al. (2002), who combined the allosteric and inhibitor functions of Figure 3 ▶ into a single ribozyme that they termed TRAP (targeted ribozyme-attenuated probe; Fig. 3C ▶). Their ribozyme incorporates an “attenuator” strand appended to the hammerhead sequence. This attenuator binds to the catalytic core and thus inhibits hammerhead activity. Because the attenuator binds to the catalytic core residues and not the substrate binding site, this strategy is more closely related to that of classical noncompetitive (rather than competitive) inhibition. Attenuator binding to the hammerhead core was disrupted by duplex formation between an exogenous oligonucleotide effector and the nucleotides connecting the ribozyme and attenuator. Thus the oligonucleotide effector up-regulated ribozyme activity by several 100-fold, which could be raised to >103 by inclusion of “anti-attenuator” sequence on the oligonucleotide. This study is an excellent example of a careful rational design approach that has significant potential for application to other RNA enzyme systems, particularly because it does not require very specific knowledge of ribozyme structure or chemistry.
Finally, Sen and coworkers have developed an approach that they term “expansive” regulation (Fig. 3D ▶; Wang and Sen 2001; Wang et al. 2002a). In this strategy, an oligonucleotide effector binds to both ribozyme and substrate. This restores a largely Watson–Crick binding arm of the enzyme–substrate complex, which increases ribozyme activity 20–30-fold. Significantly, this up-regulation occurs via modulation of the substrate binding step, rather than the chemical step of catalysis. The overall strategy of modulating the substrate binding step of enzyme activity was subsequently used to develop ATP and FMN-responsive ribozymes and deoxyribozymes (Wang et al. 2002b). The authors point out that because substrate binding is affected, this approach should be widely applicable to other nucleic acid enzymes that recognize their substrates via Watson–Crick base pairs, which, at present, describes all of them.
NATURE’S USE OF RNA AS A METABOLIC SWITCH
Although much of the research on ribozymes modulated by allostery or other means has been directed towards development of (artificial) biosensors, there has always been the intriguing possibility that nature might have evolved “riboswitches” as control elements. Very recently, Breaker and coworkers identified regions of two mRNAs that interact directly with small-molecule metabolites, thiamine pyrophosphate (TPP) and coenzyme B12. Significantly, the biosynthesis genes of these metabolites are encoded by those same mRNAs (Nahvi et al. 2002; Winkler et al. 2002). This confirmed earlier predictions that nature uses metabolite–mRNA interactions for translational control of proteins involved in metabolite biosynthesis (Nou and Kadner 1998; Gelfand et al. 1999; Stormo and Ji 2001). It is reasonable to expect that more examples of natural metabolite–mRNA translational switches will soon be discovered.
OUTLOOK FOR THE FIELD OF RNA SWITCHES AND SENSORS
The question mark in this review’s title initially suggests that RNA switches and sensors might be cobbled-together contraptions with little elegance. However, this is utterly incorrect; the work of many investigators shows that finely tuned RNA “machines” can be engineered through judicious combination of rational design and in vitro selection approaches. The resulting RNA switches and sensors are feats of ingenuity, with significant potential for real-life applications as biosensors (Seetharaman et al. 2001; Breaker 2002; Rajendran and Ellington 2002) or other uses. Such practical applications must overcome challenges associated with nuclease sensitivity, cost of oligonucleotide synthesis, and interference by nonspecific RNA-binding proteins in serum or other biological fluids, but these challenges will certainly be met. Artificial RNA switches and sensors have a noble pedigree, in that their molecular cousins have recently been identified in nature’s repertoire of mRNA–metabolite interactions. There are other facets to this story. For instance, this review has not even touched on the small noncoding RNAs (ncRNAs) that participate in transcriptional and translational regulation and can reasonably be defined as switches (Lau et al. 2001; Tuschl 2001; Gottesman 2002; Hannon 2002; Storz 2002), nor have RNA aptamers that inhibit proteins intracellularly (“intramers”) been discussed (Famulok and Verma 2002). RNA researchers are very much looking forward to the continued development and identification of RNA switches and sensors of all kinds, both artificial and natural in origin.
NOTE ADDED IN PROOF
As this manuscript was going to press, two additional reports of naturally occurring riboswitches appeared. Winkler et al. (2002) found a riboswitch for FMN, and Mironov et al. (2002) identified riboswitches for TPP and FMN. (1) Winkler, W.C., Cohen-Chalamish, S., and Breaker, R.R. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. 99: 15908–15913. (2) Mironov, A.S., Gusarov, I., Rafikov, R., Lopez, L.E., Shatalin, K., Kreneva, R.A., Perumov, D.A., and Nudler, E. 2002. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111: 747–756.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2200903.
REFERENCES
- Araki, M., Okuno, Y., Hara, Y., and Sugiura, Y. 1998. Allosteric regulation of a ribozyme activity through ligand-induced conformational change. Nucleic Acids Res. 26: 3379–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, M., Hashima, M., Okuno, Y., and Sugiura, Y. 2001. Coupling between substrate binding and allosteric regulation in ribozyme catalysis. Bioorg. Med. Chem. 9: 1155–1163. [DOI] [PubMed] [Google Scholar]
- Breaker, R.R. 2002. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotech. 13: 31–39. [DOI] [PubMed] [Google Scholar]
- Burke, D.H., Ozerova, N.D., and Nilsen-Hamilton, M. 2002. Allosteric hammerhead ribozyme TRAPs. Biochemistry 41: 6588–6594. [DOI] [PubMed] [Google Scholar]
- Famulok, M. and Verma, S. 2002. In vivo-applied functional RNAs as tools in proteomics and genomics research. Trends Biotechnol. 20: 462–466. [DOI] [PubMed] [Google Scholar]
- Famulok, M., Mayer, G., and Blind, M. 2000. Nucleic acid aptamers—from selection in vitro to applications in vivo. Acc. Chem. Res. 33: 591–599. [DOI] [PubMed] [Google Scholar]
- Frauendorf, C. and Jaschke, A. 2001. Detection of small organic analytes by fluorescing molecular switches. Bioorg. Med. Chem. 9: 2521–2524. [DOI] [PubMed] [Google Scholar]
- Gelfand, M.S., Mironov, A.A., Jomantas, J., Kozlov, Y.I., and Perumov, D.A. 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 15: 439–442. [DOI] [PubMed] [Google Scholar]
- Gottesman, S. 2002. Stealth regulation: Biological circuits with small RNA switches. Genes & Dev. 16: 2829–2842. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Hartig, J.S. and Famulok, M. 2002. Reporter ribozymes for real-time analysis of domain-specific interactions in biomolecules: HIV-1 Reverse transcriptase and the primer-template complex. Angew Chem. Int. Ed. 41: 4263–4266. [DOI] [PubMed] [Google Scholar]
- Hartig, J.S., Najafi-Shoushtari, S.H., Grune, I., Yan, A., Ellington, A.D., and Famulok, M. 2002. Protein-dependent ribozymes report molecular interactions in real time. Nature Biotechnol. 20: 717–722. [DOI] [PubMed] [Google Scholar]
- Hermann, T. and Patel, D.J. 2000. Adaptive recognition by nucleic acid aptamers. Science 287: 820–825. [DOI] [PubMed] [Google Scholar]
- Hesselberth, J., Robertson, M.P., Jhaveri, S., and Ellington, A.D. 2000. In vitro selection of nucleic acids for diagnostic applications. J. Biotechnol. 74: 15–25. [DOI] [PubMed] [Google Scholar]
- Hoffman, D., Hesselberth, J., and Ellington, A.D. 2001. Switching nucleic acids for antibodies. Nature Biotechnol. 19: 313–314. [DOI] [PubMed] [Google Scholar]
- Jayasena, S.D. 1999. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 45: 1628–1650. [PubMed] [Google Scholar]
- Jenne, A., Gmelin, W., Raffler, N., and Famulok, M. 1998. Real-time characterization of ribozymes by fluorescence resonance energy transfer (FRET). Angew Chem. Int. Ed. Engl. 38: 1300–1303. [DOI] [PubMed] [Google Scholar]
- Jhaveri, S., Rajendran, M., and Ellington, A.D. 2000a. In vitro selection of signaling aptamers. Nature Biotechnol. 18: 1293–1297. [DOI] [PubMed] [Google Scholar]
- Jhaveri, S.D., Kirby, R., Conrad, R., Maglott, E.J., Bowser, M., Kennedy, R.T., Glick, G., and Ellington, A.D. 2000b. Designed signaling aptamers that transduce molecular recognition to changes in fluorescence intensity. J. Am. Chem. Soc. 122: 2469–2473. [Google Scholar]
- Jose, A.M., Soukup, G.A., and Breaker, R.R. 2001. Cooperative binding of effectors by an allosteric ribozyme. Nucleic Acids Res. 29: 1631–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertsburg, A. and Soukup, G.A. 2002. A versatile communication module for controlling RNA folding and catalysis. Nucleic Acids Res. 30: 4599–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi, M., Soukup, G.A., Kerr, J.N., and Breaker, R.R. 1999. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nature Struct. Biol. 6: 1062–1071. [DOI] [PubMed] [Google Scholar]
- Komatsu, Y., Yamashita, S., Kazama, N., Nobuoka, K., and Ohtsuka, E. 2000. Construction of new ribozymes requiring short regulator oligonucleotides as a cofactor. J. Mol. Biol. 299: 1231–1243. [DOI] [PubMed] [Google Scholar]
- Kuwabara, T., Warashina, M., Tanabe, T., Tani, K., Asano, S., and Taira, K. 1998. A novel allosterically trans-activated ribozyme, the maxizyme, with exceptional specificity in vitro and in vivo. Mol. Cell 2: 617–627. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Levy, M. and Ellington, A.D. 2002. ATP-dependent allosteric DNA enzymes. Chem. Biol. 9: 417–426. [DOI] [PubMed] [Google Scholar]
- Miller, W.A. and Silver, S.L. 1991. Alternative tertiary structure attenuates self-cleavage of the ribozyme in the satellite RNA of barley yellow dwarf virus. Nucleic Acids Res. 19: 5313–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi, A., Sudarsan, N., Ebert, M., Zou, X., Brown, K., and Breaker, R. 2002. Genetic control by a metabolite binding mRNA. Chem. Biol. 9: 1043. [DOI] [PubMed] [Google Scholar]
- Nou, X. and Kadner, R.J. 1998. Coupled changes in translation and transcription during cobalamin-dependent regulation of btuB expression in Escherichia coli. J. Bacteriol. 180: 6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J. and Woodson, S.A. 1998. Folding intermediates of a self-splicing RNA: Mispairing of the catalytic core. J. Mol. Biol. 280: 597–609. [DOI] [PubMed] [Google Scholar]
- Perutz, M. 1994. Mechanisms of cooperativity and allosteric regulation in proteins. Cambridge University Press, New York. [DOI] [PubMed]
- Piganeau, N., Jenne, A., Thuillier, V., and Famulok, M. 2000. An allosteric ribozyme regulated by doxycycline. Angew Chem. Int. Ed. 39: 4369–4373. [DOI] [PubMed] [Google Scholar]
- Piganeau, N., Thuillier, V., and Famulok, M. 2001. In vitro selection of allosteric ribozymes: Theory and experimental validation. J. Mol. Biol. 312: 1177–1190. [DOI] [PubMed] [Google Scholar]
- Porta, H. and Lizardi, P.M. 1995. An allosteric hammerhead ribozyme. Biotechnology 13: 161–164. [DOI] [PubMed] [Google Scholar]
- Rajendran, M. and Ellington, A.D. 2002. Selecting nucleic acids for biosensor applications. Comb. Chem. High Throughput Screen 5: 263–270. [DOI] [PubMed] [Google Scholar]
- Robertson, M.P. and Ellington, A.D. 1999. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nature Biotechnol. 17: 62–66. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Design and optimization of effector-activated ribozyme ligases. Nucleic Acids Res. 28: 1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. In vitro selection of nucleoprotein enzymes. Nature Biotechnol. 19: 650–655. [DOI] [PubMed] [Google Scholar]
- Robertson, M.P., Hesselberth, J.R., and Ellington, A.D. 2001. Optimization and optimality of a short ribozyme ligase that joins non-Watson–Crick base pairings. RNA 7: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargueil, B., Pecchia, D.B., and Burke, J.M. 1995. An improved version of the hairpin ribozyme functions as a ribonucleoprotein complex. Biochemistry 34: 7739–7748. [DOI] [PubMed] [Google Scholar]
- Seetharaman, S., Zivarts, M., Sudarsan, N., and Breaker, R.R. 2001. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nature Biotechnol. 19: 336–341. [DOI] [PubMed] [Google Scholar]
- Soukup, G.A. and Breaker, R.R. 1999a. Design of allosteric hammerhead ribozymes activated by ligand-induced structure stabilization. Structure 7: 783–791. [DOI] [PubMed] [Google Scholar]
- ———. 1999b. Engineering precision RNA molecular switches. Proc. Natl. Acad. Sci. 96: 3584–3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1999c. Nucleic acid molecular switches. Trends Biotechnol. 17: 469–476. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Allosteric nucleic acid catalysts. Curr. Opin. Struct. Biol. 10: 318–325. [DOI] [PubMed] [Google Scholar]
- Soukup, G.A., Emilsson, G.A., and Breaker, R.R. 2000. Altering molecular recognition of RNA aptamers by allosteric selection. J. Mol. Biol. 298: 623–632. [DOI] [PubMed] [Google Scholar]
- Stojanovic, M.N., de Prada, P., and Landry, D.W. 2001. Catalytic molecular beacons. Chembiochem. 2: 411–415. [DOI] [PubMed] [Google Scholar]
- Stormo, G.D. and Ji, Y. 2001. Do mRNAs act as direct sensors of small molecules to control their expression? Proc. Natl. Acad. Sci. 98: 9465–9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, G. 2002. An expanding universe of noncoding RNAs. Science 296: 1260–1263. [DOI] [PubMed] [Google Scholar]
- Tang, J. and Breaker, R.R. 1997. Rational design of allosteric ribozymes. Chem. Biol. 4: 453–459. [DOI] [PubMed] [Google Scholar]
- ———. 1998. Mechanism for allosteric inhibition of an ATP-sensitive ribozyme. Nucleic Acids Res. 26: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl, T. 2001. RNA interference and small interfering RNAs. Chembiochem 2: 239–245. [DOI] [PubMed] [Google Scholar]
- Vaish, N.K., Dong, F., Andrews, L., Schweppe, R.E., Ahn, N.G., Blatt, L., and Seiwert, S.D. 2002. Monitoring post-translational modification of proteins with allosteric ribozymes. Nature Biotechnol. 20: 810–815. [DOI] [PubMed] [Google Scholar]
- Wang, D.Y. and Sen, D. 2001. A novel mode of regulation of an RNA-cleaving DNAzyme by effectors that bind to both enzyme and substrate. J. Mol. Biol. 310: 723–734. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Rationally designed allosteric variants of hammerhead ribozymes responsive to the HIV-1 Tat protein. Comb. Chem. High Throughput Screen 5: 301–312. [DOI] [PubMed] [Google Scholar]
- Wang, D.Y., Lai, B.H., Feldman, A.R., and Sen, D. 2002a. A general approach for the use of oligonucleotide effectors to regulate the catalysis of RNA-cleaving ribozymes and DNAzymes. Nucleic Acids Res. 30: 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, D.Y., Lai, B.H., and Sen, D. 2002b. A general strategy for effector-mediated control of RNA-cleaving ribozymes and DNA enzymes. J. Mol. Biol. 318: 33–43. [DOI] [PubMed] [Google Scholar]
- Wilson, D.S. and Szostak, J.W. 1999. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 68: 611–647. [DOI] [PubMed] [Google Scholar]
- Winkler, W., Nahvi, A., and Breaker, R.R. 2002. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956. [DOI] [PubMed] [Google Scholar]