Abstract
The α-actinin gene has a pair of alternatively spliced exons. The smooth muscle (SM) exon is repressed in most cell types by polypyrimidine tract binding protein (PTB). CELF (CUG-BP and ETR3-like factors) family proteins, splicing regulators whose activities are altered in myotonic dystrophy, were found to coordinately regulate selection of the two α-actinin exons. CUG-BP and ETR3 activated the SM exon, and along with CELF4 they were also able to repress splicing of the NM (nonmuscle) exon both in vivo and in vitro. Activation of SM exon splicing was associated with displacement of PTB from the polypyrimidine tract by binding of CUG-BP at adjacent sites. Our data provides direct evidence for the activity of CELF proteins as both activators and repressors of splicing within a single-model system of alternative splicing, and suggests a model whereby α-actinin alternative splicing is regulated by synergistic and antagonistic interactions between members of the CELF and PTB families.
Keywords: Polypyrimidine tract binding protein; PTB; CUG-BP, Bruno-like proteins, hnRNP
INTRODUCTION
Alternative splicing allows the generation of different proteins from a single gene (Black 2000; Smith and Valcárcel 2000; Graveley 2001; Caceres and Kornblihtt 2002). Between one-third and two-thirds of human genes are alternatively spliced, and in some spectacular cases, single genes can generate thousands of protein isoforms (for review, see Maniatis and Tasic 2002; Modrek and Lee 2002; Roberts and Smith 2002). Alternative splicing is usually regulated in a cell-type- or developmental-stage-specific manner, and is therefore a fundamental control point for gene expression, essential for the full complexity of the expressed proteome.
Understanding the mechanisms of alternative splicing has been the subject of intensive study over a number of years. These efforts have built upon an understanding of the recognition of constitutive splice site elements by factors such as U1 and U2 snRNPs, U2AF, and SF1 (for review of spliceosome assembly and splice site recognition, see Burge et al. 1999; Reed 2000). Regulation of splicing involves the action of specific cis-acting elements, which can either be variants of the consensus splice site elements or distinct enhancer or silencer elements (Lopez 1998; Black 2000; Blencowe 2000; Smith and Valcárcel 2000; Cartegni et al. 2002). Various trans-acting factors that influence alternative splicing have also been identified. In Drosophila, dedicated cell-specific regulators such as tra and sxl are sufficient to switch splicing patterns of target genes (Lopez 1998). Although some cell-specific mammalian splicing regulators have been identified (e.g., Nova1; Jensen et al. 2000), most work in mammalian systems has tended to support a view whereby regulatory decisions are made by the combinatorial action of more widespread factors (Smith and Valcárcel 2000; Maniatis and Tasic 2002). There are two broad classes of such regulatory factors, RS-domain proteins (Fu 1995; Graveley 2000) and hnRNP proteins (Krecic and Swanson 1999). Most notable among the RS-domain proteins is the SR family of proteins, which play multiple roles in both constitutive and alternative splicing, including mediation of exon enhancer-dependent activation of splicing (Fu 1995; Blencowe 2000; Graveley 2000).
HnRNP proteins are a diverse group of RNA-binding proteins with various roles in pre-mRNA and mRNA functions (McAfee et al. 1997; Krecic and Swanson 1999). They package pre-mRNA and can antagonize or assist splice site selection. For example hnRNP A1 antagonizes the effects of SR proteins on alternative splicing (Caceres et al. 1994), and when recruited to splicing silencers can induce exon skipping (e.g., Caputi et al. 1999; Del Gatto Konczak et al. 1999; Zhu et al. 2001). Polypyrimidine tract binding protein (PTB or hnRNP-I) also acts as a repressor of a number of alternatively spliced exons (for review, see Valcarcel and Gebauer 1997; Wagner and Garcia-Blanco 2001). It binds pyrimidine-rich sites often containing optimal UCUU motifs (Perez et al. 1997). These sites are frequently in the 3′-splice-site-associated polypyrimidine tract, and, in some cases, PTB-mediated repression can be explained by simple binding competition with U2AF65 (e.g., Lin and Patton 1995; Singh et al. 1995). However, additional binding sites are often found in other locations in the region of alternatively spliced exons, and cooperative PTB binding may be important for exon repression (Chou et al. 2000). The role of PTB as a splicing repressor has been demonstrated most emphatically by depletion and add-back experiments in vitro (Southby et al. 1999; Chou et al. 2000; Wollerton et al. 2001) and by RNAi-mediated knockdown in vivo (Wagner and Garcia-Blanco 2002). PTB represses the alternative N1 exon of c-src by cooperative binding to sites flanking the exon (Chou et al. 2000). Repression is relieved in some neuronal cells where PTB is replaced by nPTB (also known as brPTB; Polydorides et al. 2000), a closely related paralog that can bind RNA, but is less repressive upon the N1 exon (Markovtsov et al. 2000). In most other cases, it is not clear how repression by PTB is relieved.
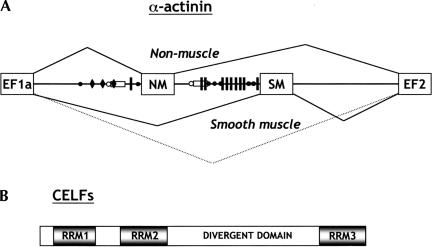
The CELF (CUG-BP and ETR3-like factors) proteins (Ladd et al. 2001), also known as Bruno-like proteins (Good et al. 2000), are also emerging as important hnRNP-like splicing regulators. They are of particular interest as increased activity of CUG-BP, and possibly other CELF family members, in myotonic dystrophy (DM1) leads to misregulated alternative splicing of insulin receptor (Savkur et al. 2001) and chloride channel pre-mRNA (Charlet et al. 2002b; Mankodi et al. 2002), causing the insulin resistance and myotonia that are characteristic symptoms of DM1. The six members (CUG-BP, ETR3, and CELFs 3–6; Good et al. 2000; Ladd et al. 2001) consist of three RRM-like RNA-binding domains and a divergent linker domain between RRMs 2 and 3 (Fig. 1B ▶). They are variably expressed, with CUG-BP, ETR3, and CELF4 being expressed in a variety of tissues, but CELF3 and CELF5 being brain-restricted (Choi et al. 1999; Lu et al. 1999; Good et al. 2000; Takahashi et al. 2000; Ladd et al. 2001). The founder family member, CUG-BP, was characterized as a protein that could bind to a (CUG)8 oligonucleotide, which was a model for the CUG repeat expansion in the 3′UTR of the DMPK gene in DM1 (Timchenko et al. 1996a, 1996b). CELF proteins have also been shown to play a role in regulation of cardiac troponin T (cTnT) splicing (Cooper 1998; Ladd et al. 2001), in which ETR3 activates exon 5 by antagonizing the repressive effect of PTB (Charlet et al. 2002a), and of two exons within the NMDA receptor A1 pre-mRNA (Zhang et al. 1999, 2002). However, to date there is limited in vitro evidence for the activity of CELF proteins as splicing regulators (Charlet et al. 2002a).
FIGURE 1.
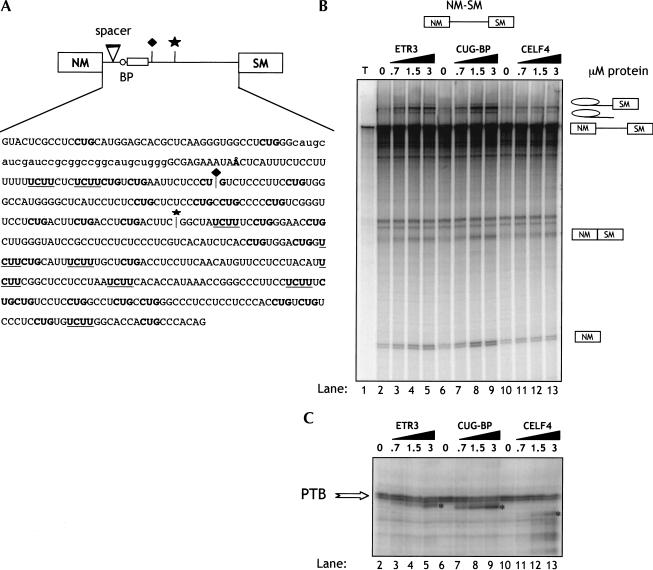
(A) Organization and alternative splicing of α-actinin exons. The NM (nonmuscle) and SM (smooth muscle) exons of α-actinin gene are not usually spliced together because of the close proximity of the SM exon branch point and pyrimidine tract (open circle and rectangle) to the 5′ splice site of the NM exon. The SM exon is repressed in most NM cells, and this repression involves multiple PTB-binding sites (UCUU motifs indicated by the vertical lines) between the exon and its upstream branch point. In brain, the major isoform contains both NM and SM exons (Kremerskothen et al. 2002), and direct splicing of EF1a to EF2 is also observed in differentiated SM cells (C. Gooding and C.W.J. Smith, unpubl. observations). Back circles indicate clusters of CUG motifs, arbitrarily defined as three or more CUGs within 18 nt. Taking into account the base composition of the intron, there are nearly 3× the number of CUG motifs as expected randomly. Black diamonds represent clusters of UG motifs, which are also potential high affinity sites for CELF proteins (Takahashi et al. 2000). (B) CELF proteins share a domain structure with three RRMs and a divergent linker region of unknown function between RRM2 and RRM3.
We have used the rat α-actinin (Act) gene as a model system of alternative splicing (Fig. 1A ▶; Southby et al. 1999; Wollerton et al. 2001). Actinin contains a pair of exons, one of which is selected predominantly in smooth muscle (SM) cells (Waites et al. 1992). In adult brain, the major isoform contains both NM (nonmuscle) and SM exons (Kremerskothen et al. 2002), whereas in smooth muscle both exons are skipped in a small proportion of the mRNA (C. Gooding and C.W.J. Smith, unpubl. observations). Inclusion of the NM exon, alone or in combination with the SM exon, leads to production of a Ca2+-binding EF hand domain, whereas SM inclusion or skipping of both exons leads to a nonfunctional domain (Waites et al. 1992). The branch point of the SM exon is 386 nt upstream of the exon, and is close enough to the NM exon to make NM-SM splicing relatively inefficient but not absolutely mutually exclusive (Southby et al. 1999). The SM exon is also repressed by PTB binding to multiple sites between the exon and the upstream branchpoint (Fig. 1A ▶; Southby et al. 1999). It is not yet known how this repression is relieved in SM or brain. However, the density of CUG motifs between the polypyrimidine tract and 3′ splice site is 3× the number expected based upon nucleotide composition (Fig. 1A ▶). This suggested a possible role for CELF proteins in regulation of actinin splicing.
We have investigated the role of the three widely expressed CELF proteins (CUG-BP, ETR3, and CELF4) in the regulation of α-actinin alternative splicing. Our in vivo and in vitro results demonstrate that CUG-BP and ETR3 can act as both activators and repressors of a pair of coregulated exons. They both promoted inclusion of the SM exon but skipping of the NM exon, and thus have the potential to impose concerted regulation on this pair of exons. In contrast, CELF4 was a repressor of both exons. Activation of SM exon selection by CUG-BP was achieved by a derepression mechanism involving the displacement of the repressor PTB from the polypyrimidine tract. Analysis of PTB and CUG-BP binding to short intron fragments, and to patch-labeled RNA, showed that there is a crucial binding competition between the two factors at adjacent sites at the 3′ end of the polypyrimidine tract. These data provide a mechanistic explanation for the activation of splicing by CELFs and support a model in which regulation of actinin exon selection is controlled by the synergistic and antagonistic influences of members of two groups of hnRNP proteins.
RESULTS
Overexpression of CELFs in vivo
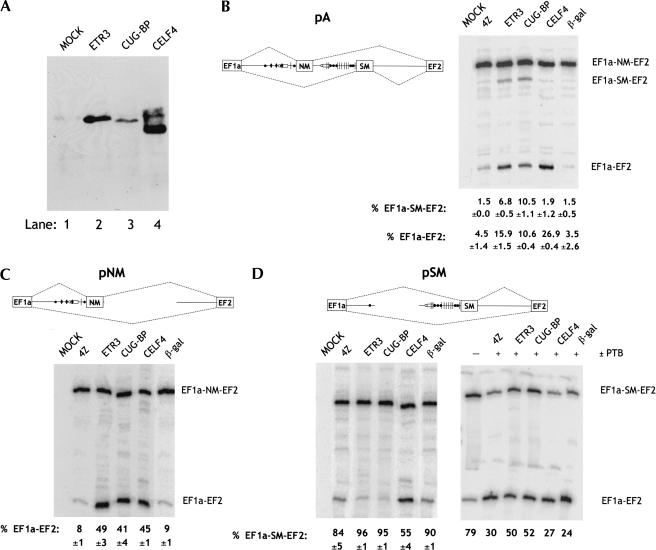
In view of the presence of multiple CUG repeats adjacent to the actinin regulated exons and the established role of CELFs as splicing regulators, we decided to test whether CELF proteins might act as regulators of actinin splicing. Vectors for ETR3, CUG-BP, and CELF4 were cotransfected with various reporter constructs into fibroblast L cells (Fig. 2 ▶). Overexpression of N-terminal epitope-tagged CELF proteins was successfully obtained in transfected cells as determined by Western blot using AntiXpress antibody (Fig. 2A ▶). CELF4 was always expressed at higher levels than the other two proteins when an equivalent amount of expression plasmid was transfected. We next looked at the effects of CELF overexpression upon alternative splicing in L cells of an α-actinin minigene construct containing the NM and SM mutually exclusive exons as well as the flanking exons EF1a and EF2 and complete introns (Fig. 2B ▶, construct pA). In the absence of cotransfected CELFs, the major splicing pathway was NM exon inclusion (Fig. 2B ▶; 4Z and βGal negative cotransfection controls). The most obvious effect upon overexpression of all three CELF proteins was an increase in the skipped EF1a-EF2 splicing product, with CELF4 having the most dramatic effect. This splicing pathway is observed in a proportion of actinin transcripts in differentiated SM cells (C. Gooding and C.W.J. Smith, unpubl. observations). Interestingly, as well as inducing EF1a-EF2 splicing, ETR3 and CUG-BP proteins also increased SM exon inclusion by four- to sevenfold, whereas CELF4 had no effect. The differential effects of CELF4 compared to the other two proteins are probably specific rather than being due to its higher expression levels, as subsequent in vitro experiments, in which the protein concentrations could be precisely controlled, also indicated it was also a less effective activator of the SM exon but an equally active repressor of the NM exon (see below). Similar effects have been recently reported upon cotransfection of zebrafish ETR3 and CUG-BP with the pA construct (Suzuki et al. 2002).
FIGURE 2.
Switching of actinin splicing by overexpression of CELF proteins. (A) Western blot analysis for overexpressed CELF proteins in L cells transiently transfected with expression vectors for ETR3, CUG-BP, CELF4, or mock-transfected (MOCK). Proteins were detected using AntiXpress-HRP conjugated antibody (Invitrogen) against the N-terminal AntiXpress tag in the expression plasmid. (B–D) L cells were transiently transfected with 0.2 μg of the pA (B), pNM (C) or pSM (D) α-actinin reporters together with 0.8 μg of pGEM4Z (4Z), ETR3, CUG-BP, CELF4, or β-gal expression constructs. In the right-hand section of D, 0.1 μg of expression plasmid for PTB4 was also cotransfected. RNA was harvested after 48 h, and RT-PCR analysis was performed to determine the extent of NM exon skipping and SM exon inclusion. In MOCK lanes, no DNA was transfected into the cells. Values below each lane give the percentage of EF1a-EF2 or EF1a-SM-EF2 splicing and represent mean ±SD for three experiments, except in the experiment with PTB4 cotransfection (D, right panel) where they represent the values for the experiment shown. In each case, the percentage of each splice product was calculated as a total of all (two or three) possible spliced products in the lane. The data show that ETR3 and CUG-BP promote SM exon inclusion and NM exon skipping, whereas CELF4 causes skipping of both NM and SM exons.
The effects of the overexpressed CELF proteins could involve inhibition or activation of either the NM or SM exons. To distinguish between the various possiblilities, we cotransfected CELF proteins with two further actinin constructs. Construct pNM has a deletion of the SM exon and contains the NM exon with its intronic sequences surrounded by EF1a and EF2 exons. In various cell lines, pNM splices with almost exclusive NM exon selection and a small proportion of exon skipping (Fig. 2C ▶). Cotransfection of each of the three CELF proteins with pNM induced a five- to sixfold increase in NM exon skipping and formation of EF1a-EF2 spliced product (Fig. 2C ▶). Thus, overexpression of the CELF proteins modified the splicing of pNM toward the SM splicing pathway by causing exclusion of the NM exon.
Construct pSM (Fig. 2D ▶) has a deletion of the NM exon and contains the SM exon surrounded by its intronic sequences and exons EF1a and EF2. pSM showed 16% SM exon skipping in L cells (Fig. 2D ▶, left panel). Thus, in the absence of the NM exon, repression of the SM exon is not sufficient to prevent its selection in the majority of transcripts. In contrast to their effect upon NM exon selection, ETR3 and CUG-BP promoted a small further increase in SM exon inclusion. In contrast, CELF4 increased SM exon skipping. The effects of ETR3 and CUG-BP were relatively modest because of the high basal level of inclusion of the SM exon. We therefore tested the effects of the CELF proteins in the presence of cotransfected PTB, which represses the SM exon and enhances the level of SM exon skipping in transfected cells (Fig. 2D ▶, right panel; Southby et al. 1999; Wollerton et al. 2001). In the experiment of Figure 2D ▶, cotransfection of PTB with pSM decreased the level of SM incorporation from 79% to 30%. Cotransfection of both ETR3 and CUG-BP increased the level of SM inclusion by 20%, whereas CELF4 had little or no effect. Thus, ETR3 and CUG-BP were able to induce skipping of the NM exon and inclusion of the SM exon, and to antagonize the activity of PTB, which is a known repressor of the SM exon. In contrast, CELF4 acted as a repressor of both exons, and so was functionally distinct from ETR3 and CUG-BP. To ascertain whether the effects of CELF proteins upon actinin splicing were direct and specific, we turned to in vitro splicing assays.
CELF proteins inhibit NM and activate SM exon splicing in vitro
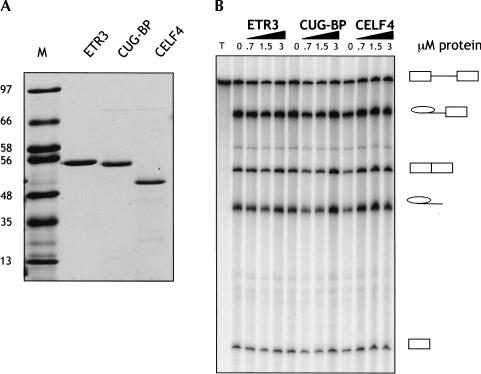
Recombinant CELF proteins were obtained (Fig. 3A ▶) by expression with N-terminal His-tag in combination with a C-terminal intein/chitin-binding domain tag, which is removed by self cleavage after purification (Materials and Methods). The activity of the recombinant CELF proteins was first tested on an unregulated splicing substrate comprising αTM exons 2 and 3 (GC + DX; Scadden and Smith 1995). None of the proteins had any significant effect upon splicing of this RNA (Fig. 3B ▶).
FIGURE 3.
(A) Coomassie-stained 12.5% SDS-PAGE of purified N-terminally His-tagged CELF proteins; the C-terminal intein/chitin-binding protein tag was cleaved away during purification. The sizes of the lane M protein markers (in kD) are indicated on the left. (B) Recombinant CELF proteins have no effect upon splicing of an unregulated control RNA. The indicated amounts of full-length CELF proteins were added to 10-μL splicing reactions containing the GC + DX pre-mRNA (Scadden and Smith 1995). Splicing precursor, intermediates, and products are indicated to the right. None of the CELF proteins had an effect upon GC + DX splicing.
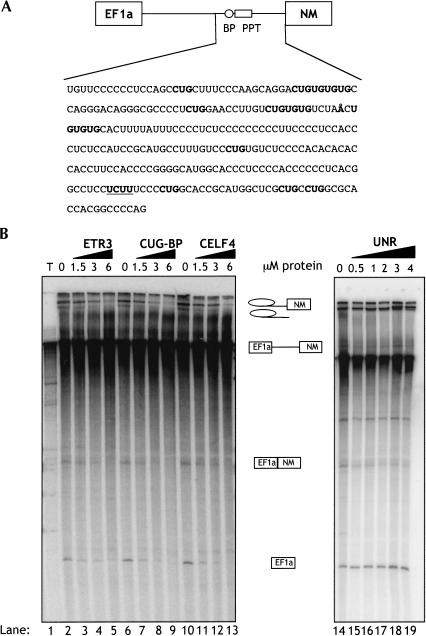
Transcripts containing the EF1a, NM, SM, and EF2 exons are not spliced detectably in vitro. We therefore used single intron constructs that have previously been used as reporters for NM and SM exon splicing (Southby et al. 1999). To investigate the activity of the CELF proteins upon in vitro splicing of the NM exon we used an RNA containing the EF1a and NM exons with the complete 954-nt intron between. Addition of increasing amounts (0.5–6 μM) of recombinant CELF proteins resulted in repression of EF1a-NM splicing, as indicated by the gradual disappearance of EF1a-NM spliced product, 5′ exon, lariat intermediate, and lariat intron (Fig. 4B ▶, lanes 2–13). Addition of an unrelated RNA-binding protein, UNR (Hunt et al. 1999), did not cause inhibition of EF1a-NM splicing (lanes 14–19). These results are consistent with the in vivo repression of the NM exon by CELF proteins (Fig. 2B,C).
FIGURE 4.
CELF proteins inhibit NM exon splicing in vitro. (A) Schematic representation of EF1a-NM RNA, with the NM branch point and pyrimidine tract indicated by the circle and rectangle. The sequence from the NM 3′ splice site to 296 nt upstream is shown with the branch point (indicated by the Å) 191 nt upstream. CUG and UG repeats are shown in bold, and the single UCUU motif is bold underlined. (B) 32P-labeled EF1a-NM α-actinin transcript was spliced in HeLa nuclear extracts for 2 h in the presence of 0–6 μM CELF proteins or 0–4 μM of the unrelated RNA-binding protein UNR (Hunt et al. 1999). RNA species were resolved on 6% polyacrylamide gels. The initial transcripts, splicing intermediates, and products are indicated next to the gels. ETR3, CUG-BP, and CELF4 inhibited NM exon splicing.
To test the effects of CELF proteins upon SM exon splicing in vitro, we used α-actinin reporter transcript NM-SM. This contains the NM and SM exons and intervening intron, with a 30-nt neutral spacer that reduces steric hindrance between the NM 5′ splice site and SM branchpoint (Fig. 5A ▶). It does not contain any sequences from the 191-nt region between the NM exon branch point and 3′ splice. Splicing of NM-SM RNA in HeLa nuclear extracts is repressed by the endogenous PTB, which acts via sites between the SM exon and its branch point (Southby et al. 1999). Therefore, this transcript is a reporter for the regulation of SM exon splicing.
FIGURE 5.
ETR3 and CUG-BP activate SM exon splicing by displacing PTB. (A) Sequence of the actinin NM-SM in vitro splicing substrate. A spacer element (lowercase) inserted between the NM 5′ splice site and SM branch point relieves the steric interference that usually prevents NM-SM splicing. The major branch point is Å. CUG motifs are shown in bold and UCUU motifs (optimal PTB-binding sites; Perez et al. 1997) are bold underlined. The diamond above the cartoon and sequence indicates the position of the junction in the ligated RNA in Figure 6A ▶, and the star indicates the position at which the NM1–4 transcripts were truncated in Figure 7D ▶. (B) 32P-labeled full-length α-actinin transcript (NM-SM) was spliced for 3 h in HeLa nuclear extract in the presence of increasing concentrations of CELF proteins and analyzed by denaturing 12% PAGE. Splicing precursor, intermediates, and products are indicated to the right. (C) Identical reactions to those in B were incubated for 30 min before UV cross-linking was carried out. Cross-linked proteins were analyzed by SDS PAGE and autoradiography. Cross-linked PTB is indicated by the arrow and CELF proteins by the asterisks. Lane numbers in C are equivalent to those in B, with the omission of lane 1, which contained RNA alone.
In striking contrast to their inhibition of EF1a-NM splicing, addition of ETR3 and CUG-BP resulted in activation of NM-SM splicing, indicated by the appearance of NM-SM spliced product, 5′ exon, lariat intermediate, and intron lariat (Fig. 5B ▶, lanes 2–5, 6–9). Interestingly, CELF4 was almost inactive, despite its equal inhibitory activity upon EF1a-NM splicing (Fig. 4B ▶). To study the interaction of endogenous splicing factors and recombinant proteins with the RNA, UV cross-linking was carried out after 30 min incubation in HeLa extract (Fig. 5C ▶). UV cross-linking of the NM-SM RNA in HeLa extract alone (lanes 2,6,10) produced a prominent PTB doublet of ∼58 kD. The identity of the PTB doublet has been confirmed by immunoprecipitation with both polyclonal and monoclonal PTB antibodies (Southby et al. 1999; J. Southby and C.W.J. Smith, unpubl. observations). Addition of increasing amounts of ETR3 and CUG-BP resulted in cross-linking of these proteins (indicated by the asterisks, lanes 3–5,7–9), which correlated with their activation of splicing (Fig. 5 ▶, cf. B and C). In the case of CUG-BP, this was accompanied by a reproducible decrease of ∼20% in the intensity of the PTB cross-link (lanes 6–9). In contrast to ETR3 and CUG-BP, CELF4 did not cross-link efficiently. In addition it caused less decrease in PTB cross-linking.
PTB displacement by CUG-BP at the polypyrimidine tract
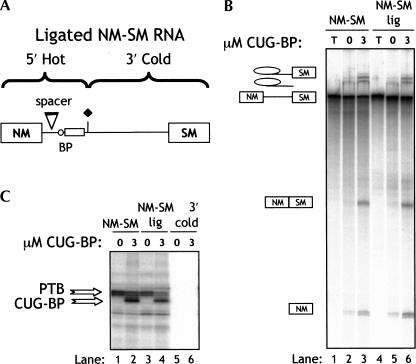
The previous data indicated that CUG-BP was the most potent activator of SM exon splicing and also reduced PTB cross-linking more effectively than the other CELF proteins. However, due to the large number of PTB-binding sites in the transcript, it was not clear at which sites CUG-BP was displacing PTB. To further investigate this point, we used a patch-labeled ligated NM-SM RNA in which radiolabel was only incorporated into the 5′ region, which contains the polypyrimidine tract (Fig. 6A ▶). Only the polypyrimidine tract region of the labeled fragment of RNA cross-links to PTB (see below, Fig. 7 ▶); all downstream PTB-binding sites in the ligated RNA were unlabeled. Splicing of the normal body-labeled and patch-labeled NM-SM was compared in the presence and absence of CUG-BP (Fig. 6B ▶). As expected, splicing of the two substrates, which are chemically identical, was the same, and both were activated similarly by CUG-BP (Fig. 6B ▶). The body-labeled NM-SM RNA once more showed a clear decrease in cross-linking of PTB in the presence of 3 μM CUG-BP (Fig. 6C ▶, 36% reduction between lanes 1 and 2). With the ligated RNA, a proportionately larger decrease in PTB cross-linking was observed upon addition of of 3 μM CUG-BP (55% decrease between lanes 3 and 4). These data indicate that the activation of NM-SM splicing by CUG-BP correlates with a competitive displacement of PTB at the polypyrimidine tract.
FIGURE 6.
Cross-linking of PTB and CUG-BP to the polypyrimidine tract in patch-labeled NM-SM RNA. (A) Patch-labeled NM-SM RNA was prepared using radiolabeled 5′ end RNA and trace-labeled 3′ end RNA. The junction between the labeled and unlabeled fragments is indicated by the diamond (see also Fig. 5A ▶ for junction sequence). (B) Body labeled and patch-labeled NM-SM RNA was incubated in nuclear extract for 3 h in the presence or absence of 3 μM CUG-BP. Lanes T: unprocessed transcript. (C) Cross-linking of body-labeled (lanes 1,2) and patch-labeled (lanes 3,4) NM-SM RNA after 30 min incubation in HeLa nuclear extract alone (lanes 1,3) or supplemented with 3 μM CUG-BP. (Lanes 5,6) Controls carried out with the trace-labeled 3′ RNA used to synthesize the patch-labeled RNA. The reduction in PTB cross-linking upon CUG-BP addition was 36% with the body-labeled RNA (lanes 1,2) and 55% for the patch labeled RNA (lanes 3,4).
FIGURE 7.
Antagonistic binding of PTB and CUG-BP to sites at the 3′ end of the polypyrimidine tract. (A) The sequence of RNA probes 1–4. Lowercase letters represent the partial overlap between adjacent probes. The branch point is represented by Å, CUG motifs are bold, and UCUU motifs are bold underlined. (B) Probes 1–4 were UV cross-linked to recombinant PTB and CUG-BP. In each case the strongest cross-link was seen to RNA 3, although PTB also cross-linked weakly to RNA 4 and CUG-BP to RNA 1. (C) UV cross-linking of HeLa nuclear extract containing increasing concentrations of CUG-BP to wild-type RNA 3 (WT) and RNA 3 with point mutations in the UCUU motifs (ΔPTB) or adjacent CUG motifs (ΔCUG). Mutated nucleotides are indicated in lowercase, CUG motifs in bold, and UCUU motifs in bold underline. Mutation of the CUG motifs inhibits CUG-BP cross-linking and prevents competition of PTB binding, whereas mutation of the UCUU motifs decreased PTB cross-linking with no effect upon CUG-BP. (D) Wild-type (WT) or ΔCUG NM1–4 RNA was incubated in HeLa nuclear extract for 25 min in the presence of increasing concentrations of CUG-BP (0, 0.75, 1.5, 3 μM, lanes 1–4 and 5–8, respectively). Splicing activation by CUG-BP was approximately twofold greater with the wild-type NM1–4 RNA than with the ΔCUG mutant (based upon phosphorimager analysis of intensity of the 5′ exon and precursor bands).
To define more precisely the site of functional antagonism between PTB and CUG-BP, we synthesized four partially overlapping RNA fragments (1–4) encompassing the 5′ end of the NM-SM RNA (Fig. 7A ▶). Radiolabeled probes were cross-linked to recombinant PTB and CUG-BP (Fig. 7B ▶). Only RNA 3, which contains the polypyrimidine tract, cross-linked efficiently to PTB. RNA 4, which is also pyrimidine-rich, cross-linked much more weakly to PTB. CUG-BP also cross-linked more efficiently to RNA 3 than to any other fragment, despite the fact that it contains fewer CUG motifs than RNA 4. Fragment 3 is of particular interest because it contains the SM exon polypyrimidine tract and so is a likely site for regulation of splicing. We next analyzed cross-linking to RNA 3 in HeLa nuclear extract in the presence of increasing concentrations of CUG-BP (Fig. 7C ▶, lanes 1–4). Although the probe cross-linked only to PTB in the nuclear extract alone (lane 1), increasing concentrations of CUG-BP (lanes 2–4) led to a strong cross-linked CUG-BP band with a concomitant decrease in PTB cross-linking even at the lowest concentrations (0.5 μM, lane 2). We next focused on the region of RNA 3 that contains the UCUU motifs and an adjacent CUGUCUG motif, which is conserved between rat and human α-actinin genes. We investigated the effects of mutations in these elements upon competition between HeLa PTB and recombinant CUG-BP (Fig. 7C ▶). Whereas wild-type RNA 3 showed competition between PTB and CUG-BP binding (lanes 2–4), ΔCUG RNA 3, containing two G-to-A mutations within the CUGs, showed a marked reduction in cross-linking to CUG-BP (lanes 6–8), indicating that these mutations have identified the major site of CUG-BP within RNA 3. The PTB cross-link to this probe was highly resistant to addition of CUG-BP. In contrast, the ΔPTB RNA 3, which contains four C-to-U transitions showed a marked reduction in PTB cross-linking in HeLa extract (lane 9) with the appearance of two new unidentified cross-linked bands. This confirms that the two closely spaced UCUU motifs consititute the major PTB binding site within the SM polypyrimidine tract. Moreover, the residual PTB cross-link in the ΔPTB RNA 3 was effectively competed by CUG-BP (lanes 10–12), which cross-linked as strongly to this probe as to the wild-type probe. These data show that there is mutually competitive binding between PTB and CUG-BP at the adjacent UCUU and CUG motifs at the 3′ end of the polypyrimdine tract.
We tested whether this binding competition might have functional consequences by looking at the effect of the ΔCUG mutation upon the ability of CUG-BP to activate splicing of NM-SM RNA. In the context of full-length NM-SM RNA, the ΔCUG mutation did not have an obvious effect upon splicing (data not shown). This could be related to the presence of additional downstream binding sites for CUG-BP in the full-length RNA. If CUG-BP binding is cooperative, the effect of the ΔCUG mutation in RNA3 might be expected to be less significant in the context of the full-length RNA. We therefore tested the effects of the ΔCUG mutation in the context of RNA1–4, which is truncated after nt 221 of the intron (indicated by the star in Figs. 5A, 7A ▶ ▶). This RNA retains the SM branch point and polypyrimidine tract and some adjacent CUG clusters, but removes seven out of nine UCUU motifs, leaving only the two within the polypyrimidine tract. The removal of these sequences activates step 1 of splicing (Southby et al. 1999). However, by analyzing splicing after 25 min, CUG-BP could be seen to accelerate the appearance of splicing intermediates (Fig. 7D ▶). Introduction of the ΔCUG mutation into the RNA substrate had a small effect upon this activity of CUG-BP. Quantitation of the phosphorimager data (by measuring the intensity of the 5′ exon compared to the precursor RNA) indicated that activation of splicing by CUG-BP was reproducibly reduced approximately twofold by the ΔCUG mutation. This correlates with the reduced ability of CUG-BP to displace PTB from ΔCUG RNA3 in cross-linking assays (Fig. 7 ▶, cf. C and D). Therefore, mutually antagonistic binding of PTB and CUG-BP to adjacent sites appears to explain the activation of actinin SM exon splicing by CUG-BP.
DISCUSSION
Our results highlight the importance and versatility of the CELF family of proteins as splicing regulators. We show that individual CELF proteins can exert concerted regulation of the two members of a mutually exclusive exon pair both by activating the actinin SM exon and repressing the NM exon (Figs. 2, 4, 5 ▶ ▶ ▶). We also demonstrate a mechanism whereby they can activate splicing by displacing the known repressor protein PTB (Figs. 5–7 ▶ ▶ ▶). Moreover, we show that different family members can have opposite effects upon selection of a single exon (Fig. 2 ▶). There have been a number of recent reports upon the activites of CELFs as splicing regulators (Ladd et al. 2001; Savkur et al. 2001; Charlet et al. 2002a, 2002b; Suzuki et al. 2002; Zhang et al. 2002). Our findings extend significantly beyond these reports in a number of respects, in particular with the direct in vitro demonstration of both splicing activation and repression activity of CUG-BP and ETR3, and with the concerted regulation of a mutually exclusive exon pair.
Recently, Suzuki et al. (2002) showed that zebrafish ETR3 and CUG-BP were able to switch splicing of the rat pA actinin transfection reporter (Fig. 2B ▶) toward inclusion of the SM exon. They analyzed the role of UG repeat elements that resemble optimal binding sites for CELF proteins (Takahashi et al. 2000; Suzuki et al. 2002), and found that mutation of the UG repeats flanking the NM exon branchpoint (Fig. 4A ▶) reduced the ability of ETR3 and CUG-BP to alter splicing. This is consistent with our demonstration of CELF-mediated inhibition of NM exon splicing both in vivo (Fig. 2B,C) and in vitro (Fig. 4 ▶). We also tested the effects of the same mutations in the branch-point-associated UG repeats upon the in vitro splicing of EF1a-NM transcripts, but saw only marginally reduced sensitivity to inhibition by CELFs (data not shown). The reason for this apparent discrepancy is not currently clear, but is probably related to differences between the assays in which the mutations were tested; the in vitro assay used a single intron construct, whereas the in vivo assays involved competing splicing pathways.
Our results provide further insight into α-actinin splicing regulation with the finding of independent activation of SM exon splicing by the CELF proteins (Figs. 2, 5–7 ▶ ▶ ▶ ▶). CUG-BP was the best activator of SM exon splicing in both the in vivo (Fig. 2 ▶) and in vitro (Fig. 5 ▶) assays. CUG-BP and ETR3 have been shown previously to act as activators of splicing (Philips et al. 1998; Ladd et al. 2001; Charlet et al. 2002a; Zhang et al. 2002). Here we provide a mechanistic explanation of how CELFs are able to achieve activation by antagonism of a splicing repressor. PTB inhibits the actinin SM exon by binding to multiple sites between the exon and its distant upstream branch point, including sites within the branch-point-associated pyrimidine tract (Southby et al. 1999). Activation of SM splicing by CUG-BP, and to a lesser extent ETR3, correlated with decreased binding of PTB to the RNA (Figs. 5–7 ▶ ▶ ▶).
Cross-linking to the patch-labeled ligated RNA indicated that CUG-BP displaced PTB from the polypyrimidine tract (Fig. 6 ▶), whereas targeted mutations demonstrated competitive binding of PTB and CUG-BP to adjacent sites at the 3′ end of the pyrimidine tract (Fig. 7C ▶). Mutation of the CUG-BP-binding site slightly decreased the ability of CUG-BP to activate splicing, but only in the context of the NM1–4 RNA, which is already activated by 3′ end truncation (Fig. 7D ▶). In the full-length RNA this mutation had no obvious effect. Similarly, the ΔPTB mutation has no dramatic effect upon splicing of the full-length NM-SM RNA (J. Southby, A.J. Matlin, and C.W.J. Smith, in prep.). The lack of effect of these mutations can be attributed to the presence of several interspersed redundant binding sites for both proteins between the polypyrimidine tract and SM exon. We have dissected the entire intron into several fragments and have identified additional downstream regions where both proteins bind (A.J. Matlin and C.W.J. Smith, unpubl. observations). Experiments with ligated RNAs show that PTB binding to the pyrimidine tract is enhanced by the presence of additional downstream sites (J. Southby, A.J. Matlin, and C.W.J. Smith, in prep.), which is consistent with either “RNA looping” or an extended “zone of silencing” model for PTB-mediated repression (Wagner and Garcia-Blanco 2001; Cartegni et al. 2002). It is probable that mutation of several sites will be required to fully impair the activity of either PTB or CUG-BP in the context of the full-length intron. The functional antagonism between PTB and CUG-BP may prove to involve competition between two sets of cooperative binding events. Such a scenario may allow the system to respond to small fluctuations in the relative levels of either class of protein. An obvious question raised by this model is why binding of PTB but not CUG-BP is repressive. One possibility is that the high affinity CUG-BP binding site is slightly further from the branch point, and so binding here may not interfere with events such as U2AF65 binding that might be directly competed by PTB binding upstream.
The model for regulation of actinin SM exon splicing has interesting similarities and differences with the activation of cTnT exon 5 (Ladd et al. 2001; Charlet et al. 2002a). Direct binding competition between ETR3 and PTB is observed at adjacent sites within the MSE2 element on the downstream side of cTnT exon 5. However, PTB also binds to the polypyrimidine tract on the opposite side of the exon, where there are no ETR3 binding sites. The common theme linking both systems is that there are extended regions containing a series of binding sites for each class of regulatory protein. In actinin, these sites are within the extended region between the branch point and exon, whereas in cTnT the sites are on both sides of the exon.
In contrast to their effects upon the actinin SM exon, all three tested CELFs were found to repress splicing of the NM exon. Whereas the first reports of CELF-regulated splicing events involved activation (Philips et al. 1998; Ladd et al. 2001; Charlet et al. 2002a), more recently examples of CELF-mediated splicing repression have been reported. These include repression by CUG-BP of exon 11 of the insulin receptor (Savkur et al. 2001), of exon 5 of NMDA receptor R1 by ETR3 (Zhang et al. 2002), and of intron 2 of CIC-1 pre-mRNA (Charlet et al. 2002b). Of interest, NMDA-R1 pre-mRNA contains exons that are either repressed (exon 5) or activated (exon 21) by CELFs, although regulation of these two exons is not as tightly coupled as the actinin mutually exclusive exons. The mechanism of repression by the CELFs is not yet clear. UV cross-linking assays using the EF1a-NM RNA were not informative, as the recombinant CELF proteins did not cross-link efficiently, and there were no obvious changes in cross-linking of HeLa proteins upon addition of CELF proteins (data not shown). However, the abrogation of the effect of transfected CELFs by mutation of the UG repeats associated with the NM branch point suggests that inhibition could be achieved by blocking access of splicing factors SF1 or U2 snRNP to the branch point (Suzuki et al. 2002). In ClC-1 pre-mRNA the UG repeats are in the polypyrimidine tract, so competition with U2AF65 is a plausible mechanism.
A prediction following from our observations is that alterations in the absolute or relative levels of members of the CELF and PTB families should be observed in cells where actinin splicing is regulated. Preliminary investigation of expression of ETR3 and CUG-BP in fully differentiated, and dedifferentiated rat aorta SM cells found that both genes were expressed in the fully differentiated cells, but at lower levels than in the dedifferentiated cells. However, the differentiated cells also have much less PTB. Antagonism between ASF/SF2 and hnRNPA1 relies on the ratio of the two proteins rather than their absolute concentrations (Caceres et al. 1994). Thus, it is possible that in differentiated SM cells a decrease in CUG-BP and/or ETR3 is more than offset by the decrease in PTB concentration. Another important factor is that in brain, PTB is replaced by nPTB/brPTB, whereas differentiated rat SM cells express a novel PTB paralog distinct from PTB, nPTB/brPTB and ROD1, with approximately 70% amino acid identity to the other family members (C. Gooding, P. Kemp, and C.W.J. Smith, in prep.). It is an attractive possibility, that the change in PTB paralogs may modulate the competition between CELFs and PTBs sufficiently to activate SM exon splicing. The distinct activity of CELF4 compared with CUG-BP or ETR3 upon SM exon splicing also emphasizes that shifts in the balance of CELF family members could be an additional important influence. CELF4 and CELF3 were also shown to be functionally distinct from CUG-BP and ETR3 as more potent activators of exon selection mediated by multimers of the cTnT MSE2 element (Ladd et al. 2001). Finally another factor that may modulate the competition is alternative splicing of CUG-BP and ETR3. The alternatively spliced PTB1 and 4 isoforms have distinct activities in some assays (Wollerton et al. 2001). Differential activity of alternatively spliced CELF isoforms is an interesting possibility.
Control of alternative splicing has often been viewed in terms of the modulation by regulatory factors of splice site selection by the general splicing machinery. Our results emphasize the point that antagonistic interactions between families of regulators that are not themselves essential splicing factors can also be important determinants in controlling alternative splicing.
MATERIALS AND METHODS
Constructs
Constructs for in vitro transcription and transient transfections were prepared by standard cloning techniques (Sambrook et al. 1989). The pA construct contained nt 25–2951 of the actinin genomic sequence (Southby et al. 1999). pNM was derived from pA and contained a BglI-HindIII deletion (nt 1319–1991) removing the SM exon and some flanking intronic sequences. pSM was also derived from pA by a deletion from BstEII to the end of the NM exon (nt 875–1277) removing the NM exon and some upstream intronic sequence. EF1a-NM and NM-SM in vitro constructs were described in Southby et al. (1999).
The individual fragments 1, 2, 3, and 4 (Fig. 6A ▶) were made as PCR templates containing a T7 RNA polymerase promoter using the following oligonucleotides:
1F: 5′-CGTAATACGACTCACTATAGGATCACTCCGGCACGTT, 1R: 5′-CTGGGGGTCGTTGCCAA;
2F: 5′-CGTAATACGACTCACTATAGGTACTCGCCTCCTGC, 2R: 5′-TGAGTTATTTCTCGCCC;
3F: 5′-CGTAATACGACTCACTATAGGCGAGAAATAACTCATT TC, 3R: 5′-TGAGCCCCATGGCCCAC;
4F: 5′-CGTAATACGACTCACTATAGGGCTCATCCTCTCCTG, 4R: 5′-GAAGTCAGAGGTCAGAAGT.
PCR products were treated with T4 DNA polymerase (New England Biolabs) to remove 3′ overhangs and gel purified.
The ΔPTB and ΔCUG-BP mutations of fragment 3 (Fig. 6D ▶) were generated by site-directed mutagenesis.
For in vivo expression, the ORFs of the CELFs were cloned into the pcDNA3.1HisC vector (Invitrogen) in frame with the N-terminal Xpress epitope tag. For expression of recombinant CELFs in Escherichia coli, the ORF of each protein was amplified by PCR using an upstream primer containing 6×HIS tag and downstream primer designed to remove the termination codon. The products were cloned as NdeI/SapI fragments into the pTYB1 expression vector (New England Biolabs), creating N-terminal fusions with the intein/chitin-binding domain tag.
Cell culture and transfections
Mouse L cells were grown under standard conditions in DMEM supplemented with 10% fetal calf serum and 1% (v/v) L-glutamine. Transient transfections were performed in 35-mm plates using lipofectamine (GIBCO BRL). Two to 5 μL of lipofectamine were preincubated with 1–2 μg of DNA (or a mixture of cotransfected DNAs) in the presence of 0.1 mL of Opti-MEM I Reduced Serum Medium for 30 min at room temperature to form DNA–liposome complexes. Complexes were diluted by addition of 0.8 mL of Opti-MEM and applied to the cells for 5–6 h at 37°C, after which the transfection mixture was replaced by complete medium, and cells were harvested after another 12–18 h.
Isolation and analysis of cellular RNA and protein
Transiently expressed cytoplasmic RNA was harvested using RNA/DNA/protein isolation TRI-reagent (Molecular Research Centre Inc.), and analyzed by RT-PCR as previously described (Wollerton et al. 2001; Gromak and Smith 2002), and quantitated using a Molecular Dynamics Phosphorimager (Gooding et al. 1994; Smith et al. 1996). Expression of Xpress epitope-tagged CELF proteins was analyzed by Western blot of total cellular protein using AntiXpress antibody conjugated to horseradish peroxidase (Invitrogen) and enhanced chemiluminescence assay.
In vitro transcription and splicing reactions
Capped pre-mRNA transcripts were transcribed by T7 or SP6 RNA polymerase using [α32P]-UTP (Gooding et al. 1998; Southby et al. 1999) and gel purified if necessary. Splicing reactions typically contained 5 ng of 32P RNA transcript in a 10-μL reaction, 2 mM MgCl2, 0.5 mM ATP, 20 mM creatine phosphate, 1.2 U/μL RNasin, 2.6% polyvinyl alcohol, 25%–30% nuclear extract. Recombinant proteins were usually added to the nuclear extract prior to assembling the remainder of the splicing reaction mixture. Splicing reactions were typically incubated at 30°C for 30 min–3 h and then subjected to proteinase K digestion and phenol extractions. RNAs were precipitated in 96% ethanol on dry ice for 1 h, and then the products of the splicing reaction were analyzed on 6%–12% gels containing 8 M urea, followed by autoradiography.
Ligation of RNA fragments
5′ and 3′ PCR templates for RNA ligation experiments were generated using the following primers (underlined residues carry a 2′-OMe group):
T7: 5′-TAATACGACTCACTATAGGG;
5′R-(OMe)2: 5′-AGGGAGAATTCAGACAG;
3′F: 5′-GCTCTAATACGACTCACTATAGTCTCCCTTCCTGTG;
3′R: 5′-GATCCTCTAGAGTCCATGTTG.
For the production of patch-labeled transcripts (Moore and Query 1998), 1360 fmoles high specific activity 5′ RNA were combined with 6600 fmoles gel-purified trace-labeled 3′ RNA (uncapped). These substrates were annealed to the bridging DNA oligonucleotide (splint 6: 5′-CCATGGCCCACAGGAAGGGAGA CAGGGAGAATTCAGACAGA; 4480 fmoles) in a 30-μL reaction containing 4 μL 10× T4 DNA ligase buffer by heating to 75°C for 2 min, followed by a 5 min incubation at room temperature. A mixture of 4 μL high concentration T4 DNA ligase, 2 μL RNA guard, and 4 μL T4 DNA ligase buffer was then added and the reaction was placed for 4 h at 30°C . Finally, the bridging oligonucleotide was digested by treatment with DNAse RQ1 for 30 min at 37°C. Products were separated from unligated substrates by gel purification.
UV cross-linking
For UV cross-linking, 10-μL splicing reactions (some without PVA) containing 20 fmoles of high specific activity [α32P]-UTP-labeled RNA probes were incubated for 30 min at 30°C. The reaction mixtures were placed on ice and exposed to 2 × 960 mJ UV light. Following irradiation, the samples were treated with 4 μL of ribonuclease solution (10 mg/mL RNase A, 100 U/μL RNase T1, in 10× RNase dilution buffer, 100 mM Tris-HCl at pH 7.5, 10 mM MgCl2, 1 M KCl) and the labeled cross-linked proteins were resolved by SDS polyacrylamide gel electrophoresis followed by autoradiography.
The UV cross-linking reactions on individual fragments contained 10 fmoles high specific activity [α32P]-UTP-labeled RNA probes, 1× BB (50 mM HEPES at pH 7.9, 15 mM MgCl2, 25% (v/v) glycerol, 5 mM DTT), KCl to a concentration of 100 mM and 0.5 μg rRNA nonspecific competitor in the presence of recombinant CELF proteins. For the competiton studies, each reaction was supplemented with 1 μL of nuclear extract. Reactions were incubated for 25 min at 30°C followed by UV irradiation, RNase digestion, and SDS electrophoresis procedure as described above.
Expression and purification of the recombinant double-tagged proteins
BL21(DE3)pLysS (Stratagene) competent cells were transformed and used for protein expression. A single colony was grown overnight at 37°C in LB medium with appropriate antibiotics (Amp and Cm), diluted 1:50, and grown at 37°C until A600 reached 0.7–0.9. Then 0.3 mM IPTG were added to the culture and it was further incubated overnight at 15°C before harvesting the cells. The cell pellet was resuspended in 50 mL of SB buffer (50 mM NaPO4 at pH 8, 300 mM NaCl, 5% glycerol, 1 mM PMSF, 1 tablet of EDTA-free protease inhibitor cocktail) and lysed in a FRENCH™ pressure cell press (SLM Instruments, Inc.). The total cell lysate was centrifuged for 10 min at 8000 rpm. The supernatant was mixed with 0.8–1 mL of the Ni-NTA resin and left rotating for 1.5 h at 4°C. Following four washes in 10 mL of SB + 20 mM imidazole, recombinant proteins were applied to Biorad disposable columns and eluted from the Ni2+ beads using a step gradient of 100–500 mM imidazole at 4°C. Fractions containing proteins of interest were pooled, and buffer exchange was carried out over PD-10 columns (Amersham Biosciences), eluting in 3.5 mL of SB buffer. Then protein preparations were incubated with 0.8–1 mL of chitin resin (New England Biolabs) for 4–5 h at 4°C. After washing with 20 volumes of SB, the resin was applied to a Biorad column, flushed with 3 volumes of SB + 50 mM DTT and incubated overnight at 4°C to allow cleavage of the chitin-binding domain. Full-length proteins were subsequently eluted with 5× 0.5 volume of SB followed by exchange into buffer E (20 mM HEPES at pH 7.9, 20% glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM DTT) over a PD-10 column. The final protein preparations were frozen in aliquots on dry ice and stored at −80°C. Protein concentrations were estimated by running alongside BSA standards on SDS-PAGE gels stained with Coomassie and followed by densitometry analysis.
Acknowledgments
We thank Justine Southby for constructs and advice on RNA ligation, Matthew Wollerton for constructs, and Emma Brown and Richard Jackson for UNR protein. This work was supported by Wellcome Trust Programme Grant 059879 and Project Grant 052968 (C.W.J.S.) and NIH Grant HL45565 (T.A.C.). N.G. was supported by a scholarship from the Darwin Trust of Edinburgh University, and A.J.M. by an MRC studentship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2191903
REFERENCES
- Black, D.L. 2000. Protein diversity from alternative splicing: A challenge for bioinformatics and post-genome biology. Cell 103: 367–370. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J. 2000. Exonic splicing enhancers: Mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25: 106–110. [DOI] [PubMed] [Google Scholar]
- Burge, C., Tuschl, T., and Sharp, P. 1999. Splicing of precursors to mRNAs by the spliceosomes. In The RNA world, 2nd ed. (eds. R.F. Gestetland et al.), pp. 525–560. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Caceres, J.F. and Kornblihtt, A.R. 2002. Alternative splicing: Multiple control mechanisms and involvement in human disease. Trends Genet. 18: 186–193. [DOI] [PubMed] [Google Scholar]
- Caceres, J.F., Stamm, S., Helfman, D.M., and Krainer, A.R. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265: 1706–1709. [DOI] [PubMed] [Google Scholar]
- Caputi, M., Mayeda, A., Krainer, A.R., and Zahler, A.M. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 18: 4060–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni, L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285–298. [DOI] [PubMed] [Google Scholar]
- Charlet, B.N., Logan, P., Singh, G., and Cooper, T.A. 2002a. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9: 649–658. [DOI] [PubMed] [Google Scholar]
- Charlet, B.N., Savkur, R.S., Singh, G., Philips, A.V., Grice, E.A., and Cooper, T.A. 2002b. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell 10: 45–53. [DOI] [PubMed] [Google Scholar]
- Choi, D.K., Ito, T., Tsukahara, F., Hirai, M., and Sakaki, Y. 1999. Developmentally regulated expression of mNapor encoding an apoptosis-induced ELAV-type RNA binding protein. Gene 237: 135–142. [DOI] [PubMed] [Google Scholar]
- Chou, M.-Y., Underwood, J.G., Nikolic, J., Luu, M.H.T., and Black, D.L. 2000. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell 5: 949–957. [DOI] [PubMed] [Google Scholar]
- Cooper, T.A. 1998. Muscle-specific splicing of a heterologous exon mediated by a single muscle-specific splicing enhancer from the cardiac troponin T gene. Mol. Cell. Biol. 18: 4519–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Gatto Konczak, F., Olive, M., Gesnel, M.C., and Breathnach, R. 1999. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 19: 251–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X.D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1: 663–680. [PMC free article] [PubMed] [Google Scholar]
- Good, P.J., Chen, Q., Warner, S.J., and Herring, D.C. 2000. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J. Biol. Chem. 275: 28583–28592. [DOI] [PubMed] [Google Scholar]
- Gooding, C., Roberts, G.C., Moreau, G., Nadal Ginard, B., and Smith, C.W.J. 1994. Smooth muscle-specific switching of α-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 13: 3861–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding, C.G., Roberts, G.C., and Smith, C.W.J. 1998. Role of an inhibitory pyrimidine-element and general pyrimidine-tract binding proteins in regulation of α-tropomyosin alternative splicing. RNA 4: 85–100. [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. . 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17: 100–107. [DOI] [PubMed] [Google Scholar]
- Gromak, N. and Smith, C.W. 2002. A splicing silencer that regulates smooth muscle specific alternative splicing is active in multiple cell types. Nucleic Acids Res. 30: 3548–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, S.L., Hsuan, J.J., Totty, N., and Jackson, R.J. 1999. Unr, a cellular cytoplasmic RNA-binding protein with five cold shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes & Dev. 13: 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, K.B., Dredge, B.K., Stefani, G., Zhong, R., Buckanovich, R.J., Okano, H.J., Yang, Y.Y., and Darnell, R.B. 2000. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability [see comments]. Neuron 25: 359–371. [DOI] [PubMed] [Google Scholar]
- Krecic, A.M. and Swanson, M.S. 1999. hnRNP complexes: Composition, structure, and function. Curr. Opin. Cell. Biol. 11: 363–371. [DOI] [PubMed] [Google Scholar]
- Kremerskothen, J., Teber, I., Wendholt, D., Liedtke, T., Böckers, T.M., and Bernekow, A. 2002. Brain-specific splicing of a-actinin 1 mRNA. Biochem. Biophys. Res. Commun. 295: 678–681. [DOI] [PubMed] [Google Scholar]
- Ladd, A.N., Charlet-B.N., and Cooper, T.A. 2001. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 21: 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C.H. and Patton, J.G. 1995. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA 1: 234–245. [PMC free article] [PubMed] [Google Scholar]
- Lopez, A.J. 1998. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32: 279–305. [DOI] [PubMed] [Google Scholar]
- Lu, X., Timchenko, N.A., and Timchenko, L.T. 1999. Cardiac elav-type RNA-binding protein (ETR-3) binds to RNA CUG repeats expanded in myotonic dystrophy. Hum. Mol. Genet. 8: 53–60. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. and Tasic, B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418: 236–243. [DOI] [PubMed] [Google Scholar]
- Mankodi, A., Takahashi, M.P., Jiang, H., Beck, C.L., Bowers, W.J., Moxley, R.T., Cannon, S.C., and Thornton, C.A. 2002. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell 10: 35–44. [DOI] [PubMed] [Google Scholar]
- Markovtsov, V., Nikolic, J.M., Goldman, J.A., Turck, C.W., Chou, M.-Y., and Black, D.L. 2000. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 20: 7463–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAfee, J.G., Huang, M., Soltaninassab, S., Rech, J.E., Iyengar, S., and Lesturgeon, W.M. 1997. The packaging of pre-mRNA. In Eukaryotic mRNA processing (ed. A.R. Krainer), pp. 68–102. Oxford University Press, Oxford/New York.
- Modrek, B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30: 13–19. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. and Query, C.C. 1998. Uses of site-specifically modified RNAs constructed by RNA ligation. In RNA protein interactions: A practical approach (ed. C.W.J. Smith), Vol. 192, pp. 75–108. Oxford University Press, Oxford.
- Perez, I., Lin, C.-H., McAfee, J.G., and Patton, J.G. 1997. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA 3: 764–778. [PMC free article] [PubMed] [Google Scholar]
- Philips, A.V., Timchenko, L.T., and Cooper, T.A. 1998. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy [see comments]. Science 280:737–741. [DOI] [PubMed] [Google Scholar]
- Polydorides, A.D., Okano, H.J., Yang, Y.Y., Stefani, G., and Darnell, R.B. 2000. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. 97: 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell. Biol. 12: 340–345. [DOI] [PubMed] [Google Scholar]
- Roberts, G.C. and Smith, C.W. 2002. Alternative splicing: Combinatorial output from the genome. Curr. Opin. Chem. Biol. 6: 375–383. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Savkur, R.S., Philips, A.V., and Cooper, T.A. 2001. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 29: 40–47. [DOI] [PubMed] [Google Scholar]
- Scadden, A.D.J. and Smith, C.W.J. 1995. Interactions between the terminal bases of mammalian introns are retained in inosine-containing pre-mRNAs. EMBO J. 14: 3236–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R., Valcarcel, J., and Green, M.R. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268: 1173–1176. [DOI] [PubMed] [Google Scholar]
- Smith, C.W.J. and Valcárcel, J. 2000. Alternative pre-mRNA splicing: The logic of combinatorial control. Trends Biochem. Sci. 25: 381–388. [DOI] [PubMed] [Google Scholar]
- Smith, C.W.J., Gooding, C., Roberts, G.C., and Scadden, A.D.J. 1996. Analyzing patterns of alternative splicing. In A laboratory guide to RNA. Isolation, analysis and synthesis (ed. P.A. Kreig), pp. 411–440. John Wiley & Sons, Inc., New York.
- Southby, J., Gooding, C., and Smith, C.W. 1999. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutally exclusive exons. Mol. Cell. Biol. 19: 2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., Jin, Y., Otani, H., Yasuda, K., and Inoue, K. 2002. Regulation of alternative splicing of α-actinin transcript by Bruno-like proteins. Genes Cells 7: 133–141. [DOI] [PubMed] [Google Scholar]
- Takahashi, N., Sasagawa, N., Suzuki, K., and Ishiura, S. 2000. The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system [In Process citation]. Biochem. Biophys. Res. Commun. 277: 518–523. [DOI] [PubMed] [Google Scholar]
- Timchenko, L.T., Miller, J.W., Timchenko, N.A., DeVore, D.R., Datar, K.V., Lin, L., Roberts, R., Caskey, C.T., and Swanson, M.S. 1996a. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 24: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko, L.T., Timchenko, N.A., Caskey, C.T., and Roberts, R. 1996b. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: Implications for myotonic dystrophy. Hum. Mol. Genet. 5: 115–121. [DOI] [PubMed] [Google Scholar]
- Valcarcel, J. and Gebauer, F. 1997. Post-transcriptional regulation: The dawn of PTB. Curr. Biol. 7:R705–R708. [DOI] [PubMed] [Google Scholar]
- Wagner, E.J. and Garcia-Blanco, M.A. 2001. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 21: 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, E.J. and Garcia-Blanco, M.A. 2002. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell 10: 943–949. [DOI] [PubMed] [Google Scholar]
- Waites, G.T., Graham, I.R., Jackson, P., Millake, D.B., Patel, B., Blanchard, A.D., Weller, P.A., Eperon, I.C., and Critchley, D.R. 1992. Mutually exclusive splicing of calcium-binding domain exons in chick α-actinin. J. Biol. Chem. 267: 6263–6271. [PubMed] [Google Scholar]
- Wollerton, M., Gooding, C., Robinson, F., Brown, E., Jackson, R., and Smith, C. 2001. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein. RNA 7: 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Liu, W., and Grabowski, P.J. 1999. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA 5: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Liu, H., Han, K., and Grabowski, P.J. 2002. Region-specific alternative splicing in the nervous system: Implications for regulation by the RNA-binding protein NAPOR. RNA 8: 671–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J., Mayeda, A., and Krainer, A.R. 2001. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell 8: 1351–1361. [DOI] [PubMed] [Google Scholar]