Abstract
Genome-wide genetic approaches have proven useful for examining pathways of biological significance in model organisms such as Saccharomyces cerevisiae, Drosophila melanogastor, and Caenorhabditis elegans, but similar techniques have proven difficult to apply to mammalian systems. Although manipulation of the murine genome has led to identification of genes and their function, this approach is laborious, expensive, and often leads to lethal phenotypes. RNA interference (RNAi) is an evolutionarily conserved process of gene silencing that has become a powerful tool for investigating gene function by reverse genetics. Here we describe the delivery of cassettes expressing hairpin RNA targeting green fluorescent protein (GFP) using Moloney leukemia virus-based and lentivirus-based retroviral vectors. Both transformed cell lines and primary dendritic cells, normally refractory to transfection-based gene transfer, demonstrated stable silencing of targeted genes, including the tumor suppressor gene TP53 in normal human fibroblasts. This report demonstrates that both Moloney leukemia virus and lentivirus vector-mediated expression of RNAi can achieve effective, stable gene silencing in diverse biological systems and will assist in elucidating gene functions in numerous cell types including primary cells.
Keywords: Retrovirus, hairpin RNA, siRNA, knockdown, dendritic cells
INTRODUCTION
Genetic analysis through targeted gene deletion in mammals has been limited principally to murine models. The discovery that the RNA interference (RNAi) pathway is active in mammals (Elbashir et al. 2001) raised the possibility that reverse genetic approaches using RNAi could be applied to mammalian systems. Like targeted gene deletion, RNAi-based silencing permits analysis of gene function in primary cells (McCaffrey et al. 2002; Novina et al. 2002). Compared to knockout genetics, RNAi-based silencing is rapid, cost effective, and can be easily adapted to study homologous gene function in a wide variety of organisms.
RNAi depends upon the formation of double-strand RNA (dsRNA) whose antisense strand is complementary to the transcript of a targeted gene. Two distinct steps are involved (for review see Hutvagner and Zamore 2002; McManus and Sharp 2002). In the first, the enzyme Dicer cleaves long dsRNA into short interfering RNA (siRNA) molecules of 21–23 bp in length. In the second, a multicomponent RNA-induced silencing complex (RISC) uses siRNA to guide the sequence-specific cleavage of the RNA transcripts of the target gene. Gene silencing by this mechanism has been achieved by transfection of chemically synthesized siRNA into host cells, by-passing the “dicing” step, to silence (“knockdown”) gene expression (McManus and Sharp 2002).
Several recently developed (Brummelkamp et al. 2002a; McCaffrey et al. 2002; McManus et al. 2002; Paddison et al. 2002; Paul et al. 2002; Sui et al. 2002; Xia et al. 2002; Yu et al. 2002) vector-based strategies that contain stem-loop constructs encoding hairpin RNAs lead to the intracellular generation of siRNA-like species. Dicer cleavage of hairpin RNA can generate small dsRNA that silence expression of genes whose transcripts are complementary to one of the two strands of the hairpin RNA (for review, see Paddison and Hannon 2002). However, most vector-based hairpin expression systems have demonstrated only transient knockdown of gene expression. More recently, knockdown of gene expression has been achieved using retroviral vector constructs that express hairpin RNAs within vector-infected cells and produce long-term knockdown within cultured cell lines (Barton et al. 2002;Brummelkamp et al. 2002b; Devroe and Silver 2002; Paddison and Hannon 2002).
To facilitate stable, long-term knockdown in populations of cells refractory to transfection-based gene transfer techniques, we designed a retroviral vector that permits delivery of stem-loop cassettes. We report the development of a versatile system of retrovirus-based vectors that make it possible to achieve durable, high efficiency siRNA-dependent gene silencing in a wide variety of cells.
RESULTS
Stable gene silencing in HeLa-GFP cells
Several studies have demonstrated that the cellular machinery required for RNAi exists in mammalian cells. To study gene function in primary mammalian cells in vitro using RNAi-based gene silencing technology, we designed a vector system (Fig. 1A ▶) in which the RNA polymerase III (pol-III) promoter of the U6 small nuclear RNA gene (U6) was used to drive high levels of expression of a small hairpin RNA. An inverted repeat sequence (a stem-loop, sl) with stem sequences complementary to the mRNA specifying green fluorescent protein (GFP) was cloned 3′ of the U6 promoter and a five thymidine repeat sequence was added to serve as a strong pol-III termination signal (Gunnery et al. 1999), which results principally in hairpin RNA species containing four uridines. However, it is unclear from our analysis at which uridine transcription terminates. Because stable gene silencing is affected by retrovirus-delivered hairpin RNA, the vector system reported here is called Retrohair. Assuming that four uridines are incorporated into the transcript from the five thymidine pol-III transcription termination signal, Retrohair encodes a 52-nt hairpin RNA (Fig. 1B ▶).
FIGURE 1.
Sequence-specific knockdown of GFP expression by retrovirus-delivered GFP hairpin RNA expression. (A) Schematic presentation of Retrohair-slGFP, a replication incompetent retrovirus that contains a pol-III promoter (U6), a stem-loop cassette followed by a 5T pol-III termination signal, cloned 3′ to a truncated gag lacking a translation start sequence (ATG). Retrohair can be selected by addition of puromycin. (B) The stem-loop encoded hairpin RNA has sequence identity to a 21-nt region of GFP mRNA. (C) FACs analysis of GFP expression in HeLa-GFP infected with (1 and 3) empty vector controls, (2) Retrohair-slGFP1, (4) Retrohair-slGFP2, or (5) Retrohair-slGFP2mut. Overlay histograms in panels 2, 4, and 5 demonstrate the level of knockdown in cells infected with Retrohair-containing stem-loop cassettes (red) compared to cells infected with empty vector controls (green). (D) Northern blot analysis of GFP expression in HeLa-GFP cells described in C using a GFP-specific probe. Northern blot with a β-actin probe served as a loading control.
To ensure rapid reductions in protein levels in response to decreases in mRNA levels, we used an enhanced GFP protein with the PEST domain at the carboxy-terminus, resulting in a fusion protein with a half-life of approximately 1 h. We developed a HeLa cell line that stably expresses this fusion protein (HeLa-GFP) to provide a sensitive, rapidly responding reporter system to examine gene silencing and its kinetics.
To examine the efficiency of retrovirus-delivered RNAi-based knockdown, HeLa-GFP were infected with virus expressing either wild-type or point mutant hairpin RNA sequences directed against GFP (Fig. 1C ▶). Fluorescence-activated cell sorting (FACS) analysis of GFP expression in HeLa-GFP cells infected with wild-type Retrohair-slGFP1 demonstrated a 10-fold reduction in mean fluorescence intensity (MFI: 691 → 76) relative to empty vector controls (containing the U6 promoter but lacking slGFP1; Fig. 1C ▶). In contrast, a construct containing a point mutation in the stem sequence (Retrohair-slGFP2mut) resulted in a twofold reduction in gene expression (MFI: 620 → 349). Northern blot analysis demonstrated decreased GFP mRNA levels only in the samples infected with wild-type viruses but not in mutant or empty vector controls (Fig. 1D ▶). Taken together, these data indicate that the hairpin RNA-based knockdown was specific and that silencing correlated with decreased GFP mRNA levels. Direct comparison demonstrated that Retrohair-slGFP1 was more efficient at silencing than Retrohair-slGFP2 (MFI = 76 vs. 158). Therefore, we used the Retrohair-slGFP1 vector for subsequent studies.
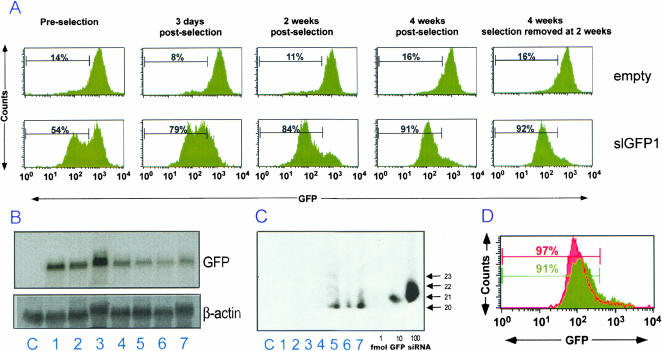
To determine the stability of the retrovirus-delivered RNAi, we infected HeLa-GFP with the empty vector control or with Retrohair-slGFP1. After infection, cells were selected by the addition of puromycin and GFP expression was analyzed (Fig. 2 ▶). GFP expression was not reduced in cells infected with the empty vector control, but was markedly decreased in cells infected with Retrohair-slGFP1 (Fig. 2A ▶). Following Retrohair-slGFP1 infection, the population exhibited a bimodal distribution and the overall MFI was 342 compared to the average MFI of the control population (859). Following 2 wk of selection, the population became more homogenous and the MFI decreased further to 89. Similar results were obtained after selection of cells infected with virus containing Retrohair-slGFP2 (data not shown).
FIGURE 2.
Time course of selection of GFP knockdown in RNAi-stable HeLa-GFP cells. (A) FACS analysis of GFP expression in HeLa-GFP cells infected with Retrohair-lacking a stem-loop (empty, top panels) or with Retrohair-slGFP1 (-slGFP1, bottom panels). GFP expression was measured in HeLa-GFP either preselection, with 3 d, 2 wk, or 4 wk of puromycin selection or in HeLa-GFP after 2 wk of growth with puromycin selection followed by 2 wk of growth without puromycin selection. (B) Northern blot analysis of GFP expression in HeLa (lane C), in HeLa-GFP infected with Retrohair at (lane 1) 3 d (lane 2) 2 wk, (lane 3) 4 wk, or infected with Retrohair-slGFP1 at (lane 4) 3 d (lane 5) 2 wk, (lane 6) 4 wk, or at (lane 7) 2 wk with selection and 2 wk without puromycin selection. β-Actin expression served as a loading control. (C) Modified Northern blot analysis of the samples described in B using an RNA probe (sense strand) with complementarity to the antisense strand of the GFP hairpin RNA. Samples were normalized by total RNA content. Titration of GFP siRNA was used as a size marker and for quantitation by densitometry of small RNAs. (D) FACS analysis of GFP expression in HeLa-GFP infected with Retrohair-slGFP1 4 wk after selection on puromycin (green) or infected with Retrohair-slGFP1 4 wk after selection on puromycin and then superinfected with Retrohair-slGFP2 (red).
To determine whether continuous selective pressure was necessary to maintain gene knockdown, puromycin was removed from the cells 2 wk after selection had been initiated. Analysis of GFP expression demonstrated that the MFIs were identical to the cells that were continuously selected (Fig. 2A ▶), indicating that constant selective pressure was not required to maintain hairpin RNA expression and silencing of GFP expression for 4 wk. Northern blot analysis of these cells revealed that loss of GFP expression correlated with loss of GFP mRNA (Fig. 2B ▶). Although constant selective pressure may not be needed to maintain silencing of genes with a neutral or growth-promoting phenotype, it is possible that continuous selective pressure may be required for stable silencing of genes whose loss have a negative effect on cell growth or are essential for cell survival.
RNAi is mediated by short, 21–23-bp dsRNA. Processing of the GFP1 hairpin RNA into GFP1 siRNA was demonstrated by the presence of multiple small RNA products in modified Northern blot analysis (Fig. 2C ▶). The major RNA cleavage product comigrated with a 20-nt species and minor products comigrated with 21-, 22-, and 23-nt species. In this analysis, 11.5 μg total RNA corresponding to approximately 7.6 × 105 cell equivalents was used (Fig. 2C ▶). If densitometry signals from each cleavage product are added, approximately 8 fmoles of GFP siRNA were present in cells following 2 wk of selection. As a conservative estimate of silencing (assuming that the approximately 85% of the HeLa-GFP that were actively silencing GFP expression were responsible for hairpin RNA expression), an estimated 8.6 × 10−21 moles siRNA per cell or approximately 5 × 103 molecules of siRNA derived from hairpin RNA precursors were associated with each of the actively silencing HeLa-GFP. It is not known which of the cleavage product(s) is the active species.
We noted that a small percentage of the HeLa-GFP remained GFP-positive following several weeks of selection. Superinfection of these cells with Retrohair-slGFP2 resulted in further reduction of GFP expression in already actively silencing HeLa-GFP previously infected with RetrohairslGFP1 (Fig. 2D ▶). These results suggest that cells containing high levels of GFP are not inherently resistant to RNAi and that some GFP-positive cells were infected (because they were puromycin resistant) but expressed insufficient siRNA for GFP silencing. Taken together, these studies indicate that (1) infection of HeLa-GFP with Retrohair-slGFP1 reduced GFP gene expression, (2) selection enriched for HeLa-GFP with higher expression levels of the hairpin RNA, and (3) continuous expression of the GFP hairpin RNAs and maintenance of the knockdown phenotype did not require continuous selective pressure.
Gene silencing in primary dendritic cells
To extend these studies to primary mammalian cells that are refractory to gene transfer (Esslinger et al. 2002), we turned to dendritic cells (DC). DC are the most potent antigen-presenting cells (APC) known. Capable of activating naïve B and T cells, they play an important role in initiating host immune responses (Steinman and Pope 2002). DC also play a critical role in maintenance of tolerance (Steinman and Nussenzweig 2002). Thus, DC are under intensive study for their potential therapeutic uses. To determine if these cells are susceptible to retrovirus-delivered siRNA-directed gene silencing, we examined bone marrow-derived (BM) DC from a GFP transgenic mouse infected ex vivo with Retrohair-slGFP1 vector or an empty vector. GFP fluorescence microscopy indicated that GFP expression was lower in cells infected with the slGFP1-containing vector than in cells infected with the empty vector control (Fig. 3A ▶). Consistent with this observation, 33% of DC-GFP infected with Retrohair-slGFP1 were observed in the GFP-negative gate compared to 11% of DC-GFP infected with the empty vector control (Fig. 3B ▶, panels 1 and 2 ). The DC-GFP population demonstrates a wide range of GFP expression. Furthermore, the MFI of the entire population of Retrohair-slGFP1-infected DC-GFP was about 80% lower than that of the control vector-infected DC-GFP population (Fig. 3B ▶, panel 3 ). The overall reduction in MFI suggested that the majority of DC-GFP had been infected by the Retrohair-slGFP1 virus. It may well be that the cells with the greatest reduction in fluorescence contained the highest level of hairpin RNA expression. High levels of hairpin RNA expression may be due to the favorable sites of proviral integration and/or to the number of proviral integration sites per cell.
FIGURE 3.
Knockdown of GFP expression in DC-GFP. (A) Photomicroscopy of DC-GFP visualized by white light or by fluorescent light after infection with Retrohair or with Retrohair-slGFP1(slGFP1). (B) FACS of GFP expression in DC-GFP infected with the Retrohair or Retrohair-slGFP1 retrovirus. A bar graph depicts the percent decrease in GFP expression as measured by MFI. Error bars represent the average (±SD) of six experiments. (C) Modified Northern blot analysis of Retrohair and Retrohair-slGFP1-infected DC-GFP using an RNA probe (sense strand) with complementarity to the antisense strand of the GFP hairpin RNA. Samples were normalized by total RNA content. Titration of GFP siRNA was used as size marker and for quantitation by densitometry of small RNAs.
Primary cells have been shown to possess the machinery to silence gene expression using siRNA (McCaffrey et al. 2002; Novina et al. 2002). Our data show that the hairpin is processed indicating that primary cells contain the RNAi machinery required to generate siRNA from the hairpin RNA (Fig. 3C ▶). Similar to the HeLa-GFP experiment, several small processed RNA species with relative mobility of oligonucleotides 20–23 bp in length were observed in DC-GFP by modified Northern blot analysis (Fig. 3C ▶). In this experiment, 1.65 μg total RNA derived from 2 × 106 infected DC was analyzed. Densitometry signals from each of the Retrohair-slGFP1 cleavage products were added and amounted to 5 fmoles of GFP-siRNA. As a conservative estimate of silencing (assuming that 33% of the DC actively silencing GFP expression were responsible for hairpin RNA expression), an estimated 7.5 × 10−21 moles siRNA per cell or approximately 4.5 × 103 molecules of siRNA were derived from hairpin RNA precursors in each actively silencing DC. This value is in agreement with the estimated number of siRNA molecules in GFP-silencing HeLa-GFP (see above).
Lentiviral delivery of stable gene silencing
Moloney-based vector systems permit gene transduction of proliferating cells but are incapable of productively infecting nondividing cells, limiting its usefulness in primary cells both in vitro and in vivo. Therefore, we constructed a lentiviral vector containing the U6 driven stem-loop directed against GFP. Because stable gene silencing is affected by lentivirus-delivered hairpin RNA, the vector system is called Lentihair (Fig. 4A ▶). HeLa-GFP were infected with Lentihair-slGFP1 and GFP expression was analyzed by FACS 3 d later (Fig. 4B ▶). GFP expression was reduced more than 10-fold relative to nonspecific stem-loop lentiviral control infections.
FIGURE 4.
Knockdown of GFP expression by lentivirus-delivered RNAi. (A) Schematic presentation of the lentiviral construct. Lentihair is a replication incompetent, self-inactivating retrovirus generated by cloning the U6 promoter and stem-loop cassette targeted to luciferase (Lentihair-slLUC) or GFP (Lentihair-slGFP1) 5′ of the cPPT. A puromycin resistance gene was cloned 3′ of the human hPGK promoter. (B) FACS analysis of GFP expression in HeLa-GFP cells (1) uninfected or infected with (2) Retrohair, (3) Retrohair-slGFP1, (4) Lentihair-slLUC or (5) Lentihair-slGFP. (C) FACS analysis of GFP expression in (1) uninfected, (2) Lentihair-slLUC, or (3) Lentihair-slGFP infected DC-GFP.
Primary DC-GFP were also infected with LentihairslGFP1 and resulted in a greater knockdown of GFP than was observed with Retrohair-slGFP1, 50% knockdown versus 33% (Fig. 4C ▶). GFP expression in DC-GFP was reduced (MFI: 1174 → 87) in cells infected with Lentihair-slGFP1 compared to uninfected controls. We also noted that there was a modest knockdown of GFP expression (>3-fold) in DC-GFP infected with a lentivirus that targeted luciferase (Lentihair-slLuc), suggesting that there is some nonspecific knockdown in these cells. Interestingly, the lentivirus-delivered RNAi was more efficient in both HeLa-GFP and DC-GFP compared to Moloney leukemia virus-delivered RNAi. The Moloney-based vector contains a transcriptionally active LTR whereas the lentivirus is transcriptionally inactive due to mutations in the U3 region of the LTR. Therefore, differences between Moloney- and lentivirus-delivered gene silencing may be due to different vector configurations.
DISCUSSION
Utilizing several biological systems, we demonstrate retroviral delivery and intracellular processing of hairpin RNA in a variety of cell types, including primary mammalian cells. We have extended the analysis of retrovirus-delivered stable gene silencing to normal human cells, and the vector system reported was used to silence medically relevant genes such as p53 (data can be accessed at www2.dfci.harvard.edu/hahnlab). This data demonstrates that a complete knockout is not required to produce a biological phenomenon. By understanding and using retrovirus-delivered knockdown in primary cells, the function of human genes in biological and disease processes may be determined.
Although retroviral delivery of hairpin RNA has been shown to result in the knockdown of targeted proteins in human primary fibroblasts (Barton and Medzhitov 2002), we have demonstrated hairpin RNA processing into siRNA in bone marrow-derived primary DC. The level of RNA hairpin expression and its processing into siRNAs in a target cell will likely determine the extent of gene knockdown. It is possible that different cell types vary in their ability to process long dsRNA and hairpin RNA into smaller cleavage products. We found that HeLa-GFP and DC-GFP contained similar levels of processed RNAs. The slightly higher levels of small RNAs found in HeLa-GFP may be due to higher levels of hairpin RNA processing or, alternatively, to the amount of hairpin RNA expressed from the integrated provirus(es). In the latter instance, superinfection of cells already infected with a virus encoding one stem-loop with another virus having either the same sequence or a sequence directed against a different region of the same gene may yield greater knockdown of that gene. Indeed, we observed that superinfection of HeLa-GFP resulted in greater knockdown, suggesting that gene expression can be further silenced by retroviral vectors targeting different sequences in the same mRNA.
DC are critical for initiating primary immune responses (Stark et al. 1998; Jung et al. 2002). To initiate a primary immune response, the DC undergo a series of maturation steps ultimately producing a fully differentiated immunocompetent cell poised to function as an activated APC. Efforts to genetically manipulate these cells (without activating them) by conventional transfection methods have proven ineffectual (Gasperi et al. 1999; Esslinger et al. 2002). Adeno-vectors are efficient gene transducers (Song et al. 1997; Klein et al. 2000) but they activate immune responses (Marshall 1999). Moreover, preexisting immunity to adenoviral vectors (Yang et al. 1994) potentially limits their use in vivo. Previous studies have indicated that lentiviral infection of DC does not activate these cells (Esslinger et al. 2002) raising the possibility that stable gene silencing by retroviral vector delivery may be able to modulate host immune responses. Lentihair-slGFP1 infection of HeLa-GFP and DC-GFP indicates that this vector configuration is highly effective in gene silencing.
The principal advantage of the lentiviral system is that it will allow gene silencing in nondividing cells and will therefore expand the usefulness of the RNAi-based gene silencing system. It is interesting to note that we observed some nonspecific knockdown in DC-GFP when we used Lentihair-slLUC, an effect not observed in HeLa-GFP. The nonspecific knockdown in DC-GFP may be due to activation of the interferon pathway in these cells. Recognition of dsRNA by DC result in their maturation and the DC-GFP may have demonstrated some degree of maturation following lentivirus infection (Le Bon and Tough 2002). Preliminary data suggest that limiting lentiviral infection of DC to one infection rather than two results in fewer cells exhibiting knockdown (32% vs. 50%). In addition, nonspecific knockdown with the Lentihair-slLUC was reduced to twofold. Therefore, changing the kinetics or dose of infection may limit the nonspecific knockdown. Our data indicate that the system for retrovirus delivery of RNAi-based gene silencing can be adapted to use in lentiviral systems, thereby opening opportunities to analyze gene function in diverse cells, dividing or not, under physiological conditions.
RNAi provides plants with effective antiviral defenses (for review, see Waterhouse et al. 2001). A natural role for mammalian RNAi in antiviral defenses has not been shown. It is possible that RNAi-like antiviral defenses may assist the host immune response against viral infection, including the retroviral vector used to deliver the stem-loop-containing cassette. However, our data indicate that productive infection can occur and that the retroviral RNA can be produced and packaged, and is unlikely to be targeted for degradation by an RNAi-like mechanism despite the presence of complementary sequences in the provirus. Thus, our system demonstrates a proof of principle that retroviral delivery of RNAi can be used for the elucidation of gene function in a wide variety of cell types, including differentiated, nondividing cells.
MATERIALS AND METHODS
Vector design
The U6 promoter (a gift from Nouria Hernandez, Cold Spring Harbor Laboratoy, Cold Spring Harbor, New York) was cloned into pBABEpuro (pBp) with the addition of an ApaI site at the transcription start site (Morgenstern and Land 1990). The pBpU6 was digested with ApaI, blunted, and digested with EcoRI. The following oligomers were used: GFP1 forward 5′P-GGCTACGTC CAGGAGCGCACCCTCGAGGGTGCGCTCCTGGACGTAGCCT TTTTG-3′ and GFP1 reverse 5′P-AATTCAAAAAGGCTACGTC CAGGGCGACCCTCGAGGGTGCGCTCCTGGACGGAGCC-3′ and GFP2 forward 5′P-CGCAAGCTGACCCTGAGTTCATTCAA GAGATGAACTTCAGGGTCAGCTTGCTTTTTG-3′ and GFP2 back 5′P-AATTCAAAAAGCAAGCTGACCCTGAAGTTCATCT CTTGAATGAACTTCAGGGTCAGCTTGCGGGCC-3′. In the process of screening GFP2 clones, one contained a point mutation (mutation is underscored: 5′-CGCAAGCTGACCCTGAAGTT TATTCAAGAGATGAACTTCAGGGTCAGCTTGCTTTTTG-3′ ).
The Lentihair-slLUC lentiviral construct contains the U6 promoter and a stem-loop directed against the luciferase gene (LUC; Paddison et al. 2002) or GFP cloned 5′ of the central polypurine tract (cPPT) from pRRL-cPPT-hPGKEsin (Dull et al. 1998). A cPPT was inserted 5′ of the human phosphoglycerate kinase promoter in the pRRL-hPGKsin vector. The puromycin-resistant gene was cloned 3′ of the human phosphoglycerate kinase (hPGK) promoter.
Cell culture
HeLa cells, grown in Dulbecco Modified Eagle medium (DME) plus 10% heat-inactivated fetal calf serum (FCS) containing penicillin and streptomycin (pen/strep), were transfected with pd1EGFP-N1 (Clontech), selected with 500 μg/mL G418, and single-cell cloned.
Isolation, culture, and infection of BM-DC
Isolation, culture, and infection of BM-DC has been described (Inaba et al. 1992). Bone marrow was flushed from the femur and tibia of C57BL/6-EGFP mice (Dr. Okabe, Osaka University; Okabe et al. 1997), red blood cells were lysed, and the remaining cells were plated in RPMI1640 (plus 10% FCS, 2 mM L-glutamine, 10 mM HEPES, 50 μM 2-mercaptoethanol and pen/strep) at 1 × 106 cells/mL/well. The medium was supplemented (1:25) with culture supernatant from J5 cells expressing GM-CSF.
Viral production and infections
The pBp-amphotrophic viruses were produced by cotransfection of 293T cells with the pBp constructs and pCL10A1. The pBp-VSV-G viruses were produced by cotransfecting pBp constructs, pUMVC3, and pCMV-VSV-G (at a 9:1 ratio). Lentihair-slLUC and Lentihair-slGFP1 were produced by cotransfection with pHCMVG (Burns et al. 1993) and pCMVΔR8.20vpr (An et al. 1999). Transfections were carried out using Fugene 6 (Roche). Virus was harvested at 48 and 72 h posttransfection and infections were carried out in the presence of 10 μg/mL of polybrene and 10 mM HEPES. Following transduction, cells were selected with 1 μg/mL puromycin. DC were spin infected on days 1 and 2 at 2.5K for 1.5 h. Virus-containing supernatant was removed after 2 h.
Flow cytometry and microscopy
Flow cytometry was carried out using FACScalibur and Cellquest software. For imaging, DC were mounted onto microscope slides and images were collected on an Axioplan 2 microscope (Zeiss) using Axiovision Viewer 3 software (Zeiss).
Northern blot
Total RNA was prepared using the RNeasy mini kit. Northern blot was performed using the Northern Max Northern blotting kit. The GFP probe was prepared by PCR amplification of a 497-nt fragment from pd1EGFP-N1 (5′-CCTACGGCAAGCTGACCCTGA AGTTCA-3′ [forward] and 5′-GGACTGGGTGCTCAGGTAGTG GTTGT-3′ [reverse]) that was purified using the Qiaquick gel extraction kit (Qiagen). The purified GFP fragment and the β-actin control (DECA template mouse β-actin control DNA) were labeled by random prime labeling using the Decaprime II DNA labeling kit (Ambion).
Modified Northern blot
Total RNA was prepared by extraction with Trizol (Molecular Research Center, Inc.) as recommended by the manufacturer. The resulting homogenate was centrifuged for 15 min at 20,000g (4°C), precipitated, and reextracted as above. The DC went through one round of extraction. The precipitated RNA pellets were washed in 70% ethanol, air-dried, and resuspended in nuclease-free H2O. A total of 11.5 μg HeLa-GFP total RNA or 1.65 μg DC total RNA were run on a 15% sequencing gel, electroblotted to Hybond N membrane (Amersham), UV cross-linked and prehybridized overnight at 37°C in 5× SSPE, 2× Denhardt’s solution, and 0.1× SDS containing 0.1 mg/mL denatured salmon sperm DNA and 0.1 mg/mL denatured yeast tRNA. The sense strand (100 pmoles) of GFP1 siRNA (5′P-GGCUACGUCCAGGAGCGCACC-3′) was end labeled with polynucleotide kinase and [γ32P]-ATP (150 μCi), purified on a G-25 MicroSpin Column (Amersham), heated for 5 min to 65°C, added to the prehybridization buffer, and hybridized overnight. Blots were washed at room temperature 2 × 5 min in 2× SSC and 0.1% SDS, 3 × 10 min in 0.1× SSC and 0.1% SDS and exposed to film.
Acknowledgments
We thank Helen Cargill for assistance in preparation of the figures, Dr. Herman Eisen for the critical reading of the manuscript, and members of the WI-MIT-Dana-Farber RNAi Consortium for encouragement. We thank Dr. Hernandez for the U6 promoter. This work was supported by NCI F32 CA93033-01 (S.A.S.), NCI R01CA78461-04, Temin Award K01CA94223 (W.C.H.), NIH F32 AI10523 (C.D.N.), NIH MERIT grant R37-GM34277, R01-AI32486, NCI grant P01-CA42063, P30-CA14051 (P.A.S.), and NCI Cancer Center Support (Core) grant P30-CA14051 (P.A.S.) and grants from Merck/MIT and Co, Inc. (R.A.W.), a Doris Duke Charitable Foundation Clinical Scientist Development Award (W.C.H.), and a Kimmel Scholar Award (W.C.H.), and a Stressgen postdoctoral fellowship (D.P.). The work was supported by the Fidelity Investments Foundation (D.M.S.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2192803.
REFERENCES
- An, D.S., Morizono, K., Li, Q.X., Mao, S.H., Lu, S., and Chen, I.S. 1999. An inducible human immunodeficiency virus type 1 (HIV-1) vector which effectively suppresses HIV-1 replication. J. Virol. 73:7671–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton, G.M. and Medzhitov, R. 2002. Retroviral delivery of small interfering RNA into primary cells. Proc. Natl. Acad. Sci. 99: 14943–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002a. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- ———. 2002b. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. Published online. [DOI] [PubMed]
- Burns, J.C., Friedmann, T., Driever, W., Burrascano, M., and Yee, J.K. 1993. Vesicular stomatitis virus C glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. 90: 8033–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devroe, E. and Silver, P.A. 2002. Retrovirus-delivered siRNA. BMC Biotechnol. 2: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull, T., Zufferey, R., Kelly, M., Mandel, R.J., Nguyen, M., Trono, D., and Naldini, L. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72: 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Esslinger, C., Romero, P., and MacDonald, H.R. 2002. Efficient transduction of dendritic cells and induction of a T-cell response by third-generation lentivectors. Hum. Gene Ther. 13: 1091–1100. [DOI] [PubMed] [Google Scholar]
- Gasperi, C., Rescigno, M., Granucci, F., Citterio, S., Matyszak, M.K., Sciurpi, M.T., Lanfrancone, L., and Ricciardi-Gastagnoli, P. 1999. Retroviral gene transfer, rapid selection, and maintenance of the immature phenotype in mouse dendritic cells. J. Leukoc. Biol. 66: 263–267. [DOI] [PubMed] [Google Scholar]
- Gunnery, S., Ma, Y., and Mathers, M.B. 1999. Termination sequence requirements vary among genes transcribed by RNA polymerase III. J. Mol. Biol. 286: 745–757. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G. and Zamore, P.D. 2002. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 12: 225–232. [DOI] [PubMed] [Google Scholar]
- Inaba, K., Inaba, M., Romani, N., Aya, H., Deguchi, M., Ikehara, S., Muramatsu, S., and Steinman, R.M. 1992. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 176: 1693–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, S., Unutmaz, D., Wong, P., Sano, G., De los Santos, K., Sparwasser, T., Wu, S., Vuthoori, S., Ko, K., Zavala, F., et al. 2002. In vivo depletion of CD11c(+) dendritic cells abrogates priming of CD8(+) T cells by exogenous cell-associated antigens. Immunity 17: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, C., Bueler, H., and Mulligan, R.C. 2000. Comparative analysis of genetically modified dendritic cells and tumor cells as therapeutic cancer vaccines. J. Exp. Med. 191: 1699–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bon, A. and Tough, D.F. 2002. Links between innate and adaptive immunity via type I interferon. Curr. Opin. Immunol. 14: 432–436. [DOI] [PubMed] [Google Scholar]
- Marshall, E. 1999. Gene therapy death prompts review of adenovirus vector. Science 286: 2244–2245. [DOI] [PubMed] [Google Scholar]
- McCaffrey, A.P., Meuse, L., Pham, T.T., Conklin, D.S., Hannon, G.J., and Kay, M.A. 2002. RNA interference in adult mice. Nature 418: 38–39. [DOI] [PubMed] [Google Scholar]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3: 737–747. [DOI] [PubMed] [Google Scholar]
- McManus, M.T., Petersen, C.P., Haines, B.B., Chen, J., and Sharp, P.A. 2002. Gene silencing using micro-RNA designed hairpins. RNA 8: 842–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern, J.P. and Land, H. 1990. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18: 3587–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novina, C.D., Murray, M.F., Dykxhoorn, D.M., Beresford, P.J., Riess, J., Lee, S.K., Collman, R.G., Lieberman, J., Shankar, P., and Sharp, P.A. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8: 681–686. [DOI] [PubMed] [Google Scholar]
- Okabe, M., Ikawa, M., Kominami, K., Nakanishi, T., and Nishimune, Y. 1997. “Green mice” as a source of ubiquitous green cells. FEBS Lett. 407: 313–319. [DOI] [PubMed] [Google Scholar]
- Paddison, P.J. and Hannon, G.J. 2002. RNA interference: The new somatic cell genetics? Cancer Cell 2: 17–23. [DOI] [PubMed] [Google Scholar]
- Paddison, P.J., Caudy, A.A., Bernstein, E., Hannon, G.J., and Conklin, D.S. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 16: 948–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, C.P., Good, P.D., Winer, I., and Engelke, D.R. 2002. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 20: 505–508. [DOI] [PubMed] [Google Scholar]
- Song, W., Kong, H.L., Carpenter, H., Torii, H., Granstein, R., Rafii, S., Moore, M.A., and Crystal, R.G. 1997. Dendritic cells genetically modified with an adenovirus vector encoding the cDNA for a model antigen induce protective and therapeutic antitumor immunity. J. Exp. Med. 186: 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, G.R., Kerr, I.M., Williams, B.R., Silverman, R.H., and Schreiber, R.D. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67: 227–264. [DOI] [PubMed] [Google Scholar]
- Steinman, R.M. and Nussenzweig, M.C. 2002. Avoiding horror autotoxicus: The importance of dendritic cells in peripheral T cell tolerance. Proc. Natl. Acad. Sci. 99: 351–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman, R.M. and Pope, M. 2002. Exploiting dendritic cells to improve vaccine efficacy. J. Clin. Invest. 109: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui, G., Soohoo, C., Affar, el B., Gay, F., Shi, Y., and Forrester, W.C. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. 99: 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. 2001. Gene silencing as an adaptive defence against viruses. Nature 411: 834–842. [DOI] [PubMed] [Google Scholar]
- Xia, H., Mao, Q., Paulson, H.L., and Davidson, B.L. 2002. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 20: 1006–1010. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Nunes, F.A., Berencsi, K., Furth, E.E., Gonczol, E., and Wilson, J.M. 1994. Cellular immunity to viral antigens limits E1-deleted adenoviruses for gene therapy. Proc. Natl. Acad. Sci. 91: 4407–4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.Y., DeRuiter, S.L., and Turner, D.L. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. 99: 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]