Abstract
Nuclear factor 110 (NF110) belongs to the nuclear factor 90 (NF90) family of double-stranded RNA (dsRNA) binding proteins that regulate gene expression at the transcriptional level in vertebrates. The proteins are identical at their N terminus, which functions as a negative regulatory region, but have distinct C termini as a result of alternate splicing. Maximal transcriptional activity of NF110 requires its C-terminal domain and a central domain that contains a nuclear localization signal and two dsRNA-binding motifs (dsRBMs). We find that dsRNA binding is reduced by RGG and GQSY motifs present in the C-terminal region. To directly evaluate the role of RNA binding in transactivation, we conducted site-directed mutagenesis to substitute conserved residues in one or both of the dsRBMs. The mutations reduced the ability of NF110 to stimulate gene expression to an extent that paralleled the mutants’ reduced ability to bind dsRNA. Full activity was restored when the dsRBM-containing region of NF110 was replaced with the RNA-binding region of the protein kinase PKR. Finally, NF110-mediated transactivation was inhibited by cotransfection of a plasmid encoding an artificial highly structured RNA. These data suggest that NF110 and its homologs are regulated by cis-acting domains present in some of the protein isoforms, and via interactions with RNAs that bind to their dsRBMs. We propose a model in which structured RNAs regulate gene expression by modulating transcription through interactions with members of the NF90 protein family.
Keywords: NF90, ILF3, dsRNA binding protein, dsRNA, gene expression
INTRODUCTION
The innate immune system is a first line of defense against microbial invasion, relying on the recognition of foreign molecular cues characteristic of pathogens. One such cue is double-stranded RNA (dsRNA), which can be generated within cells by viral replicative processes. The presence of dsRNA in the cytoplasm of cells is sensed by cellular proteins including, as a well-studied example, the dsRNA-activated protein kinase PKR. Activation of this interferon-induced kinase results in the phosphorylation of eukaryotic initiation factor 2 (eIF2) and blocks protein synthesis at the level of initiation with consequent inhibition of viral replication (Kaufman 2000). Although PKR is a critical component of the cellular antiviral response, PKR-knockout mice remained surprisingly sensitive to the effects of dsRNA, and their interferon response to viral infection was only mildly reduced (Yang et al. 1995; Abraham et al. 1999; Zhou et al. 1999). These observations suggest that other mechanisms, possibly involving other dsRNA-binding proteins, exist and play a role in the antiviral defense system.
The nuclear factor 90 (NF90) family consists of a group of closely related proteins generated through alternate splicing. Members of this family in humans include NF90 itself (also known as DRBP76 and NFAR1), a longer isoform termed NF110 (interleukin enhancer binding factor 3, ILF3, or NFAR2), and translational control protein 80 (TCP80). These proteins are identical in their N terminal and central regions but diverge at their C termini (Xu and Grabowski 1999; Duchange et al. 2000; Saunders et al. 2001a). The central region contains a functional nuclear localization signal and an RNA-binding domain with two dsRNA binding motifs (dsRBMs; Fig. 1A ▶). An additional alternate splicing event inserts four amino acids (NVKQ) into the spacer region between the two dsRBMs, separating NF90 and NF110 into two further isoforms: NF90b and NF110b bear the NVKQ insert, which is absent from NF90a and NF110a (Duchange et al. 2000; T.W. Reichman, A.M. Parrott, I. Fierro-Monti, P.N. Kao, C.G. Lee, H. Li, and M.B. Mathews, in prep.). The original NF90 cDNA (Kao et al. 1994), here referred to as NF90c, appears to contain a two base-pair insertion that results in a frameshift and gives rise to a protein with a shorter C terminus lacking both the RGG and GQSY domains. In contrast, NF90a and NF90b both have the RGG motif but not the GQSY region. It is not known whether NF90c is an artifact of cloning or if it is a bona fide, naturally occurring variant.
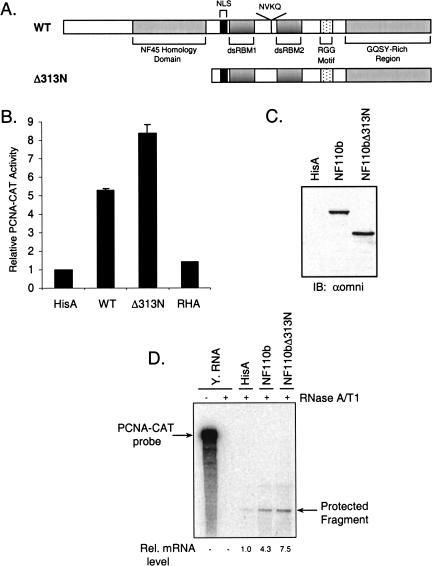
FIGURE 1.
The RNA-binding and C-terminal domains of NF110b are sufficient for activation of gene expression. (A) Schematic representation of wild-type (WT) NF110b and N-terminal deletion mutant. (B) Transient expression assays were conducted in HeLa cells with vectors expressing wild-type NF110b, the N-terminal deletion mutant, or RHA and PCNA-CAT reporter plasmid. HeLa cells were transiently transfected with 200 ng PCNA(−1265)-CAT reporter plasmid, 200 ng of the indicated expression plasmid or empty vector control (HisA), and 600 ng HisA. After 48 h, cells were harvested and assayed for CAT activity. Results were plotted as fold activation relative to the control (HisA), which was assigned the arbitrary value of 1. The error bars represent the standard deviation (n=3). (C) Western blot analysis of extract (40 μg) from transfected cells using the omni probe (αomni). (D) RNase protection assays were carried out for CAT transcripts in RNA from HeLa cells transiently expressing the PCNA-CAT reporter plasmid and the vectors indicated, or yeast RNA as a control (−, untreated; +, RNase-treated). Protected RNA fragments were quantified and their relative levels are shown.
The dsRBM is a modular nucleic-acid–binding motif, 65–70 amino acids long, which characteristically binds the A form of duplexed RNA with little affinity for dsDNA, ssDNA, and ssRNA (Fierro-Monti and Mathews 2000). In addition to authentic dsRNA, it binds to several single-stranded but highly structured RNA molecules such as the adenovirus VA RNAs. Exceptionally, NF90 and its homologs also bind to ssRNA (at high concentrations), DNA–RNA hybrids, and both ssDNA and dsDNA (Bass et al. 1994; Corthésy and Kao 1994; Orford et al. 1998; Satoh et al. 1999; Parker et al. 2001a). Most of the interactions between the dsRBM and dsRNA involve the phosphodiester backbone and 2‘-OH groups, accounting for the dsRBM’s general lack of RNA sequence specificity. However, sequence-dependent structures in RNAs might dictate which dsRNA-binding proteins they interact with and could be a source of specificity (Bycroft et al. 1995; Kharrat et al. 1995; Bevilacqua and Cech 1996; Ryter and Schultz 1998). The dsRBM can also mediate protein-protein interactions between dsRNA-binding proteins (Cosentino et al. 1995; Patel et al. 1995a; Parker et al. 2001a).
Several lines of evidence link the NF90 family of nuclear dsRNA-binding proteins to antiviral defense mechanisms. First, NF90 and its heteromeric partner, NF45, were isolated as a complex that binds to the antigen recognition response element 2 (ARRE-2) in the interleukin 2 (IL-2) promoter and participates in the upregulation of IL-2, a cytokine required for T-cell proliferation (Corthésy and Kao 1994). Second, NF90 and its relatives interact with, regulate, and act as substrates for PKR (Langland et al. 1999; Coolidge and Patton 2000; Parker et al. 2001a,b; Saunders et al. 2001b). Third, NF90 binds preferentially to adenovirus VA RNAII (Liao et al. 1998), an abundant RNA similar to VA RNAI, which is well characterized as a viral countermeasure to the PKR-mediated interferon-induced antiviral response (Mathews and Shenk 1991). Finally, human osteosarcoma cells overexpressing NF90 contain elevated levels of mRNAs for several interferon-regulated genes (Krasnoselskaya-Riz et al. 2002).
NF90 and its homologs selectively regulate gene expression in mammalian cells, both positively and negatively (Saunders et al. 2001b; Krasnoselskaya-Riz et al. 2002; Reichman et al. 2002). The dsRNA-binding region is essential for these regulatory actions but the role of RNA has not been directly established. Similarly, a role for RNA binding has been inferred for the Xenopus homologs of NF110 and NF90, CBTF122 and CBTF98. These proteins (also known as 4F.2 and 4F.1; Bass et al. 1994) bind to the CAATT box element present in the GATA-2 gene and presumably coordinate its expression (Orford et al. 1998; Brzostowski et al. 2000). The CBTF complex is sequestered in the cytoplasm of immature zygotes via the dsRBMs of CBTF122 and translocates to the nucleus upon digestion of cytoplasmic RNA prior to the midblastula transition or after ribonuclease injection (Brzostowski et al. 2000).
We show here, by mutagenesis and dsRNA expression studies, that RNA binding regulates the transcriptional activity of NF110b. The protein’s N terminus, which is homologous to NF45, reduces its ability to activate reporter gene expression, while its C terminus is required for maximal activity. The C-terminal region, unique to NF110, moderates dsRNA binding and appears to confer a degree of specificity upon the interaction. These data suggest a model in which protein interactions and structured cellular RNA(s) modulate the ability of NF110b to stimulate transcription.
RESULTS
Reporter gene activation by NF110b
NF90 proteins regulate gene expression driven by several promoters in mammalian cells, and both positive and negative effects have been observed (Corthésy and Kao 1994; Saunders et al. 2001b; Reichman et al. 2002). Among the five isoforms tested, NF110b was found to be the most potent activator (T.W. Reichman, A.M. Parrott, I. Fierro-Monti, P.N. Kao, C.G. Lee, H. Li, and M.B. Mathews, in prep.). Maximal stimulation of gene expression in transfected cells by NF90c required the nuclear localization signal (NLS) and two dsRBMs in its C-terminal region, whereas the protein’s N terminus tempered this activity (Reichman et al. 2002). Conversely, removal of the NLS region from NF90c by deleting the N terminus resulted in a loss of transcriptional activation by the protein (Reichman et al. 2002). Similarly, as shown in Figure 1B ▶, full-length NF110b protein (wild-type, WT) stimulated expression of the PCNA-CAT reporter by approximately fivefold and deletion of the N-terminal 312 amino acids (NF110bΔ313N; Fig. 1A ▶) gave rise to a more potent activator (approximately eightfold). Western blots with antibody directed against the epitope tag confirmed that the proteins were expressed at equivalent levels (Fig. 1C ▶). RNA helicase A (RHA), another nuclear dsRNA-binding protein with two dsRBMs and a GQSY-rich region, did not affect PCNA-CAT expression (Fig. 1B ▶), indicating that the effect of NF110b is specific.
RNA analysis was carried out to determine whether NF110b enhances reporter gene expression at the mRNA level. RNA was isolated from HeLa cells transfected with the PCNA-CAT construct and subjected to ribonuclease protection assay with antisense RNA complementary to CAT mRNA. Cotransfection with a plasmid encoding the NF110b protein resulted in a four- to fivefold increase in PCNA-CAT mRNA as compared to the empty vector (Fig. 1D ▶). Expression of the NF110b mutant NF110bΔ313N resulted in a seven- to eightfold increase in PCNA-CAT mRNA levels. We conclude that NF110b regulates gene expression by increasing the level of mRNA, presumably by altering the rate of gene transcription. This result is consistent with previous data, from RNase protection and nuclear run-on assays (Saunders et al. 2001b; Reichman et al. 2002) and in vitro transcription experiments (Corthésy and Kao 1994), which indicated a transcriptional role for NF90 in regulating gene expression. Furthermore, the effect appears to be promoter-specific as NF110b is able to stimulate expression of a reporter gene from some, but not all, promoters (T.W. Reichman, A.M. Parrott, I. Fierro-Monti, P.N. Kao, C.G. Lee, H. Li, and M.B. Mathews, in prep.).
dsRBM requirement for reporter gene activation
To assess the role of the dsRBMs in mediating the activator function of NF110b, mutations were introduced into one or both of the dsRBMs (Fig. 2A,B ▶). Mutations were created in highly conserved residues using previous mutational analysis of the dsRNA-binding protein PKR as a guide. One pair of mutations changed a phenylalanine residue in each dsRBM to alanine (F432A and F559A in motifs 1 and 2, respectively). The equivalent residues were shown to be critical for the RNA-binding property of other proteins (Green et al. 1995; Krovat and Jantsch 1996; Patel et al. 1996; Brzostowski et al. 2000). A double-point mutation was also created, which contained both the F432A and the F559A mutations (F432A, F559A). In addition, a second double-point mutation was created by substituting proline for critical equivalent conserved alanine residues in the second α-helix region of the dsRBM (A458P, A588P). Mutation of these alanine residues in the dsRBMs of PKR resulted in the complete loss of RNA-binding and protein dimerization activity (Green et al. 1995; Patel et al. 1996).
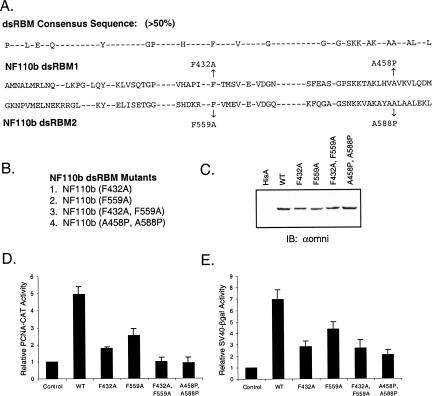
FIGURE 2.
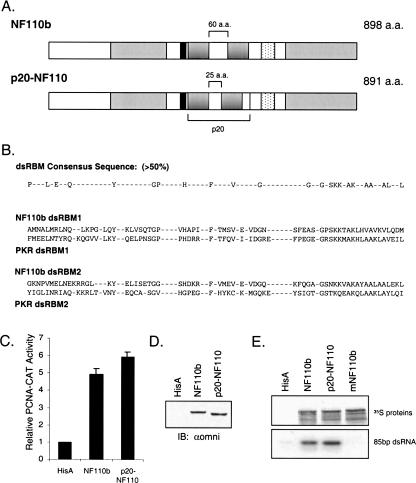
Mutagenesis of the dsRNA binding motifs of NF110b. (A) A dsRBM consensus sequence (showing residues with >50% conservation) (Fierro-Monti and Mathews 2000) is compared to the sequences of dsRBM1 and dsRBM2 of the NF90 family. Conserved amino acid residues were mutated as indicated to give rise to the four NF110b dsRBM mutants listed (B). (C) Western blot analysis of HeLa cells transiently expressing wild-type NF110b or the mutants indicated, as in Figure 1C ▶. (D) Results of transient expression assays conducted with the dsRBM mutants and the PCNA-CAT reporter, as in Figure 1B ▶ (n=3). Two-tailed student t-test indicated the difference between the double mutants and wild-type significant (P<0.01) and the difference between the two single mutants was significant at the level of P<0.1. (E) Assays similar to those of panel D were carried out with the SV40–βgal reporter (n=3).
In transient expression assays, the mutations all reduced the ability of NF110b to stimulate the PCNA promoter (Fig. 2D ▶). Whereas wild-type NF110b increased gene expression about fivefold, the stimulation was reduced to less than twofold by mutation of F432 in the first dsRBM and to ∼2.5-fold by mutation of F559 in the second dsRBM. Both of the double mutations essentially abrogated reporter gene activation by NF110b. Similar results were obtained using an SV40–βgal reporter plasmid (Fig. 2E ▶). Western blot analysis indicated that the proteins were expressed at approximately equivalent levels (Fig. 2C ▶). These findings therefore suggest that dsRNA binding is important for the induction of gene expression by NF110b, and that the first dsRBM may be more important than the second.
Mutations and C-terminal sequences influence dsRNA binding properties of NF110
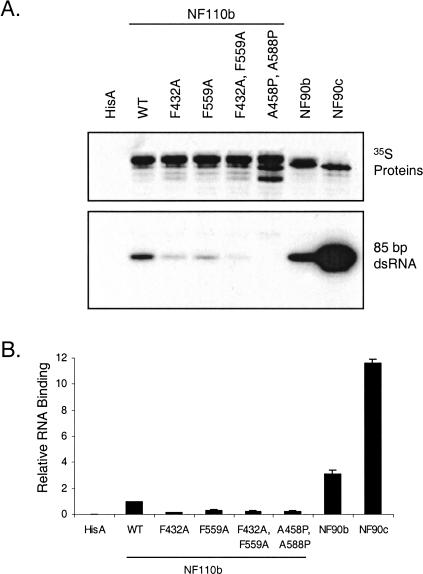
We evaluated the dsRNA-binding abilities of the NF110b mutants using a pull-down assay. The NF90 isoforms NF90b and NF90c were included for comparison. Approximately equal quantities of [35S]-labeled proteins, produced in rabbit reticulocyte lysate, were collected on antibody-Sepharose beads then incubated with synthetic [32P]-labeled dsRNA of 85 bp. The RNA:protein complexes were washed and analyzed by gel electrophoresis and autoradiography. Figure 3 ▶ shows that both single-point mutations reduced the ability of NF110b to interact with dsRNA. The two double-point mutants were almost completely defective in their ability to interact with dsRNA. These data parallel the effects of the dsRBM mutations in gene expression assays (Fig. 2 ▶). Surprisingly, NF90c and NF90b bound dsRNA considerably more efficiently than did wild-type NF110b, by about 10-fold and twofold respectively, implying that both the C-terminal RGG motif and GQSY region impede the binding of dsRNA to the NF90 family proteins.
FIGURE 3.
RNA-binding activities of NF110b dsRBM mutants and NF90 proteins. (A) The [35S]-labeled proteins indicated were synthesized in vitro and adsorbed to beads carrying an antibody (omni probe) that recognized an epitope tag present on the N terminus of all of the proteins. Extract programmed with empty vector (HisA) was a negative control. The beads were incubated with [32P]-labeled 85bp dsRNA, washed, and analyzed by SDS-gel electrophoresis. The bound [35S]-labeled proteins (top panel) and [32P]-labeled RNA (bottom panel) were detected by autoradiography. (B) Quantitation of [32P]-labeled RNA binding (panel A), relative to the 35S protein level. A value of 1.0 was assigned to wild-type NF110b (WT).
RNA binding was also assessed using the Northwestern blotting technique (Fig. 4A ▶). Epitope-tagged proteins produced in transfected HeLa cells were isolated by immunoprecipitation, resolved in a gel, and transferred to a membrane for probing with [32P]-labeled RNA. Quantitative data are shown in Figure 4B ▶. Probing with the 85 bp synthetic dsRNA (Fig. 4A, ▶top panel) showed that the dsRNA binding activities of F432A and F559A were reduced to <10% and <50% of wild-type NF110b, respectively, while no dsRNA binding was detectable with either of the double-point mutants. These results resemble those obtained above (Fig. 3 ▶) but the effect of the C-terminal region was less pronounced. In the Northwestern blot, the binding activities of NF90c and NF90b were only about twofold and 1.5-fold higher than that of NF110b. Evidently the RGG motif and GQSY region present in NF110 mask the dsRBMs less effectively in this assay, presumably because the proteins remained partially denatured on the blot despite the renaturation process. While dsRBMs derived from several proteins appear to renature efficiently (St Johnston et al. 1992; Cosentino et al. 1995; Patel et al. 1995b; Tian et al. 2000), this is not the case for all proteins or motifs. Therefore, it is possible that modulation of RNA binding by higher-order protein structures might go undetected in the Northwestern blot assay.
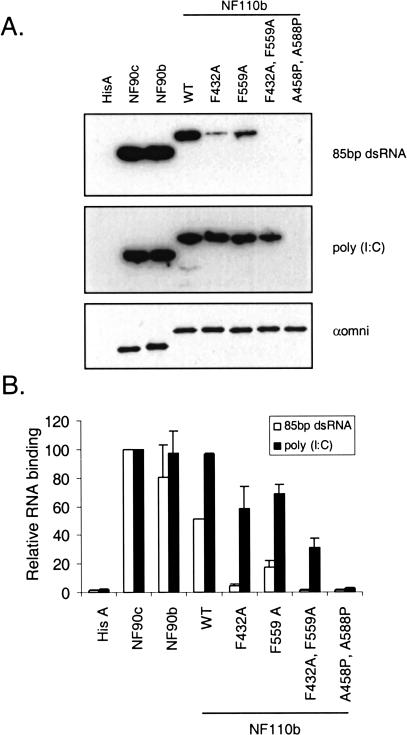
FIGURE 4.
Differential dsRNA-binding characteristics of NF90/110 proteins and mutants assessed by Northwestern blot assay. (A) The indicated proteins were immunoprecipitated from transfected HeLa cells with the omni probe antibody, transferred to nitrocellulose membranes, probed with [32P]-labeled 85-bp dsRNA (top panel) or poly (I:C) (middle panel), and exposed by autoradiography. A similar blot was probed with the omni antibody (bottom panel) to ensure equal loading of protein. Blots used in the Northwestern assay were also stained with Ponceau S stain to ensure that the proteins were not lost during the transfer (data not shown). (B) Quantitation of the data for 85 bp dsRNA (open bars) and poly (I:C) (filled bars) from two independent experiments. RNA binding is shown relative to NF90c, set at 100; and error bars represent the standard deviation.
The effects of the mutations were also apparent when the blot was probed with labeled poly (I:C), but this probe completely failed to discriminate among the three wild-type proteins (Fig. 4A ▶, middle panel, and Fig. 4B ▶). Similar results were obtained when the proteins were produced in vitro and probed with radiolabeled poly (I:C) or when tested in a poly (I:C)-Sepharose pull-down assay (data not shown). These findings suggest that the C-terminal region of NF110 confers a degree of RNA binding specificity upon the native NF110 protein.
Substitution of the dsRBMs of NF110b
Because the transcriptional and RNA-binding activities of NF110 are modulated by sequences in the protein’s N and C termini, we considered the possibility that the dsRNA-binding domain participates in regulatory interactions with surrounding regions of the protein. If this were the case, extensive alteration of the dsRNA-binding domain would be expected to abrogate the interactions, which would register as changes in NF110’s ability to stimulate gene expression and interact with RNA. We therefore replaced the dsRNA-binding domain of NF110b with the corresponding region of PKR, its N-terminal fragment (referred to as p20), which binds dsRNA strongly (Green and Mathews 1992; Patel and Sen 1994; Schmedt et al. 1995; Tian and Mathews 2001). The resulting chimera, p20-NF110, contains the two dsRBMs of PKR together with the intervening linker region in place of the equivalent region in NF110b (Fig. 5A,B ▶).
FIGURE 5.
The RNA-binding region of PKR can functionally replace that of NF110b. (A) Schematic of wild-type NF110b and the p20-NF110 chimera in which the dsRBM region of NF110b is substituted by the dsRBM region of PKR (p20). (B) Comparison of dsRBMs 1 and 2 of NF110b with PKR and the dsRBM consensus sequence. Note that both dsRBMs of NF110b are type A whereas in PKR dsRBM1 is type A and dsRBM2 is type B. (C) The p20-NF110b fusion protein transactivates the PCNA-CAT promoter as well as wild-type NF110b in transient expression assays conducted as in Figure 1B ▶ (n=3). (D) Western blot analysis of transfected cell extracts with omni probe antibody as in Figure 1C ▶. (E) NF110b, p20-NF110b, and mutant NF110b (mNF110b; A458P, A588P) were synthesized in wheat germ extract and equal amounts of protein were tested for their ability to bind to [32P]-labeled 85 bp dsRNA as described in Figure 3A ▶.
When cotransfected into HeLa cells, p20-NF110 efficiently stimulated expression from the PCNA-CAT promoter (Fig. 5C ▶). The chimera was slightly more effective than wild-type NF110b protein (approximately sixfold versus fivefold), possibly because of a small difference in protein expression (Fig. 5D ▶). In an antibody-Sepharose assay for RNA binding, p20-NF110 bound labeled 85bp dsRNA at least as well as did NF110b (Fig. 5E ▶). We conclude that the dsRBMs of NF110b are not specifically required for interactions with other regions of the protein to affect RNA binding and gene expression.
Structured RNA can modulate the activity of NF110b
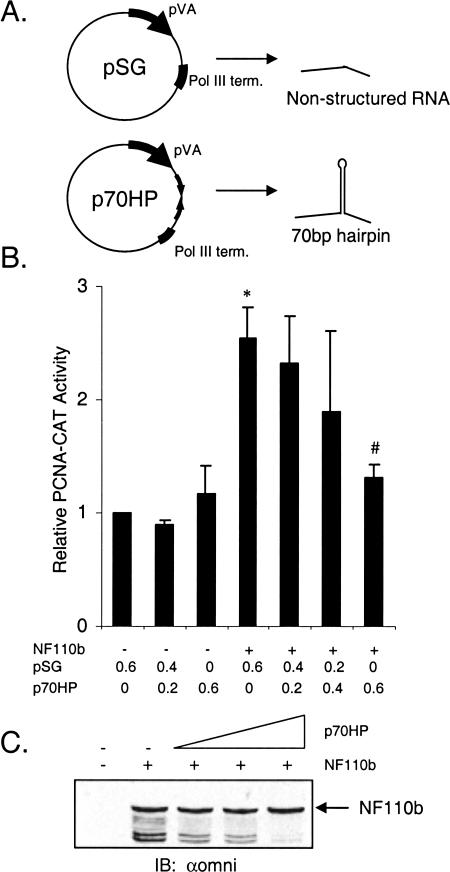
Given that the RNA-binding function of NF110b is essential for its effect on gene expression, it is likely that cellular and perhaps viral RNAs interact with the protein to influence this activity. Introduction of a nonspecific, highly structured RNA might therefore perturb NF110b function. To test this hypothesis, a plasmid was constructed that generates an RNA of 234 nucleotides including a 70-bp hairpin under the control of the strong pol III promoter of adenovirus-2 VA RNAI (Fig. 6A ▶). This plasmid was cotransfected into cells together with the PCNA-CAT promoter and a plasmid encoding NF110b, and reporter activity was monitored. As a negative control, the pSG plasmid was used, which generates an unstructured RNA of 94 nucleotides. The data indicate that the structured RNA inhibited NF110b-induced gene activation and effectively neutralized it at the highest concentration tested (Fig. 6B ▶). No significant change was detected in NF110b protein levels (Fig. 6C ▶), excluding possible nonspecific effects of the pol III vector (e.g., as a result of PKR activation). We conclude that at high concentrations, the structured RNA suppresses induction of gene activation by NF110b.
FIGURE 6.
An artificial structured RNA represses activation of gene expression by NF110b. (A) Schematic representation of the p70HP vector used to generate the hairpin-containing RNA from control sequences in pSG, which contains the strong VA RNA (pol III) promoter for high level of expression. (B) HeLa cells (0.8 × 105 cells/well) were transfected with 200 ng PCNA-CAT, 200 ng of empty vector (HisA) or NF110b expression plasmid, and increasing amounts of p70HP (0–600 ng) or empty pSG such that each transfection had a total of 600 ng pol III plasmid. The results of CAT activity are presented as in Figure 1B ▶ (n=3). A two-tailed student t test was performed and P values <0.01 were obtained for control versus NF110b (*) and NF110b versus NF110b + 600 ng p70HP (#). (C) Western blot analysis of transfected cell lysate with omni probe antibody as in Figure 1C ▶.
DISCUSSION
Indirect evidence has implied that RNA binding regulates the function of NF90 and its congeners. To test this idea, we examined the dsRNA-binding and transcriptional activation properties of NF110b. NF110b is the most potent member of the NF90 family, implicating its unique C-terminal GQSY-rich domain and NVKQ insert in its function. The N-terminal region of the protein was dispensable for its effect on gene expression, suggesting that the dsRBMs are important for NF90/NF110 function. To explore this hypothesis, we conducted site-directed and substitution mutagenesis to disrupt the ability of the dsRBMs to engage in RNA binding and protein-protein interactions. The mutant proteins were defective in their ability to enhance gene expression to a degree that paralleled the impairment in binding dsRNA. Furthermore, NF110b-mediated gene activation was inhibited by the expression of a dsRNA-like molecule, most likely by competing with cellular RNAs that bind to NF110b.
Model for NF110 function
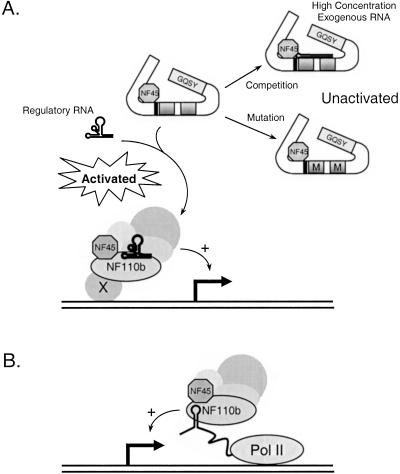
These data indicate that the dsRNA-binding function of NF110b, and presumably of other members of the NF90 family, is essential for the enhancement of gene expression directed by promoters including those of the PCNA gene and SV40. Based on findings presented here and previously (Reichman et al. 2002), we propose a model in which the dsRBMs of NF110b interact with one or more structured cellular RNA molecules that regulate its ability to associate with its targets and influence transcription (Fig. 7A ▶). Such RNAs may control the nucleation of an NF90/NF110 complex, or its recruitment to the promoter, to enhance or inhibit transcription. NF110b proteins in which the dsRBMs are mutated (denoted by M in Fig. 7 ▶) are unable to bind to the putative regulatory RNAs and are therefore not recruited to the promoter region. These mutant proteins do not act as dominant negatives even when expressed at levels higher than the wild-type protein (Fig. 2 ▶; data not shown). Assuming that NF110 is the limiting regulatory factor and that other components are present in excess, this observation would suggest that the association of NF110b with other factors that mediate its effect is RNA-dependent. The interaction of NF110b with exogenous-structured, hairpin-like RNA possibly results in mistargeting of NF110b. Alternatively, NF110b might associate with nascent mRNAs and facilitate transcription possibly via effects on transcriptional elongation (Fig. 7B ▶). A precedent for this alternative, which is compatible with results of run-on transcription assays (Saunders et al. 2001b), is furnished by the HIV-1 promoter. The viral transactivator protein Tat binds to nascent RNA generated from the TAR region of the HIV long-terminal repeat and increases transcription through interactions with the elongation factor P-TEFb (Zhu et al. 1997; Price 2000).
FIGURE 7.
Models for the regulation of NF110 function by structured RNA. The NLS and dsRBMs are designated by a black bar and shaded gray boxes, respectively. In (A), the transcription stimulation function of NF110b is activated by binding to a putative structured cellular regulatory RNA. This is prevented by the binding of a nonspecific structured RNA (shown as a hairpin) or mutation of the dsRBMs of NF110b (indicated by M). As an alternative, the interaction of NF110b with nascent transcripts is envisioned to regulate transcription elongation (B). See text for details.
The modulation of NF90/NF110 by RNA appears to be independent of NF45 binding. Previous data showed that the N terminus of NF90/NF110 reduces the protein’s ability to activate reporter gene expression and that the association of NF45 with NF90 enhances its ability to activate transcription to a level that is almost equivalent to deletion of the N terminus (Reichman et al. 2002). This led to the view that NF45 binding to NF90/NF110 alters its structure in such a way as to relieve the inhibitory effect of the N terminus (Reichman et al. 2002). Deletion of the N terminus of NF90 does not appear to enhance dsRNA binding, however (Parker et al. 2001a; Saunders et al. 2001b). Thus, it appears that NF110b’s interaction with cellular RNA(s) acts as a second step in regulating NF90/NF110 function, independent of NF45.
RNA binding
The dsRBM, which is present in two copies in all NF90 family members, is generally considered to be a nonspecific and modular motif although evidence for some specificity in dsRBM:RNA interactions is accumulating (Ma and Mathews 1993; Liao et al. 1998; Ramos et al. 2000). In the case of NF110b, specificity appears to be imparted by the protein’s C terminus rather than by the dsRBMs themselves. This region is also responsible for the greater activity of the NF110 isoforms relative to their NF90 counterparts (T.W. Reichman, A.M. Parrott, I. Fierro-Monti, P.N. Kao, C.G. Lee, H. Li, and M.B. Mathews, in prep.). The C-terminal domain decreased binding to dsRNA, and the binding of the dsRNA analog poly (I:C) was less affected than a synthetic 85 bp dsRNA (Figs. 3, 4 ▶ ▶). Conceivably, the C terminus may increase the affinity for the natural target RNA species. Strikingly, the NF110 C terminus contains two putative nucleic-acid–binding domains, the RGG domain and the GQSY domain. The RGG domain, shared with NF90b but not NF90c, is found in several RNA-binding proteins where it binds avidly to single-stranded nucleic acids (Ghisolfi et al. 1992; Darnell et al. 2001). The GQSY region, present in the NF110 isoforms but not in NF90, is an RNA-binding module; a similar domain in RHA has a strong preference for binding to ssRNAs (Zhang and Grosse 1997). Although it is an attractive idea that these domains confer specificity via interactions with RNA, protein-mediated interactions must also be considered. The GQSY domain has protein-binding activity and the TLS and SMN proteins were recently isolated in a yeast two-hybrid screen using the GQSY region as bait. TLS and SMN play roles in splicing and ribonucleoprotein complex biogenesis, respectively (Saunders et al. 2001b), but the significance of these interactions for NF110 function has not been defined.
Further support for the conclusion that RNA-binding specificity is conveyed by the C terminus of NF110 comes from experiments in which its dsRBMs were replaced by those of PKR. Even though NF110 and PKR both have two closely spaced dsRBMs, they differ structurally and in their RNA-binding properties. For example, NF90/NF110 has high affinity for VA RNAII but low affinity for VA RNAI, whereas PKR has much higher affinity for VA RNAI as compared to VA RNAII (Ma and Mathews 1993; Liao et al. 1998). In addition, the NF90/NF110 dsRBMs are both of type A, whereas PKR has one type A and one type B motif. Type A motifs conform to the consensus sequence along their entire length, and accordingly have a much higher affinity for dsRNA than type B motifs, which are homologous only in their C termini. Despite these differences, the chimeric protein p20-NF110 was equivalent to wild-type NF110b in RNA binding and transcriptional activator functions (Fig. 5 ▶), emphasizing the importance of the C-terminal RGG and GQSY motifs in dictating the specificity and function of the dsRBMs.
NF90/NF110 function
Our findings that RNA binding modulates the ability of NF110 to enhance gene expression, and that this activity can be abrogated by an artificial structured RNA containing a 70-bp hairpin, strongly suggest that NF110 interacts with cellular regulatory RNA(s). Noncoding RNAs are known to regulate diverse functions such as transcription and translation. For example, 7SK RNA modulates the function of transcription elongation factor P-TEFb (Nguyen et al. 2001; Yang et al. 2001) and SRA is a regulatory RNA that acts as a coactivator of steroid receptor-mediated transcriptional regulation (Lanz et al. 1999). U1 snRNA was recently shown to regulate transcriptional initiation by associating with TFIIH (Kwek et al. 2002). Functions remain to be assigned to numerous noncoding RNAs, including a recently cloned class of short, 22-bp dsRNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). These miRNAs appear to be related, in structure and possibly in function, to the let-7 and lin-4 RNAs discovered in genetic screens by virtue of their requirement in nematode development (Lee et al. 1993; Reinhart et al. 2000), and also to siRNAs, which are part of the RNA interference pathway that regulates gene-specific silencing (Hannon 2002). We speculate that these or other RNAs, both cellular and viral, bind to NF90 family proteins and regulate their function.
In addition to regulating cellular transcription, the NF90 family members may function in the antiviral defense pathway. The ability of an infected cell to detect the presence of foreign dsRNA is a key element of the innate immune system’s defenses against viral pathogens. The expression by viruses of dsRNA or other large dsRNA-like molecules might compete with the natural ligand of NF110b, thereby inhibiting its activity and blocking gene expression. Furthermore, small structured RNAs such as the VA RNAs may also bind to and modulate the activity of NF110b. Interestingly, a VA RNAI–/VA RNAII– mutant adenovirus replicated less well than the VA RNAI– virus, and viral production was also delayed (Thimmappaya et al. 1982; Bhat and Thimmappaya 1984 Bhat and Thimmappaya 1985; Bhat et al. 1985). It is possible that NF110 binds viral dsRNA during infection with the VA RNAII– deficient virus, resulting in reduced expression of genes critical for viral production. It may be significant that RHA, another VA RNAII-interacting protein, also interacts with NF90/NF110 (T.W. Reichman, A.M. Parrott, I. Fierro-Monti, P.N. Kao, C.G. Lee, H. Li, and M.B. Mathews, in prep.), suggesting that VA RNAII targets and prevents the inactivation of a common cellular pathway (Liao et al. 1998). Indications that NF90 might play a role in regulating the expression of genes critical for the cell cycle and cell proliferation include the observations that it is phosphorylated during M-phase of the cell cycle, and that NF90 and its mRNA are expressed at high levels in some tumor cell lines (Matsumoto-Taniura et al. 1996; Fung et al. 2000; Parker et al. 2001b). Identification of NF90/NF110 interacting proteins and the cellular genes that they regulate will be essential for understanding the functions of this protein family. These discoveries, and the characterization of NF90/NF110-associated RNAs, will expand our knowledge of regulatory RNAs and how they serve to regulate gene expression and normal cell function.
MATERIALS AND METHODS
Cloning, plasmid construction, and mutagenesis
The cDNAs encoding NF110b, NF90b, and NF90c (T.W. Reichman, A.M. Parrott, I. Fierro-Monti, P.N. Kao, C.G. Lee, H. Li, and M.B. Mathews, in prep.) were cloned into pcDNA3.1 and pcDNA3.1-HisB (Invitrogen) for expression of untagged and His-tagged versions of the proteins, respectively. His-NF110b was mutated using the QuikChange Site-directed Mutagenesis Kit (Stratagene). The sense PCR primers used for mutagenesis were as follows, listed 5′-3′ with mutations indicated in boldface: F432A, CCCGTCCATGCCCCCATCGCCACCATGTCTGTGGAGGTTG ATGG; F559A, GGCAGCCACGACAAGCGCGCCGTCATGGAG GTCGAAGTGG; A458P, CGGCCAAGCTGCACGTGCCAGTTA AGGTGTTACAGG ACATGG; and A588P, GGTGGCGAAGGCC TACGCTCCCCTTGCTGCCCTAGAAAAGCA. A complementary antisense primer was included in each PCR reaction.
To create His-p20-NF110, the N terminus of NF110 was amplified using primers GCGCGGATCCGAATTCGTAAAAATGC GTCCAATG and GCGCGGTACCCATAGCCTGGGGGGGCTC. The resulting PCR fragment was digested with BamHI and KpnI. p20 was amplified from pcDNA3-PKR-K296R using primers GC GCGGTACCATGGCTGGTGATCTTTCA and GCGCAAGCTTA GCAAAAGAACCAGAGG. The PCR product was digested with KpnI and HindIII. The inserts were then ligated into pcDNA3.1-HisC. The resulting plasmid was then digested with BamHI and HindIII to release the p20-NF110 N terminus insert. The C terminus of NF110 digested with HindIII and BstXI was then ligated into pcDNA3.1-His B together with the p20-NF110 N-terminal insert. Upon sequencing, a mutation was found in the N terminus of the protein. The mutation was corrected by ligating the N terminus of NF110b (EcoRI/EcoRV fragment) and the C terminus of p20-NF110 (EcoRV/NotI fragment) into pcDNA3.1-His B (EcoRI/NotI) to create His-p20-NF110.
The plasmid p70HP encodes a synthetic 70-bp RNA hairpin flanked by 5′ and 3′ sequences from adenovirus VA RNAI. It was constructed by inserting two tandem copies of a polylinker fragment from a plasmid vector in opposite orientations into pSG. pBluescriptII-KS+ was digested with BamHI and BssHII and the gel-purified fragment was ligated into pSG digested with BamHI. pSG contains the adenovirus VA RNAI (pol III) promoter and terminator without the VA RNAI central domain required for its function as PKR inhibitor (Gunnery et al. 1999). All constructs described were verified by sequencing.
Transient expression assays
HeLa cells were seeded in 12-well plates (1.2 − 1.5 × 105 cells/well) 1 d prior to transfection. Cells were transfected with 200 ng of the proliferating cell nuclear antigen (PCNA, −1265 − +60)-CAT plasmid (Morris and Mathews 1990) plus the indicated amount of the NF90 family expression plasmid (or empty vector) using 2.5 μL of Quantum Prep Cytofectene Reagent (Biorad). Empty pcDNA3.1 vector was used to adjust the total CMV plasmid level transfected to 800 ng, unless otherwise indicated. After 48 h, cells were lysed in 150 μL 0.25 M Tris-Cl [pH 7.6] by three freeze-thaw cycles. Lysates were clarified by centrifugation and 5 − 7.5 μL of extract was assayed for CAT activity using thin-layer chromatography (TLC) and [14C] chloramphenicol (ICN Pharmaceuticals). TLC plates were analyzed using a Packard Instant Imager. For β-galactosidase assays, cells were transfected with 200 ng pSV-β-galactosidase control vector (Promega) plus 800 ng of the indicated expression plasmid. After 48 h, cells were lysed with 150 μL of reporter lysis buffer and assayed using the Galactolight reporter assay (Tropix) as previously described (Reichman et al. 2002).
RNase protection assay
HeLa cells grown as described above were seeded at 1.2 × 105 cells in a six-well dish ∼12 h prior to transfection. For transfection, 400 ng of PCNA-CAT, 400 ng of empty vector or the indicated NF110 expression plasmid, and 1.2 μg of empty His A vector were diluted in 100 μL Opti-MEM medium (Invitrogen). Cells were transfected with 6 μL of lipofectamine 2000 (Invitrogen) with serum-free medium as described. After ∼5 h, the transfection medium was removed and replaced with 2 mL of DMEM + 10% FBS.
After 32–36 h, RNA was extracted using RNAzol B (Teltest). RNA was resuspended in 20 μL TE, 7.4. Fifteen microliters of each sample was assayed for PCNA-CAT mRNA levels using the RPA III kit (Ambion). Radiolabeled probe was prepared from PCNA-CAT (SP6) plasmid (Morris and Mathews 1990) linearized with EcoRI, by in vitro transcription with SP6 polymerase (New England Biolabs) at 40°C for 90 min in the presence of [α-32P]UTP (ICN Pharmaceuticals). Following treatment with DNase I, probe RNA was purified in a 5% denaturing acrylamide gel. Protected RNAs were resolved in a 5% denaturing acrylamide gel, visualized by autoradiography and quantified using a Packard Instant Imager.
Antibody–Sepharose assay
Wild-type and mutant proteins were synthesized using a T7 RNA polymerase/rabbit reticulocyte lysate coupled system (TNT, Promega) in the presence of [35S] Translabel (ICN Pharmaceuticals) for 90 min at 30°C. Labeled proteins were immunoprecipitated by adding equal counts of each protein to 100 μL binding buffer A (25 mM HEPES-NaOH [pH 7.4], 50 mM NaCl, 3 mM MgCl2, 10% glycerol, 0.05% Triton X-100, 1 mM DTT, 0.5 mM PMSF, 0.1 mM benzamidine, plus 0.2 μg of aprotinin, 0.2 μg leupeptin, and 0.1 μg pepstatin A per milliliter) together with 2.5 μL omni probe antibody (Santa Cruz Biotechnology) and 10 μL packed protein A-Sepharose beads. After incubation for 3 h at 4°C, immunoprecipitates were washed three times with binding buffer A and resuspended in 50 μL binding buffer A (with 0.05% BSA). Radiolabeled 85 bp dsRNA (200 fmole), prepared as described previously (Manche et al. 1992), was added to each tube and samples were incubated for 35 min at room temperature, washed four times in binding buffer A, and resuspended in Laemmli sample buffer. Products were resolved in a 15% polyacrylamide-sodium dodecyl sulfate (SDS) gel and analyzed by autoradiography. Aliquots of the supernatant were analyzed by gel electrophoresis to verify that the results obtained were because of differences in dsRNA binding rather than RNA degradation. A similar procedure was performed for the p20-NF110 experiments, except that proteins were made in the wheat germ TNT system (Promega).
Northwestern blot assay
HeLa cells were plated in 60 mm dishes and transfected at 80% confluency with 2 μg of the indicated NF90 expression plasmid (or empty vector) plus 10 μL Cytofectene. After 48 h, cells were lysed in 300 μL lysis buffer (50 mM Tris-Cl [pH 7.6], 500 mM NaCl, 1% Triton X-100, 1.5 mM MgCl2, 10% glycerol, 0.5 mM DTT, 0.5 mM PMSF, 0.2 mM benzamidine, plus 0.4 μg of aprotinin, 0.4 μg leupeptin, and 0.2 μg pepstatin A per milliliter) for 30 min at 4°C with gentle rocking. After clarification by centrifugation at 14,000 rpm for 15 min at 4°C, 200 μL of each lysate was diluted in 200 μL lysis buffer plus 10 μL omni probe antibody and incubated at 4°C for 3 h. Then 10 μL packed protein A-Sepharose was added and the samples were incubated for an additional hour. Immunoprecipitates were washed four times in lysis buffer with 800 mM NaCl and twice in lysis buffer (150 mM NaCl). Pellets were resuspended in 50 μL 2× concentrated Laemmli sample buffer, and 3 μL of each extract was resolved in an 8% polyacrylamide-SDS gel. Proteins were analyzed by Western blotting using the omni probe antibody for estimation of relative protein concentrations. Equal amounts of each protein were separated in an 8% gel and transferred to a 0.45-μ nitrocellulose membrane. Proteins were allowed to renature overnight in Northwestern buffer (10 mM Tris-Cl [pH 7.6], 50 mM NaCl, 1 mM EDTA, 1× Denhardt’s reagent). The following day, blots were incubated with 1 × 105 cpm/mL of the indicated probe [poly (I:C) or 85 bp dsRNA] for 1 h at room temperature. Poly (I:C) (Amersham Pharmacia Biotech) was radiolabeled using T4 polynucleotide kinase (Invitrogen) and [γ-32P]ATP (ICN Pharmaceuticals). The 85-bp dsRNA was prepared as described above. Blots were washed 2× with Northwestern buffer (with 150 mM NaCl) for 15 min, then subjected to autoradiography and quantitation using a Packard Instant Imager. To control for protein loading, a parallel blot prepared with one-third as much protein was blotted with the omni probe antibody.
Acknowledgments
This work was supported by grant R01 AI34552 from the National Institutes of Health to M.B.M. T.W.R was the recipient of a predoctoral fellowship 12013CCRS0 from the New Jersey Commission for Cancer Research.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2181103.
REFERENCES
- Abraham, N., Stojdl, D.F., Duncan, P.I., Methot, N., Ishii, T., Dube, M., Vanderhyden, B.C., Atkins, H.L., Gray, D.A., McBurney, M.W., et al. 1999. Characterization of transgenic mice with targeted disruption of the catalytic domain of the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 274: 5953–5962. [DOI] [PubMed] [Google Scholar]
- Bass, B.L., Hurst, S.R., and Singer, J.D. 1994. Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr. Biol. 4: 301–314. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C. and Cech, T.R. 1996. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35: 9983–9994. [DOI] [PubMed] [Google Scholar]
- Bhat, R.A. and Thimmappaya, B. 1984. Adenovirus mutants with DNA sequence perturbations in the intragenic promoter of VAI RNA gene allow the enhanced transcription of VAII RNA gene in HeLa cells. Nucleic Acids Res. 12: 7377–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1985. Construction and analysis of additional adenovirus substitution mutants confirm the complementation of VAI RNA function by two small RNAs encoded by Epstein-Barr virus. J. Virol. 56: 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, R.A., Domer, P.H., and Thimmappaya, B. 1985. Structural requirements of adenovirus VAI RNA for its translation enhancement function. Mol. Cell. Biol. 5: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzostowski, J., Robinson, C., Orford, R., Elgar, S., Scarlett, G., Peterkin, T., Malartre, M., Kneale, G., Wormington, M., and Guille, M. 2000. RNA-dependent cytoplasmic anchoring of a transcription factor subunit during Xenopus development. EMBO J. 19: 3683–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft, M., Grunert, S., Murzin, A.G., Proctor, M., and St. Johnston, D. 1995. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 14: 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolidge, C.J. and Patton, J.G. 2000. A new double-stranded RNA-binding protein that interacts with PKR. Nucleic Acids Res. 28: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corthésy, B. and Kao, P.N. 1994. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J. Biol. Chem. 269: 20682–20690. [PubMed] [Google Scholar]
- Cosentino, G.P., Venkatesan, S., Serluca, F.C., Green, S.R., Mathews, M.B., and Sonenberg, N. 1995. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc. Natl. Acad. Sci. 92: 9445–9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell, J.C., Jensen, K.B., Jin, P., Brown, V., Warren, S.T., and Darnell, R.B. 2001. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107: 489–499. [DOI] [PubMed] [Google Scholar]
- Duchange, N., Pidoux, J., Camus, E., and Sauvaget, D. 2000. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene 261: 345–353. [DOI] [PubMed] [Google Scholar]
- Fierro-Monti, I. and Mathews, M.B. 2000. Proteins binding to duplexed RNA: One motif, multiple functions. Trends Biochem. Sci. 25: 241–246. [DOI] [PubMed] [Google Scholar]
- Fung, L.F., Lo, A.K., Yuen, P.W., Liu, Y., Wang, X.H., and Tsao, S.W. 2000. Differential gene expression in nasopharyngeal carcinoma cells. Life Sci. 67: 923–936. [DOI] [PubMed] [Google Scholar]
- Ghisolfi, L., Joseph, G., Amalric, F., and Erard, M. 1992. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem. 267: 2955–2959. [PubMed] [Google Scholar]
- Green, S.R. and Mathews, M.B. 1992. Two RNA binding motifs in the double-stranded RNA activated protein kinase DAI. Genes Dev. 6: 2478–2490. [DOI] [PubMed] [Google Scholar]
- Green, S.R., Manche, L., and Mathews, M.B. 1995. Two functionally distinct RNA-binding motifs in the regulatory domain of the protein kinase DAI. Mol. Cell. Biol. 15: 358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnery, S., Ma, Y., and Mathews, M.B. 1999. Termination sequence requirements vary among genes transcribed by RNA polymerase III. J. Mol. Biol. 286: 745–757. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Kao, P.N., Chen, L., Brock, G., Ng, J., Kenny, J., Smith, A.J., and Corthésy, B. 1994. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem. 269: 20691–20699. [PubMed] [Google Scholar]
- Kaufman, R. 2000. Double-stranded RNA activated protein kinase PKR. In: Translational control of gene expression (eds. N. Sonenberg et al.), pp. 503–528. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kharrat, A., Macias, M.J., Gibson, T.J., Nilges, M., and Pastore, A. 1995. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 14: 3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnoselskaya-Riz, I., Spruill, A., Chen, Y.W., Schuster, D., Teslovich, T., Baker, C., Kumar, A., and Stephan, D.A. 2002. Nuclear factor 90 mediates activation of the cellular antiviral expression cascade. AIDS Res. Hum. Retroviruses 18: 591–604. [DOI] [PubMed] [Google Scholar]
- Krovat, B.C. and Jantsch, M.F. 1996. Comparative mutational analysis of the double-stranded RNA binding domains of Xenopus laevis RNA-binding protein A. J. Biol. Chem. 271: 28112–28119. [DOI] [PubMed] [Google Scholar]
- Kwek, K.Y., Murphy, S., Furger, A., Thomas, B., O’Gorman, W., Kimura, H., Proudfoot, N.J., and Akoulitchev, A. 2002. U1 snRNA associates with TFIIH and regulates transcriptional initiation. Nat. Struct. Biol. 9: 800–805. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Langland, J.O., Kao, P.N., and Jacobs, B.L. 1999. Nuclear factor-90 of activated T-cells: A double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry 38: 6361–6368. [DOI] [PubMed] [Google Scholar]
- Lanz, R.B., McKenna, N.J., Onate, S.A., Albrecht, U., Wong, J., Tsai, S.Y., Tsai, M.J., and O’Malley, B.W. 1999. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97: 17–27. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Liao, H.J., Kobayashi, R., and Mathews, M.B. 1998. Activities of adenovirus virus-associated RNAs: Purification and characterization of RNA binding proteins. Proc. Natl. Acad. Sci. 95: 8514–8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y. and Mathews, M.B. 1993. Comparative analysis of the structure and function of adenovirus virus-associated RNAs. J. Virol. 67: 6605–6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manche, L., Green, S.R., Schmedt, C., and Mathews, M.B. 1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12: 5238–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, M.B. and Shenk, T. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65: 5657–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Taniura, N., Pirollet, F., Monroe, R., Gerace, L., and Westendorf, J.M. 1996. Identification of novel M phase phosphoproteins by expression cloning. Mol. Biol. Cell 7: 1455–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G.F. and Mathews, M.B. 1990. Analysis of the proliferating cell nuclear antigen promoter and its response to adenovirus early region 1. J. Biol. Chem. 265: 16116–16125. [PubMed] [Google Scholar]
- Nguyen, V.T., Kiss, T., Michels, A.A., and Bensaude, O. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414: 322–325. [DOI] [PubMed] [Google Scholar]
- Orford, R.L., Robinson, C., Haydon, J.M., Patient, R.K., and Guille, M.J. 1998. The maternal CCAAT box transcription factor which controls GATA-2 expression is novel and developmentally regulated and contains a double-stranded-RNA-binding subunit. Mol. Cell. Biol. 18: 5557–5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, L.M., Fierro-Monti, I., and Mathews, M.B. 2001a. Nuclear factor 90 is a substrate and regulator of the eIF2 kinase PKR. J. Biol. Chem. 276: 32522–32530. [DOI] [PubMed] [Google Scholar]
- Parker, L.M., Fierro-Monti, I., Reichman, T.W., Gunnery, S., and Mathews, M.B. 2001b. Double-stranded RNA-binding proteins and the control of protein synthesis and cell growth. Cold Spring Harbor Symposia on Quantitative Biology 66: 485–497. [DOI] [PubMed] [Google Scholar]
- Patel, R.C. and Sen, G.C. 1994. Characterization of the interactions between double-stranded RNA and the double-stranded RNA binding domain of the interferon induced protein kinase. Cell. Mol. Biol. Res. 40: 671–682. [PubMed] [Google Scholar]
- Patel, R.C., Stanton, P., McMillan, N.M., Williams, B.R., and Sen, G.C. 1995a. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc. Natl. Acad. Sci. 92: 8283–8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1995b. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc. Natl. Acad. Sci. 92: 8283–8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, R.C., Stanton, P., and Sen, G.C. 1996. Specific mutations near the amino terminus of double-stranded RNA-dependent protein kinase (PKR) differentially affect its double-stranded RNA binding and dimerization properties. J. Biol. Chem. 271: 25657–25663. [DOI] [PubMed] [Google Scholar]
- Price, D.H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20: 2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, A., Grunert, S., Adams, J., Micklem, D.R., Proctor, M.R., Freund, S., Bycroft, M., St. Johnston, D., and Varani, G. 2000. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 19: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichman, T.W., Muniz, L.C., and Mathews, M.B. 2002. The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol. Cell. Biol. 22: 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Ryter, J.M. and Schultz, S.C. 1998. Molecular basis of double-stranded RNA-protein interactions: Structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 17: 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh, M., Shaheen, V.M., Kao, P.N., Okano, T., Shaw, M., Yoshida, H., Richards, H.B., and Reeves, W.H. 1999. Autoantibodies define a family of proteins with conserved double-stranded RNA-binding domains as well as DNA binding activity. J. Biol. Chem. 274: 34598–34604. [DOI] [PubMed] [Google Scholar]
- Saunders, L.R., Jurecic, V., and Barber, G.N. 2001a. The 90– and 110-kDa human NFAR proteins are translated from two differentially spliced mRNAs encoded on chromosome 19p13. Genomics 71: 256–259. [DOI] [PubMed] [Google Scholar]
- Saunders, L.R., Perkins, D.J., Balachandran, S., Michaels, R., Ford, R., Mayeda, A., and Barber, G.N. 2001b. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 276: 32300–32312. [DOI] [PubMed] [Google Scholar]
- Schmedt, C., Green, S.R., Manche, L., Taylor, D.R., Ma, Y., and Mathews, M.B. 1995. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR). J. Mol. Biol. 249: 29–44. [DOI] [PubMed] [Google Scholar]
- St. Johnson, D., Brown, N.H., Gall, J.G., and Jantsch, M. 1992. A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. 89: 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmappaya, B., Weinberger, C., Schneider, R.J., and Shenk, T. 1982. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell 31: 543–551. [DOI] [PubMed] [Google Scholar]
- Tian, B. and Mathews, M.B. 2001. Functional characterization of and cooperation between the double-stranded RNA-binding motifs of the protein kinase PKR. J. Biol. Chem. 276: 9936–9944. [DOI] [PubMed] [Google Scholar]
- Tian, B., White, R.J., Xia, T., Welle, S., Turner, D.H., Mathews, M.B., and Thornton, C.A. 2000. Expanded CUG repeat RNAs form hairpins that activate the double-stranded-RNA-dependent protein kinase PKR. RNA 6: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.H. and Grabowski, G.A. 1999. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab. 68: 441–454. [DOI] [PubMed] [Google Scholar]
- Yang, Y.L., Reis, L.F., Pavlovic, J., Aguzzi, A., Schafer, R., Kumar, A., Williams, B.R., Aguet, M., and Weissmann, C. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14: 6095–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Zhu, Q., Luo, K., and Zhou, Q. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414: 317–322. [DOI] [PubMed] [Google Scholar]
- Zhang, S. and Grosse, F. 1997. Domain structure of human nuclear DNA helicase II (RNA helicase A). J. Biol. Chem. 272: 11487–11494. [DOI] [PubMed] [Google Scholar]
- Zhou, A., Paranjape, J.M., Der, S.D., Williams, B.R., and Silverman, R.H. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258: 435–440. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Pe’ery, T., Peng, J., Ramanathan, Y., Marshall, N., Marshall, T., Amendt, B., Mathews, M.B., and Price, D.H. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes & Dev. 11: 2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]