Abstract
Methylation of tRNA at the N-1 position of guanosine to form m1G occurs widely in nature. It occurs at position 37 in tRNAs from all three kingdoms, and the methyltransferase that catalyzes this reaction is known from previous work of others to be critically important for cell growth in Escherichia coli and the yeast Saccharomyces cerevisiae. m1G is also widely found at position 9 in eukaryotic tRNAs, but the corresponding methyltransferase was unknown. We have used a biochemical genomics approach with a collection of purified yeast GST-ORF fusion proteins to show that m1G9 formation of yeast tRNAGly is associated with ORF YOL093w, named TRM10. Extracts lacking Trm10p have undetectable levels of m1G9 methyltransferase activity but retain normal m1G37 methyltransferase activity. Yeast Trm10p purified from E. coli quantitatively modifies the G9 position of tRNAGly in an S-adenosylmethionine-dependent fashion. Trm10p is responsible in vivo for most if not all m1G9 modification of tRNAs, based on two results: tRNAGly purified from a trm10-Δ/trm10-Δ strain is lacking detectable m1G; and a primer extension block occurring at m1G9 is removed in trm10-Δ/trm10-Δ-derived tRNAs for all 9 m1G9-containing species that were testable by this method. There is no obvious growth defect of trm10-Δ/trm10-Δ strains. Trm10p bears no detectable resemblance to the yeast m1G37 methyltransferase, Trm5p, or its orthologs. Trm10p homologs are found widely in eukaryotes and many archaea, with multiple homologs in several metazoans, including at least three in humans.
Keywords: Saccharomyces cerevisiae, S-adenosylmethionine, TRM10, YOL093w, 1-methylguanosine
INTRODUCTION
tRNA molecules are extensively modified after transcription. In addition to 5′- and 3′-end maturation, CCA addition, and splicing of introns, all tRNAs contain a number of nucleoside modifications (Bjork 1995; Grosjean et al. 1997; Hopper and Phizicky 2003). In Saccharomyces cerevisiae, 25 different modifications of base or sugar moieties have been identified within the 34 tRNA species that have been characterized (Sprinzl et al. 1998), and every tRNA molecule has some subset of these modifications, altering nucleoside identity at an average of 11 positions. Some modifications such as i6A or m22G occur only at a single position in one or more tRNAs, whereas other modifications such as Ψ occur at a number of different positions in tRNAs (Sprinzl et al. 1998). The identity and position of many of these modifications are highly conserved; however, despite their widespread conservation, the role that these modifications play in the cell is not fully understood.
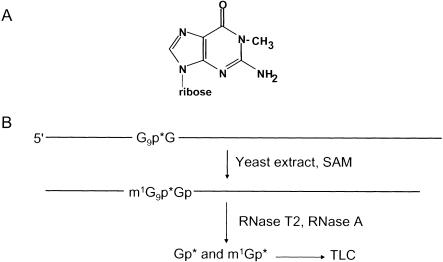
One modification of interest is methylation of guanosine (G) at the N-1 position of the base (m1G; Fig. 1A ▶). This modification occurs at two locations in tRNAs. m1G is found at position 37 in several tRNAs from all three major kingdoms. It is also found at position 9 in several cytoplasmic tRNAs from all eukaryotes that have sequenced tRNAs, in mitochondrial tRNAs from several animal species, in two vertebrate viral tRNAs, and in tRNA from at least one archaeal species (Sprinzl et al. 1998). In the yeast S. cerevisiae, m1G is found at position 9 in 10 tRNAs and at position 37 in 8 tRNAs, with one species, tRNAPro, having m1G at both positions. The methyltransferase proteins responsible for m1G37 formation are critical in both bacteria and yeast; Salmonella typhimurium trmD mutants and S. cerevisiae trm5 mutants are each severely compromised for growth (Bjork et al. 2001).
FIGURE 1.
(A) N1-Methylguanosine. (B) Assay scheme to detect m1G formation in tRNAGly uniquely labeled at position 9 (G9*Gly). This assay results in the production of either Gp* if the substrate remains unmodified, or m1Gp* if it is modified.
The goal of this work was to identify the gene and protein responsible for m1G9 formation in S. cerevisiae tRNA. It is known that the yeast m1G37 methyltransferase plays no role in m1G9 formation in yeast (Bjork et al. 2001). Furthermore, unlike the case with many other modification enzymes, there is no obvious candidate gene in the yeast database with homology to the known m1G37 methyltransferases (Bjork et al. 2001) that might be responsible for m1G9 methylation of tRNAs. Using a biochemical genomics approach, we show here that the protein encoded by ORF YOL093w catalyzes tRNA m1G methyltransferase activity in vitro at position 9 of tRNA substrates, and that this protein (Trm10p) is responsible for m1G formation at position 9 in yeast tRNAs in vivo.
RESULTS
To determine the protein responsible for catalyzing the modification of tRNA at G9 to m1G, we first developed a sensitive assay using yeast tRNAGly, a tRNA that is known to contain m1G9 in vivo (Sprinzl et al. 1998). This tRNA was synthesized in vitro and then uniquely labeled at the phosphate immediately 3′ of G9, by directing cleavage at this position with RNase H, labeling the 5′ nucleotide of the resulting 3′ RNA product, and reforming the tRNA by ligation, as described in Materials and Methods (Yu 1999). After incubation of G9*Gly tRNA with a source of m1G9 methyltransferase activity (crude yeast extracts or purified protein) in the presence of the methyl donor S-adenosylmethionine (SAM), RNA was treated with a combination of RNase T2 and RNase A, and nucleoside 3′-monophosphate products were resolved on cellulose TLC plates (Fig. 1B ▶). Because G9 was the only labeled nucleotide in this substrate, modification of the tRNA at this position is easily detected by looking for a change in mobility of the labeled spot. As shown in Figure 2A ▶ (lanes a and b), treatment of tRNA with yeast crude extract results in the quantitative conversion of Gp* to a modified product that migrates significantly more slowly in the solvent system. This modification was presumed to be m1Gp*, which will be demonstrated below.
FIGURE 2.
Identification of a yeast ORF associated with m1G9 methyltransferase activity. Reaction mixtures containing G9*Gly tRNA and protein as indicated were incubated in methyltransferase buffer at 30°C for 4 h, and then RNA was digested with nucleases to produce 3′-phosphorylated nucleotides, which were resolved by thin layer chromatography. (A) Assay of a genomic collection of pools of purified yeast GST-ORF fusion proteins for G9 methyltransferase activity. Substrate G9*Gly tRNA was incubated with 64 pools of purified GST-ORF fusion proteins, each derived from 96 yeast strains of a library of strains expressing individual GST-ORF fusion proteins, as indicated. (Lane a) Saccharomyces cerevisiae crude extract; (lane b) no extract. (B) Assay of subpools from pool 47 for tRNA G9 methyltransferase activity. (First panel) Substrate G9*Gly tRNA was incubated with pools of GST-ORFs derived from the strains in rows A–H from plate 47, as indicated. (Lane P47) plate 47; (lane a) crude extract; and (lane b) no extract. (Second panel) Substrate was incubated with GST-ORFs from columns 1–12 of plate 47. (Lanes P47,a,b,) Same as previous panel.
Identification of a yeast ORF associated with an activity that modifies position 9 of tRNAGly
To identify the protein responsible for this modification activity, we screened a library of purified yeast GST-ORF fusion proteins for the activity, as has been done for a number of other activities (Martzen et al. 1999; Alexandrov et al. 2002; Hazbun and Fields 2002; Xing et al. 2002). This library is an ordered collection of 6144 yeast strains each expressing a predicted yeast open reading frame (ORF) as a GST-ORF fusion protein. For ease of assaying more than 6000 proteins, the library of strains is grown in pools derived from each microtiter plate, such that after purification each pool contains a mixture of 96 individual GST-ORF fusion proteins. After activity is observed in one pool, further deconvolution leads to the identification of the individual fusion protein responsible for the activity. When the GST-ORF pools were screened for the modification activity that altered G9 of tRNAGly, a positive signal was observed in pool 47 (Fig. 2A ▶). To determine the individual ORF responsible for this modification, we assayed 8 subpools of 12 proteins comprising each row (A–H) of plate 47 and 12 subpools of 8 proteins comprising each column (1–12) of plate 47. Activity was detected in row E and column 6 (Fig. 2B ▶). To confirm the assignment of the activity to the strain at position E6, the corresponding strain (MRM4470) was grown, and extracts were analyzed for activity. Crude extracts made from this strain overexpress tRNA m1G9 methyltransferase activity at least fivefold relative to wild-type strains (data not shown), demonstrating that the corresponding ORF, YOL093w, is the limiting factor required for activity. The protein encoded by YOL093w has no previously identified function in yeast; we have assigned it the name TRM10 because, as we show below, its protein product is the tRNA m1G9 methyltransferase.
Trm10p is necessary for modification of position 9 of tRNAGly and has no effect on modification of position 37
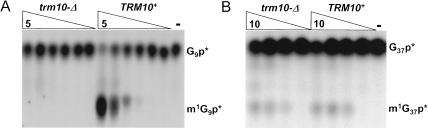
To determine if Trm10p is required for modification at position 9, we prepared extracts from the homozygous trm10-Δ/trm10-Δ strain and its isogenic TRM10+/TRM10+ parent strain, and compared their activities with the tRNAGly substrate (G9*Gly) used above. Extracts from wild-type cells were active with the G9*Gly substrate when diluted as much as 125-fold, whereas G9 modification activity was undetectable in the trm10-Δ/trm10-Δ strain even at the highest concentration of extract tested (Fig. 3A ▶). This is consistent with the identification of Trm10p as the protein responsible for G9 modification activity.
FIGURE 3.
Assay of a trm10-Δ/trm10-Δ strain for methyltransferase activity at positions 9 and 37. Extracts were prepared from isogenic wild-type (TRM10+/TRM10+) and trm10-Δ/trm10-Δ strains, and assayed for methyltransferase activites. (A) m1G9 methyltransferase activity. Assays contained G9*Gly tRNA and decreasing concentrations of crude extracts (5 mg/mL to 1.6 μg/mL by factors of 5) made from wild-type and trm10-Δ/trm10-Δ yeast strains and were carried out at 30°C for 1 h. (B) m1G37 methyltransferase activity. Assays contained G37*Leu tRNA with decreasing concentrations of crude extracts from wild-type and trm10-Δ/trm10-Δ yeast strains (2.5 mg/mL to 2.5 μg/mL by factors of 10) and were carried out at 30°C for 5 h.
Because yeast tRNA also contains m1G at position 37 of some tRNA species, we investigated the site specificity of the reaction using a tRNACAALeu substrate normally modified by Trm5p to m1G at position 37. Although our in vitro assay for Trm5p activity uses a similar uniquely labeled substrate (G37*Leu) and similar conditions, we consistently observe much lower levels of Trm5 methyltransferase activity than we observe with Trm10p and its substrate. Nonetheless, m1G37 methyltransferase activity can be observed by using longer incubation times (Fig. 3B ▶). Assay of the wild-type and trm10-Δ/trm10-Δ extracts with the G37*Leu substrate indicates no difference in m1G37 methyltransferase activity between them, as expected if the modification activity of Trm10p is specific for position 9 (Fig. 3B ▶). We note that because the modification at G37 of tRNACAALeu is known to be m1G (Bjork et al. 2001), the identical migration of the products observed with the two different substrates is consistent with the modification at position 9 also being m1G.
Trm10p is sufficient for m1G9 modification in vitro
To determine if Trm10p could catalyze modification of tRNA without additional S. cerevisiae proteins, the ORF was cloned into a pET14b-based vector for production of the protein in Escherichia coli. This plasmid, pJEJ12-3, expresses soluble His6-Trm10 fusion protein when introduced into BL21(DE3)pLysS cells and induced for expression, as observed by SDS-PAGE of extracts (Fig. 4A ▶, lanes 1,2). The prominent new band in lane 2 has a molecular mass consistent with the predicted molecular mass of 35.5 kD for the His6-Trm10 fusion protein. Assays of extracts demonstrated a >1000-fold increase in G9*Gly modification activity from cells expressing His6-Trm10p compared with cells with vector alone (Fig. 4B ▶). As shown in Figure 4B ▶, there was no background m1G9 methyltransferase activity in extracts made from cells containing the control vector; this is as expected because E. coli cells do not contain an m1G9 modification in their tRNAs.
FIGURE 4.
Overexpression and purification of Trm10p from Escherichia coli. (A) SDS-PAGE gel of Trm10p overproduction and purification. (Lane 1) Crude extract from control strain with vector only (40 μg); (lane 2) crude extract from strain with plasmid pJEJ12-3 expressing His6-Trm10p (40 μg); (lane 3) broad range MW markers; (lane 4) crude extract used for purification, from strain containing pJEJ12-3 (40 μg); (lanes 5,6) purified Trm10p (4 and 8 μg, respectively). (B) Assay of E. coli extracts for m1G9 methyltransferase activity. Assays containing G9*Gly tRNA and decreasing concentrations of either the control extract (panel A, lane 1) or Trm10p-expressing extract (panel A, lane 2) were performed at 30°C for 1 h.
The His6-Trm10 fusion protein was purified from extracts with immobilized metal-ion affinity chromatography (IMAC; Fig. 4A ▶, lanes 5,6). The purified protein is nearly homogeneous, has an increased specific activity of about twofold over the original extract (Fig. 4A ▶, lanes 4–6), and is obtained at 30% yield (data not shown), indicating that Trm10p is sufficient for catalysis in vitro in the absence of other interacting partners. The activity of the purified protein from E. coli is completely dependent on the addition of 0.5 mM SAM in the assay, although this dependence is only apparent at low enzyme concentrations, where the enzyme is required to undergo multiple turnovers during the course of the reaction (data not shown). This indicates that some amount of bound SAM copurifies with Trm10p and therefore that Trm10p is able to bind SAM with some degree of affinity.
m1G is the product of Trm10p activity
To show that the product of Trm10p activity is m1G, we modified in vitro transcribed tRNAGly with Trm10p purified from E. coli in a reaction mixture containing 10 μM tRNA and 10 μM Trm10p, and then analyzed nucleosides from the product tRNA by HPLC (Fig. 5A ▶, bottom two traces). Unmodified tRNAGly contains four main peaks for the four expected nucleosides C, U, G, and A, whereas the Trm10p-modified tRNA contains an additional peak that comigrates with and has a spectrum identical to an m1G nucleoside standard (Fig. 5A ▶, upper trace). This evidence demonstrates that m1G is the product of the Trm10p reaction. Quantification of the amount of m1G indicates 1.1 mole of m1G/mole tRNA based on the areas of the peaks observed before and after reaction with Trm10p, consistent with modification of a single nucleotide in tRNAGly to m1G (Table 1 ▶).
FIGURE 5.
Identification of m1G as the product of Trm10p activity in vitro and in vivo. (A) Trm10 protein modifies tRNAGly to produce material that matches m1G. (Upper trace) m1G chemical standard. (Middle trace) In vitro transcribed tRNAGly treated with buffer and digested to nucleosides. (Lower trace) In vitro transcribed tRNAGly treated with Trm10p and digested to nucleosides. All samples were analyzed by HPLC as described in Materials and Methods. (B) Comparison of nucleosides of tRNAGly from wild-type and trm10-Δ/trm10-Δ strains. tRNAGly was purified from wild-type and trm10 mutant strains, and its nucleosides were prepared and resolved on HPLC as described above.
TABLE 1.
Quantification of nucleoside modifications to tRNAGly
| Modified nucleoside | Number expected per mole of tRNAa | In vitro product of Trm10pb | tRNAGly from wild typeb | tRNAGly from trm10 − Δb |
| m1G | 1 | 1.1 | 0.98 | <0.01 |
| m5C | 1 | — | 1.7 | 1.8 |
| Cm | 1 | — | 1.0 | 1.0 |
| DHU | 2 | — | 0.92 | 1.4 |
| pseudoU | 4 | — | 3.9 | 3.9 |
bCalculated from areas under peaks observed upon HPLC elution of nucleosides, as described in Materials and Methods.
TRM10 is responsible for m1G9 modification of tRNAGly in vivo
To determine if Trm10p is responsible for formation of m1G9 of tRNAGly in vivo, we purified tRNAGly from both wild-type and trm10-Δ/trm10-Δ cells, and analyzed the nucleosides by the same HPLC analysis. The peak for m1G is evident in tRNAGly derived from the wild-type parent (Fig. 5B ▶, upper trace) and absent in the tRNAGly derived from the trm10-Δ/trm10-Δ strain (Fig. 5B ▶, lower trace). Other modified nucleosides known to be present in tRNAGly (m5C and Cm) were identified based on retention time and spectrum of the respective peaks and are labeled in Figure 5B ▶. Quantitation of the amount of modified nucleosides apparent in these tRNAs indicates that <1% m1G remains in tRNAGly isolated from the trm10-Δ/trm10-Δ cells, whereas all other nucleotide modifications remain essentially unchanged, except for dihydrouridine levels, which are slightly altered between the two samples but well within the variation observed because of its proximity to pseudouridine and lack of a clear absorption peak (Table 1 ▶). For tRNA from both wild-type and trm10-Δ strains, we reproducibly observe a higher than expected amount of m5C; this probably reflects an additional m5C modification in tRNAGly that has not been previously reported. Thus, Trm10p activity is the only significant in vivo source of m1G in tRNAGly, and its absence has little effect on other modifications.
TRM10 is responsible for m1G9 modification of other tRNAs in vivo
To examine the in vivo effect of Trm10p on m1G modification of other tRNA species, we have used a primer extension assay. The presence of m1G in a nucleotide sequence should result in generation of a primer extension block at the position immediately before an m1G, because the N-1 methyl group of the m1G residue impairs its ability to form the normal Watson-Crick base pair necessary for continued extension by reverse transcriptase (RT). Because primers used in this assay are specific for an individual tRNA species, this assay allows for rapid analysis of the modification status of multiple tRNAs in a single preparation of bulk RNA. We tested this method by probing tRNAGly from bulk preparations of RNA, using a primer that could hybridize to it at a region ending in the D-loop (shown in bold in Fig. 6A ▶). With tRNA from wild-type cells, a block to primer extension was observed at position G10 (corresponding to C in the sequencing ladder shown in Fig. 6A ▶), because the presence of m1G9 has prevented RT from reading past the modified nucleotide. However, with RNA from the trm10-Δ/trm10-Δ strain, this primer is extended through G9 to the 5′-end of the tRNA, as expected because of the presence of an unmodified G9 in tRNAGly from this strain (Fig. 5B ▶; Table 1 ▶).
FIGURE 6.
Deletion of TRM10 results in loss of m1G9, but not m1G37 in vivo. (A) Establishment of primer extension assay to assess m1G modification. (Left panel) Cloverleaf structure of tRNAGly. Nucleotides in bold indicate the position of the primer used for primer extension analysis of the 5′-end of the tRNA. (Right panel) Primer was used with in vitro transcribed tRNAGly and AMV Reverse Transcriptase to generate a sequencing ladder (lanes G, A, T, and C), and alone (−) or with RNA derived from wild type (wt) or trm10-Δ/trm10-Δ (Δ) strains to analyze for the presence of m1G9 in tRNAGly from each population of RNA. (B) Primer extension to assess modification status of tRNAVal, tRNATrp, tRNAICGArg, and tRNAUUULys at position G9. Primer extensions were performed with either (−) primer alone, (wt) wild-type RNA, or (Δ) trm10-Δ/trm10-Δ RNA. (C) Primer extension to assess modification status of tRNAUAGLeu and tRNAPhe at position G37. Same lanes as B.
The primer extension assay described above was used to analyze m1G9 content in 8 of the 9 other S. cerevisiae tRNA species known to contain m1G9: tRNATrp, tRNAPro, tRNAVal, tRNAiMet, tRNAIle, tRNAICGArg, and both tRNAUCUArg species. Primer extension assays with tRNAAla have been repeatedly unsuccessful for unknown reasons. With the eight tRNAs for which we were able to see primer extension products, the results were the same as that observed for tRNAGly. For example, primer extension of tRNAVal, tRNATrp, and tRNAICGArg from wild-type cells is blocked before reaching the 5′-end of the RNA, at a position consistent with the presence of m1G9 in these tRNA species (Fig. 6B ▶). This primer extension block is completely absent in tRNA from trm10-Δ/trm10-Δ cells, strongly implying a quantitative loss of m1G9 in these cells (Fig. 6B ▶). The same results were observed with the other five tRNA species examined (data not shown). In contrast, control primer extension assays with primers targeting tRNAs that do not contain m1G9 exhibit only the primer extension stop at the 5′-end of the tRNA in RNA derived from either the wild-type or the trm10-Δ/trm10-Δ strain (Fig. 6B ▶, tRNAUUULys). For tRNAICGArg (Fig. 6B ▶) and a few of the other tRNA species (tRNAiMet, tRNAIle, and both tRNAUCUArg species; data not shown), the block appeared as more of a stutter rather than as the block at a single site observed with tRNAGly (Fig. 6A ▶), tRNAVal, tRNATrp (Fig. 6B ▶), and tRNAPro (data not shown). This may reflect a sequence-specific variability in the way AMV-RT deals with the presence of m1G in various sequences; variation in the response of RT to sequences that act as blocks to primer extension has been observed previously (Lorsch et al. 1995). These results demonstrate that TRM10 is required for the catalysis of at least 9 of the 10 m1G modifications observed at position 9 in yeast. It is likely that Trm10p is also responsible for modification of G9 of tRNAAla.
Specificity of Trm10p activity in vivo
The primer extension assay has also been used to confirm that Trm10p is not involved in modification of G37 in vivo. Primers were used to target two tRNA species known to be substrates for Trm5p: tRNAUAGLeu, which contains m1G37, and tRNAPhe, which contains Y37 (wybutosine), the synthesis of which is initiated by formation of m1G37 by Trm5p (Bjork et al. 2001). Primer extension assays demonstrated that the extent of m1G37 modification is unchanged in strains lacking Trm10p (Fig. 6C ▶). This result was confirmed by HPLC analysis of nucleosides generated from another tRNA known to contain m1G37, tRNACAALeu; the m1G content of this tRNA is similar when purified from wild-type or trm10-Δ/trm10-Δ cells (data not shown).
Effects of loss of m1G9 on cell viability
As we have shown above, Trm10p is the major, if not the only, m1G9 tRNA methyltransferase acting in S. cerevisiae cells. However, trm10-Δ/trm10-Δ strains do not exhibit any obvious growth defect when compared with their isogenic TRM10+/TRM10+ parent strain in rich media and in minimal media at several different temperatures (data not shown). In contrast, a trm5-Δ strain grows extremely slowly (Bjork et al. 2001).
The Trm10 methyltransferase family is widely conserved, but is not homologous to the yeast m1G37 methyltransferase, Trm5
Iterative BLAST searches show that Trm10p is a member of a family of proteins found widely in all free-living eukaryotes (but not in the obligate parasite microsporidia, Encephalitozoon cuniculi) and half of all archaeal species sequenced (Fig. 7 ▶) but not in any Eubacteria. Indeed, this family of proteins has been described in both the PFAM database (DUF 425) and the COGs (Cluster of Orthologous Groups, COG2419) database as being conserved, albeit with no ascribed function. Unexpectedly, the database searches also revealed the presence of multiple TRM10-like homologs in each of several metazoan genomes. For instance, the human and mouse genomes harbor at least three homologs, whereas mosquito, fruit fly, and nematode genomes possess at least two. The occurrence of multiple Trm10p family members in metazoans is unexpected because Trm10p is responsible for most, if not all, of the m1G9 tRNA methylation observed in yeast (Fig. 6 ▶).
FIGURE 7.
Alignment of Trm10p homologs. The various Trm10 homologs were identified using iterative BLAST searches, and aligned using CLUSTALX. A representative alignment is shaded to a 50% consensus using MacBoxShade, and indicates residues that are similar (gray background) or identical across all species (black background). The numbers adjacent to the name of each species are the accession numbers for the proteins used. The numbers in parentheses before and after the alignment indicate the numbers of amino acids from individual sequences not included in the alignment for clarity of presentation.
The presence of the archaeal and multiple metazoan homologs prompted a phylogenetic reconstruction of the evolution of this family, presented in Figure 8 ▶. In this phylogeny, the archaeal and eukaryotic homologs clearly group separately from one another. In addition, it is apparent that at least one of the lineages found in metazoans (human #3 in Fig. 8 ▶) appears to branch separately from those that group the human homologs #1 and #2 with yeast Trm10p. This implies two possibilities: either this lineage of proteins predates the human–yeast split; or it has rapidly evolved, causing its phylogenetic placement to be artifactual. Comparison of the human–mouse orthologs for each of lineages 1, 2, and 3 indicates that all three pairs are evolving under similar evolutionary constraints, supporting the argument against artifactual placement in the tree. Therefore, we conclude that there were at least two separate lineages of Trm10-like proteins in eukaryotes, only one of which has been retained in protists, fungi, and plants, whereas metazoans continue to harbor both. One of the Caenorhabditis elegans homologs branches even earlier than the two metazoan lineages, but this may be caused by rapid evolution rather than an ancient divergence, as is the case for a number of C. elegans genes (Blaxter et al. 1998).
FIGURE 8.
Phylogeny of the Trm10p homologs. A phylogeny of the various Trm10p homologs from eukaryotes and archaea was constructed using the neighbor-joining method, rooted on the archaeal homologs. Bootstrap analysis was carried out using the PAUP* version 4.0b10, and bootstrap support (percentage of 1000 trials where a certain grouping was supported) is indicated at the respective nodes. The tree is collapsed to a 50% bootstrap support. The source species of each protein is listed beside the appropriate line, together with the corresponding accession number.
Remarkably, database searches, pairwise BLAST, and comparisons of multiple alignments do not identify even any remote homology between Trm10p and Trm5p, the S. cerevisiae m1G37 methyltransferase (Bjork et al. 2001; see Materials and Methods); thus, these two proteins are likely unrelated, despite their conserved biochemical activity (see also Discussion). We note also that Trm10p was not identified in a search for SAM-dependent methyltransferases in S. cerevisiae based on known SAM binding motifs (Niewmierzycka and Clarke 1999), and that none of our searches revealed any similarity with known methyltransferases. Because Trm10p requires SAM, this indicates that Trm10p may contain a more subtle or a different SAM-binding sequence.
DISCUSSION
We have shown that S. cerevisiae Trm10p, encoded by YOL093w, catalyzes the methylation of G residues to m1G in tRNAGly at position 9. Using a substrate that is uniquely labeled with 32P at the junction between residues 9 and 10, S-adenosylmethionine as a methyl donor, and assays of a genomic array of GST-fusion proteins, we have identified the corresponding protein and gene (Fig. 2 ▶). Cloning of this gene and subsequent overproduction of Trm10p activity in E. coli demonstrates that Trm10p alone is sufficient for catalysis of m1G9 methylation in vitro (Fig. 4 ▶). The product of the Trm10p reaction is m1G, based on comparison of the TLC mobility of the RNase T2 digestion product m1Gp with that produced by modification of G37 of tRNALeu to m1G in extracts (Fig. 3 ▶), and on comparison of the HPLC mobility and the spectrum of m1G derived from Trm10p-modified tRNAGly with a commercial source of m1G (Fig. 5 ▶).
Our data also show that Trm10p is specific for G9 of tRNA substrates. Extracts from trm10-Δ/trm10-Δ strains lack m1G9 methyltransferase activity but have m1G37 methyltransferase activity, as expected if Trm10p catalyzes modification of position 9, but not at position 37 (Fig. 3 ▶). In addition, because only 1 mole of m1G is produced during modification of tRNAGly transcript in vitro (Table 1 ▶), because the modified site is known to be position 9 with the G9*Gly substrate (Fig. 2 ▶), and because this is the only position known to be modified to m1G in tRNAGly, we conclude that Trm10p primarily modifies G9 in vitro.
Finally, we have shown that Trm10p is required in vivo for methylation of G9 in 9 of the 10 yeast tRNAs known to have this modification, based on two results. First, a comparison of tRNAGly purified from wild-type and trm10-Δ cells shows that m1G modification is dependent on Trm10p (Fig. 5 ▶). Second, RT pauses at position 10 in primer extension assays of 9 tRNAs species known to contain m1G9, but not if the tRNAs were isolated from an otherwise isogenic trm10-Δ/trm10-Δ strain (Fig. 6 ▶).
There is a clear demarcation of position specificity for m1G modification by Trm10p at G9, as shown here, and by Trm5p at G37 of substrate tRNAs (Bjork et al. 2001). This strict site specificity for what is essentially the same chemical reaction is similar to that observed with S. cerevisiae Tad1p and Tad2p/Tad3p, which are responsible for formation of I37 and I34, respectively (Gerber et al. 1998; Gerber and Keller 1999), and with Pus4p and Pus6p, which are responsible for formation of Ψ55 and Ψ31, respectively (Becker et al. 1997; Ansmant et al. 2001). Other tRNA modifications that occur at more than one position are catalyzed by proteins with region-specific modification activity, such as Pus3p, which forms Ψ38 and Ψ39 (Lecointe et al. 1998), and Trm7p, which 2′-O-methylates at positions 32 and 34 (Tsai et al. 2002). In contrast, some proteins exhibit promiscuous modification activity at multiple sites, such as Trm4p, which catalyzes m5C formation at positions 34, 40, 48, and 49 (Motorin and Grosjean 1999), and Pus1p, which catalyzes Ψ formation at up to eight widely different positions in tRNA substrates (Simos et al. 1996; Motorin et al. 1998).
The mechanistic basis for the site specificity and catalysis of m1G modification by Trm5p and Trm10p is an intriguing question that remains to be addressed by further study. A search for homologs of each of these two S. cerevisiae proteins by BLAST yields a distinct family of enzymes for each one (Fig. 7 ▶; data not shown). Independent alignments of these two predicted families of m1G methyltransferases yield core sequences of conserved amino acids within each group; however, none of these blocks of conserved sequence are shared between the two groups of enzymes. It is possible that Trm5p and Trm10p share a common chemical mechanism for m1G methylation but that the amino acids in the active site are hard to identify. Alternatively, the two enzymes may catalyze m1G modification in completely distinct ways. Further analysis of the mechanism of these two enzymes will help to address this question and may elucidate how Trm5p and Trm10p each recognize their different positions and tRNA substrates for m1G methylation.
Another intriguing specificity question is how Trm10p is able to distinguish between tRNA species to be modified and those to remain unmodified at position 9. Every eukaryotic tRNA sequenced to date contains a purine at position 9, with the exception of two tRNAHis species, one from Drosophila and one from sheep. In S. cerevisiae position 9 is occupied most often by G (in 23 out of 34 tRNAs sequenced); however, the mere presence of a G at position 9 is not the determining factor for m1G formation. In S. cerevisiae there are 10 tRNA species that contain m1G9 (tRNAGly, tRNATrp, tRNAPro, tRNAVal, tRNAICGArg, both tRNAUCUArg species, tRNAIle, tRNAiMet, and tRNAAla), but there are 12 other tRNA species that contain a G at position 9 that remains unmodified in vivo (Sprinzl et al. 1998). There is no obvious primary sequence similarity in the region of position 9 that would explain m1G modification at this site. One possibility is that m1G9 is formed either in response to, or to participate in, a common structural feature. An interaction involving the residue at position 9 has been observed in the crystal structure of tRNAPhe, in which a base-triple has been observed between an A at position 9 and the U12–A23 base pair via a reverse-Hoogsteen interaction between the non-Watson-Crick faces (N7 and N6) of A9 and A23 (Kim et al. 1974). An investigation of the sequences of the 22 G9-containing elongator tRNAs from S. cerevisiae yields no sequence pattern for the base pair at position 12–23 that covaries with the modification status of G9 that would indicate that m1G9 participates in a similar structural feature (Sprinzl et al. 1998). However, in eukaryotic initiator tRNAs position 9 is overwhelmingly found to be a G in eukaryotes (17 out of 18 tRNAs sequenced) and, moreover, that G is always modified to m1G (Sprinzl et al. 1998; Marck and Grosjean 2002). Interestingly, the base pair at position 12–23 in the m1G9-containing tRNAiMet species is invariably a G12–C23 base pair. Although the electron density of the m1G9 in the crystal structure of S. cerevisiae tRNAiMet is diffuse and therefore a specific interaction cannot be proven, it does appear to be in the correct position and orientation to interact with the G12–C23 base pair in an analogous interaction to that observed between A9–A23–U12 in tRNAPhe (Basavappa and Sigler 1991). If such an interaction does occur, it may be that m1G9, although present in other tRNA species, plays its most important role in the correct structure or function of eukaryotic initiator tRNAs.
Trm10p is a highly conserved protein that is widely found in eukaryotes (Figs. 7, 8 ▶ ▶), consistent with the widespread occurrence of m1G9 in eukaryotic tRNAs. The finding of multiple paralogous proteins in some metazoans is unexpected, and raises the question of their origin and function. The fact that these multiple homologs are no more related to each other than to other members of the Trm10p family indicates that the duplication occurred prior to the divergence of eukaryotic species, implying that organisms such as yeast have lost the duplicated members after their divergence. It seems likely that at least one of the different paralogs in humans and other organisms is responsible for G9 methylation of tRNAs. The other paralogs may have altered substrate specificity, contributing to m1G modification of specific subclasses of tRNAs or other RNAs, or they may have altered tissue specificity; alternatively, they may have evolved other functions. The presence of related archaeal homologs implies that they too may be m1G9 methyltranferases, because at least one archaeal organism, Halobacterium cutirubrum, has tRNA with this modification (Gu et al. 1983), although whether H. cutirubrum has a Trm10p homolog cannot be ascertained at this point owing to lack of sequence information.
We note that the pattern of m1G9 modification in tRNAs from other eukaryotes is also not obvious, either by the subset of G9 residues that are modified to m1G9, or by the identity of the tRNA species that are modified to m1G9 in each organism. For example, the yeast tRNAs for Ala, Gly, Ile, Pro, Arg, Val, Trp, and initiator tRNA all contain m1G9, whereas in humans only Asn, Gln, and initiator tRNAs have been shown to have this modification, and at least one sequenced tRNAGly species has an unmodified G at position 9. All told, at least six other tRNAs (those for Asp, Glu, Met, Asn, Gln, and Thr) are modified with m1G9 in some eukaryotes, but not in yeast. This indicates that the enzymes from different eukaryotes have evolved ways to distinguish between different sets of tRNAs to be modified, and may provide an explanation for why there are multiple paralogs of Trm10p in some metazoans. Cloning and analysis of other eukaryotic Trm10p homologs, particularly from species with multiple paralogs, will help to address these issues of specificity.
It is noteworthy that the extended superfamily of Trm5p homologs includes proteins that modify substrates other than tRNA, including rRNA and DNA. The finding of other methyltransferases in the Trm5 protein family, although still not identifying Trm10p, underscores their evolutionary unrelatedness. Although the Trm10 family of proteins is smaller, our phylogenetic analysis indicates that there may have been functional diversification in this family of proteins as well; further analysis of the multiple homologs present in metazoans will be required to elucidate any diverse functions.
The cellular role of m1G9 modification is unclear at present, despite the evolutionary conservation of this modification and the conservation of the protein (Figs. 7, 8 ▶ ▶). Cells lacking Trm10p have no obvious growth defects under standard growth conditions in rich or minimal media. For a number of other mutants of tRNA modification enzymes, dramatic phenotypes are only manifested in combination with other mutations that alter tRNA modification or tRNA levels (Grosshans et al. 2001; Johansson and Bystrom 2002). Further tests of this type may need to be done to clarify the role of Trm10 protein. The question of why this modification (and other modifications) is so widely conserved in eukaryotes is an important one because tRNA molecules are among the most abundant nucleic acids in the cell, and therefore a large amount of cellular energy is being spent on production of modifications. From our estimation of the number of mature tRNA molecules produced during a generation of growth in S. cerevisiae, methylation at position 9 alone requires the consumption of ∼720,000 molecules of SAM per generation.
MATERIALS AND METHODS
Preparation of specifically labeled tRNA substrates
tRNAGly specifically labeled at the phosphate following guanosine 9 (G9*Gly) was prepared as follows. In vitro transcribed tRNAGly (1 nmole) was incubated with Rnase H (a gift from Y.-T. Yu, University of Rochester Medical Center) and a chimeric 2′-O-methylated/deoxy oligonucleotide (HHMI/Keck Center, Yale) to direct cleavage 3′ of G9 (Yu 1999). After gel purification of the resulting 3′ half-molecule, 100 pmole was treated with alkaline phosphatase (Roche), and then rephosphorylated with T4 Polynucleotide kinase (PNK; Roche) in the presence of [γ-32P]ATP (ICN, 7000 Ci/mmole) to yield a labeled, 5′-phosphorylated 3′ half-molecule (beginning at G10) that was gel-purified. The labeled RNA (40–50 pmole) was ligated with a synthetic RNA comprising the first nine nucleotides of tRNAGly (100–200 pmole; Dharmacon Research), using a DNA oligonucleotide (100–200 pmole) designed to bridge the desired ligation site, 80 U of T4 DNA ligase (USB), and incubation in buffer containing 1× T4 DNA ligase reaction buffer (USB) supplemented with an additional 1 mM ATP at 37°C for 4–12 h. Ligated RNA was purified by PAGE. Typical ligation efficiencies were 50%–70% of labeled 3′ molecule ligated.
G37*Leu substrate was produced in essentially the same way, except that the chimeric oligonucleotide used in the RNase H reaction directed cleavage 3′ of G37 in in vitro transcribed mature tRNACAALeu, and the 5′ half-molecule used for the subsequent ligation was PAGE-purified from the same RNase H reaction.
Primer extension assays were performed on the purified 3′ fragments of RNA produced by RNase H to demonstrate that cleavage occurred at the correct position for each RNA. Ligated reaction products were analyzed by RNase T2 and P1 nuclease cleavage reactions to determine that the correct junction sequence was obtained after ligation.
Methyltransferase assay conditions
m1G methyltransferase activity was assayed for both G9*Gly and G37*Leu tRNA substrates, in 10-μL reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 2.5 mM MgCl2, 1 mM dithiothreitol (DTT), 50 mM ammonium acetate, 0.05 mM ethylenediaminetetraacetic acid (EDTA), 1 mM spermidine, 0.5 mM S-adenosylmethionine (SAM), 5000–10,000 cpm of the corresponding specifically labeled tRNA, and a source of m1G methyltransferase activity (crude extract or purified protein). Reactions were incubated at 30°C for 1–8 h as indicated, and then stopped by the addition of 90 μL of 0.5 M Tris-HCl (pH 8.0) containing 20 μg of carrier tRNA, followed by phenol extraction and ethanol precipitation of the RNA. RNA was resuspended in a 4-μL solution containing 20 mM ammonium acetate (pH 5.2), 1 mM EDTA, 10 μg of RNase A, and 1 U of RNase T2, and incubated at 50°C for 1 h, and then digestion products were applied to cellulose TLC plates (EM Science) and resolved in buffer containing saturated ammonium sulfate:isopropanol:water (80:18:2).
GST-ORF fusion proteins
The pools and subpools of purified GST-ORF fusion proteins were prepared as previously described (Martzen et al. 1999).
Strains and plasmids
The homozygous diploid strain deleted for YOL093w (trm10-Δ/trm10-Δ) and its isogenic parent strain (BY4743) were obtained from Invitrogen. Plasmid pJEJ12-3 (N-terminal His6-Trm10p) was constructed by PCR amplification of the yeast TRM10 ORF, followed by insertion of the DNA into a pET-derived vector specifying the N-terminal sequence Met-Ala-His6 before the first Met codon (gift of E. Grayhack, University of Rochester School of Medicine), and sequencing of the resulting insert.
Growth and preparation of crude yeast extracts
To prepare crude extracts, 250 mL of each strain was grown from single colonies in YPD media to an OD600 of 2, and cells were harvested, resuspended in 1 mL of extraction buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 4 mM MgCl2, 5 mM DTT, 10% glycerol, 1 M NaCl, with 2.5 μg/mL leupeptin and 2.5 μg/mL pepstatin, supplemented with 4 mM AEBSF and sheared with a bead-beater containing zirconium beads. Extracts were clarified by centrifugation; protein concentration was determined by BioRad protein assay, and samples were quick-frozen and stored at −80°C.
Purification of Trm10p
Plasmid pJEJ12-3 (N-terminal His6-Trm10p) was freshly transformed into E. coli BL21(DE3) pLysS cells (Novagen), and an individual transformant was grown at 37°C in 500 mL of LB containing 100 μg/mL ampicillin to OD600 = 0.4, and induced for 5 h by the addition of 1 mM isopropyl β-D-thiogalactopyranoside (IPTG). Purification of His6-Trm10p was performed essentially as previously described (Steiger et al. 2001), using TALON resin (Clontech) for immobilized metal-ion affinity chromatography (IMAC), except that the protein bound to the TALON resin was eluted using a gradient of 0–100 mM imidazole. Trm10p eluted as a single peak at ∼80–90 mM imidazole and was dialyzed into buffer containing 20 mM Tris-HCl (pH 7.5), 4 mM MgCl2, 2 mM EDTA, 55 mM NaCl, 1 mM DTT, and 50% glycerol for storage at −20°C. The purified protein (6.9 mg/mL) was estimated to be >95% pure on SDS-PAGE by Coomassie staining.
Preparation of small-molecular-weight RNA from yeast
To prepare RNA, the wild-type parent strain BY4743 (TRM10+/TRM10+) and the trm10-Δ/trm10-Δ strain were grown in 125 mL of YPD to an OD600 of 2–3. Cells were harvested, and RNA was extracted with hot phenol, precipitated twice with ethanol, and resuspended in 1 mL of ddH2O essentially as described previously (Rubin 1975) and stored at −80°C. The concentration of the final purified RNA was calculated by assuming A260 of 40 μg/mL RNA = 1.
Purification of tRNAGly from yeast
tRNAGly was purified from total yeast cellular RNA using a 5′-biotinylated oligonucleotide (Integrated DNA Technologies) complementary to the 3′-end of tRNAGly. Binding of the biotinylated oligonucleotide to streptavidin (SA) magnetic particles (Roche) was performed according to the manufacturer’s instructions. The tRNAGly was purified from 1 mg of total cellular RNA using 2 mg of bound beads according to the method of Tsurui et al. (1994). The eluted tRNA was centrifuged at 13,000 rpm for 10 min to remove any residual beads and desalted/concentrated by centrifugation using a Centricon YM-10 (Amicon) with successive concentration and dilution into ddH2O. The concentration of purified tRNA was determined using A260; ∼10–15 μg of purified tRNA was recovered per milligram of total cellular RNA. The same procedure was used to individually purify tRNACAALeu from wild-type and trm10-Δ-derived RNAs using a biotinylated oligonucleotide complementary to the 3′-end of this tRNA.
HPLC analysis of nucleosides
Purified tRNAGly isolated from yeast (1 μg) or in vitro transcribed tRNAGly (20 μg) incubated with Trm10p or buffer was treated with 4 μg of P1 nuclease in buffer containing 30 mM sodium acetate (pH 5.2) and 0.2 mM ZnCl2 at 37°C for 16 h, and then with 8 U of calf intestinal alkaline phosphatase (Roche) in 1× alkaline phosphatase reaction buffer at 37°C for 3 h. The resulting nucleosides were resolved by HPLC (Waters Alliance Model 2690, equipped with Waters 996 photodiode array detector) on a reverse phase C18 column (supelcosil LC-18-T, 25 cm × 4.6 μm; Supelco, Inc.) essentially as described (Gehrke and Kuo 1989), and individual spectra of the nucleosides were used to confirm the assignments. N-1 methylguanosine (m1G) was obtained from R.I. Chemical and analyzed in the same way.
Quantification of modified nucleosides was carried out by measuring the area under each nucleoside peak at its known maximum absorbance. The areas of the four main nucleosides (C, U, G, and A) were used to determine the total number of moles of tRNA in each injection by dividing the area observed by the known extinction coefficient for each nucleoside and then normalizing for the expected number of that nucleoside in the tRNA being analyzed. The moles of modified nucleotide in each injection were then determined by dividing the area by the known extinction coefficient for each modified nucleoside; this number was compared with the total moles of tRNA in the sample to obtain the ratio of modified nucleoside per tRNA.
Primer extension assays
Primer extension experiments were carried out using 15–20 nt primers (Integrated DNA Technologies) that were 5′-end-labeled with T4 PNK and [γ-32P]ATP (ICN, 7000 Ci/mmole), followed by centrifugation through a MicroBioSpin 6 column (BioRad) to remove unincorporated ATP. In a 5-μL reaction containing 50 mM Tris-HCl (pH 8.3), 30 mM NaCl, and 10 mM DTT, 1 pmole of each primer was annealed with 2 μg of total RNA (isolated as described above) by heating to 95°C for 3 min followed by slow cooling to 37°C. Then 2 μL of annealed RNA/primer was added to a 5-μL reaction containing 0.1–0.4 mM each dNTP (G, A, T, and C) and 0.7 U of AMV-Reverse transcriptase (Promega, high concentration) in 1× AMV-RT reaction buffer (Promega). Sequencing reactions additionally contained 0.2 mM of each ddNTP (G, A, T, or C) as indicated. After 5 min at room temperature, extensions were allowed to continue at 37°C for 30 min–1 h and then stopped by the addition of 7 μL of formamide containing 0.2 mg/mL carrier RNA, 0.1% bromophenol blue, and 0.1% xylene cyanol. Reactions were run on 10% polyacrylamide, 4 M urea gels and visualized using the PhosphorImager (Molecular Dynamics).
Bioinformatics and phylogenetic analysis
Using the yeast Trm10p as the query sequence, PSI-BLAST searches were carried out (Altschul et al. 1997). These led to the identification of archaeal homologs in the second iteration, which were also used as queries in subsequent PSI-BLAST analyses to obtain a comprehensive list of Trm10p homologs. To ascertain if there was any homology between the m1G9 and m1G37 methyltransferases, we used the Blocks database (http://blocks.fhcrc.org; Henikoff et al. 2000); preconstructed blocks corresponding to the m1G37 methyltransferases (IPB002905) were used as queries to search both the list of individual Trm10p homologs using PSI-BLAST, or consensus blocks constructed from the Trm10p homologs, using a LAMA search (Pietrokovski 1996). In neither case was any significant homology found. The Trm10p homologs were aligned using CLUSTALX (Thompson et al. 1997); the alignment was used to generate a phylogeny using the neighbor-joining program (Saitou and Nei 1987), and bootstrap analysis was carried out in the PAUP* suite (Swofford 2002).
Acknowledgments
We are grateful to Beth Grayhack, Andrei Alexandrov, and Feng Xing for extensive helpful advice and discussion during preparation of this manuscript and to Martha Wilkinson for preparation of some of the subpools of GST-ORF fusion proteins. This research was supported by grants GM52347 and HG02311 to E.M.P. from the National Institutes of Health, a Helen Hay Whitney postdoctoral fellowship to H.S.M., and NIH postdoctoral fellowship 2T32CA09363 to J.E.J.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5070303
REFERENCES
- Alexandrov, A.V., Martzen, M.R., and Phizicky, E.M. 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8: 1253–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansmant, I., Motorin, Y., Massenet, S., Grosjean, H., and Branlant, C. 2001. Identification and characterization of the tRNA:Psi 31-synthase (Pus6p) of Saccharomyces cerevisiae. J. Biol. Chem. 276: 34934–34940. [DOI] [PubMed] [Google Scholar]
- Basavappa, R. and Sigler, P. 1991. The 3 Å crystal structure of yeast indicator tRNA: Functional implications in initiator/elongator discrimination. EMBO J. 10: 3105–3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, H.F., Motorin, Y., Planta, R.J., and Grosjean, H. 1997. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res. 25: 4493–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork, G.R. 1995. Biosynthesis and function of modified nucleosides. In tRNA: Structure, biosynthesis and function (eds. D. Soll and U.L. RajBhandary), pp. 165–205. ASM Press, Washington, DC.
- Bjork, G.R., Jacobsson, K., Nilsson, K., Johansson, M.J., Bystrom, A.S., and Persson, O.P. 2001. A primordial tRNA modification required for the evolution of life? EMBO J. 20: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaxter, M.L., De Ley, P., Garey, J.R., Liu, L.X., Scheldeman, P., Vierstraete, A., Vanfleteren, J.R., Mackey, L.Y., Dorris, M., Frisse, L.M., et al. 1998. A molecular evolutionary framework for the phylum Nematoda. Nature 392: 71–75. [DOI] [PubMed] [Google Scholar]
- Gehrke, C.W. and Kuo, K.C. 1989. Ribonucleoside analysis by reversed-phase high-performance liquid chromatography. J. Chromatogr. 471: 3–36. [DOI] [PubMed] [Google Scholar]
- Gerber, A.P. and Keller, W. 1999. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286: 1146–1149. [DOI] [PubMed] [Google Scholar]
- Gerber, A., Grosjean, H., Melcher, T., and Keller, W. 1998. Tad1p, a yeast tRNA-specific adenosine deaminase, is related to the mammalian pre-mRNA editing enzymes ADAR1 and ADAR2. EMBO J. 17: 4780–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean, H., Szweykowska-Kulinska, Z., Motorin, Y., Fasiolo, F., and Simos, G. 1997. Intron-dependent enzymatic formation of modified nucleosides in eukaryotic tRNAs: A review. Biochimie 79: 293–302. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., Lecointe, F., Grosjean, H., Hurt, E., and Simos, G. 2001. Pus1p-dependent tRNA pseudouridinylation becomes essential when tRNA biogenesis is compromised in yeast. J. Biol. Chem. 276: 46333–46339. [DOI] [PubMed] [Google Scholar]
- Gu, X.R., Nicoghosian, K., Cedergren, R.J., and Wong, J.T. 1983. Sequences of halobacterial tRNAs and the paucity of U in the first position of their anticodons. Nucleic Acids Res. 11: 5433–5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun, T.R. and Fields, S. 2002. A genome-wide screen for site-specific DNA-binding proteins. Mol. Cell. Proteomics 1: 538–543. [DOI] [PubMed] [Google Scholar]
- Henikoff, J.G., Greene, E.A., Pietrokovski, S., and Henikoff, S. 2000. Increased coverage of protein families with the blocks database servers. Nucleic Acids Res. 28: 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper, A.K. and Phizicky, E.M. 2003. tRNA transfers to the limelight. Genes & Dev. 17: 162–180. [DOI] [PubMed] [Google Scholar]
- Johansson, M.J. and Bystrom, A.S. 2002. Dual function of the tRNA(m5U54)methyltransferase in tRNA maturation. RNA 8: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.H., Suddath, F.L., Quigley, G.J., McPherson, A., Sussman, J.L., Wang, A.H., Seeman, N.C., and Rich, A. 1974. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185: 435–440. [DOI] [PubMed] [Google Scholar]
- Lecointe, F., Simos, G., Sauer, A., Hurt, E.C., Motorin, Y., and Grosjean, H. 1998. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of ξ 38 and ξ 39 in tRNA anticodon loop. J. Biol. Chem. 273: 1316–1323. [DOI] [PubMed] [Google Scholar]
- Lorsch, J.R., Bartel, D.P., and Szostak, J.W. 1995. Reverse transcriptase reads through a 2′–5′ linkage and a 2′thiophosphate in a template. Nucleic Acids Res. 23: 2811–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck, C. and Grosjean, H. 2002. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8: 1189–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martzen, M.R., McCraith, S.M., Spinelli, S.L., Torres, F.M., Fields, S., Grayhack, E.J., and Phizicky, E.M. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286: 1153–1155. [DOI] [PubMed] [Google Scholar]
- Motorin, Y. and Grosjean, H. 1999. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA 5: 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin, Y., Keith, G., Simon, C., Foiret, D., Simos, G., Hurt, E., and Grosjean, H. 1998. The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA 4: 856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewmierzycka, A. and Clarke, S. 1999. S-Adenosylmethyionine-dependent methylation in Saccharomyces cerevisiae. J. Biol. Chem. 274: 814–824. [DOI] [PubMed] [Google Scholar]
- Pietrokovski, S. 1996. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 24: 3836–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G.M. 1975. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 12: 45–64. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Simos, G., Tekotte, H., Grosjean, H., Segref, A., Sharma, K., Tollervey, D., and Hurt, E.C. 1996. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 15: 2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger, M.A., Kierzek, R., Turner, D.H., and Phizicky, E.M. 2001. Substrate recognition by a yeast 2′-phosphotransferase involved in tRNA splicing and by its Escherichia coli homolog. Biochemistry 40: 14098–14105. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), Version 4. Sinauer Associates, Sunderland, MA.
- Thompson, J., Gibson, T., Plewniak, F., Jeanmougin, F., and Higgins, D. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, E.Y., Thim, S., Baena, A., Boussiotis, V.A., Reynes, J.M., Sath, S., Grosjean, P., Yunis, E.J., Goldfeld, A.E., and Pintard, L. 2002. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. Proc. Natl. Acad. Sci. 99: 7576–7581.12032325 [Google Scholar]
- Tsurui, H., Kumazawa, Y., Sanokawa, R., Watanabe, Y., Kuroda, T., Wada, A., Watanabe, K., and Shirai, T. 1994. Batchwise purification of specific tRNAs by a solid-phase DNA probe. Anal. Biochem. 221: 166–172. [DOI] [PubMed] [Google Scholar]
- Xing, F., Martzen, M.R., and Phizicky, E.M. 2002. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA 8: 370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.T. 1999. Construction of 4-thiouridine site-specifically substituted RNAs for cross-linking studies. Methods 18: 13–21. [DOI] [PubMed] [Google Scholar]