Abstract
Mammalian cells express small and large forms of the RNA editing enzyme ADAR1, referred to as ADAR1-S and ADAR1-L, respectively. Here we observed that ADAR1-L was >70-fold more active than was ADAR1-S when assayed with a substrate that could be edited in either the nucleus or cytoplasm, and was also much more active when assayed with a substrate that was generated in the cytoplasm during viral replication. In contrast, when a substrate that could only be edited within the nucleus was assayed, the activity of ADAR1-S was found to be somewhat higher than that of ADAR1-L. We show here not only that editing could occur in the cytoplasm but also that the process was extremely efficient, occurred rapidly, and could occur in the absence of translation. Consistent with the observation that editing in the cytoplasm can be very efficient, deletion of the nuclear localization signal from ADAR2 resulted in a protein with 15-fold higher activity when tested with a substrate that contained an editing site in the mature message. In addition to its potential role in an antiviral response, we propose that ADAR1-L is the form primarily responsible for editing mRNAs in which the editing site is retained after processing.
Keywords: ADAR1, adenosine deaminase, HDV, inosine, RNA editing
INTRODUCTION
The ADAR (adenosine deaminase that act on RNA) family of RNA-editing enzymes catalyzes the deamination of adenosine to inosine in perfect and imperfect duplex RNA. This family includes two members, ADAR1 and ADAR2, that have been shown to be catalytically active. Transcripts that can be edited by ADARs include pre-mRNAs for mammalian glutamate (Sommer et al. 1991;Higuchi et al. 1993;Lomeli et al. 1994) and serotonin (Burns et al. 1997) receptors, the rat-ADAR2 pre-mRNA (Rueter et al. 1999) and hepatitis delta virus (HDV) antigenomic RNA (Polson et al. 1996). When editing of these transcripts occurs within coding sequence, the resulting mRNA expresses a different form of the protein from that which is genomically encoded. Hence, selective A-to-I editing serves as a means for generating protein diversity. In several ADAR substrates, the editing site partially resides within an intron such that the spliced transcript cannot be edited. For this reason, ADAR-mediated editing is generally thought to occur in the nucleus as nascent transcripts are being synthesized.
The pre-mRNA of the glutamate receptor subunit B (GluR-B) is the best-characterized substrate of ADAR1 and ADAR2. This message encodes a subunit of a glutamate-gated ion channel. Specific editing of the transcript occurs at three main positions: the R/G, Q/R, and +60 sites. Deamination of two of these sites results in coding changes in which the genome-encoded CAG (Q) is converted to CIG (R) at the Q/R site, and AGA (R) is converted to IGA (G) at the R/G site. These changes result in proteins with altered properties. By using mice homozygously deleted for ADAR2, the GluR-B Q/R site was shown to be the most important physiological substrate of ADAR2 (Higuchi et al. 2000), whereas mice heterozygously deleted for ADAR1 die as embryos (Wang et al. 2000).
RNA editing is an essential step in the life cycle of HDV. This process allows the virus to express its two proteins, the small δ antigen (HDAg-S) and the large δ antigen (HDAg-L), from the same coding sequence. Editing of a replication intermediate, referred to as the antigenome, converts the UAG amber codon of the HDAg-S to a UIG tryptophan codon (Casey and Gerin 1995). The mRNA that results from replication and transcription of edited antigenomes has an extended ORF and enables translation of HDAg-L. Although both ADAR1 and ADAR2 are able to edit the HDV amber/W site efficiently in vivo (Sato et al. 2001), the endogenous activity in tissue culture cells responsible for editing HDV RNA during replication is ADAR1 (Wong and Lazinski 2002).
Through the use of biochemical assays that monitored deaminase activity, ADAR1 was purified and cloned from mammalian cells (Kim et al. 1994;Lai et al. 1995;O‘Connell et al. 1995). Soon thereafter, two other laboratories independently identified ADAR1: chicken ADAR1 was purified in a search for Z-DNA binding proteins (Herbert et al. 1995), and human ADAR1 was identified as an interferon inducible message (Patterson and Samuel 1995;Patterson et al. 1995). The latter investigators described two different molecular-weight forms of ADAR1: a full-length 150-kD form (ADAR1-L) and an 110-kD (ADAR1-S) form. They showed that ADAR1-L is localized to both the cytoplasm and nucleus, and its expression is induced by interferon. In contrast, ADAR1-S, which lacks the N-terminal 295 amino acids found in ADAR1-L, localizes to the nucleus and is constitutively expressed. In most cells, ADAR1-L is expressed at a much lower level than is ADAR1-S.
The N terminus of ADAR1-L has several sequences and domains absent from ADAR1-S. These include a region with potential sites for arginine dimethylation (RG repeats); a nuclear export signal (NES; Poulsen et al. 2001); and a domain (Zα) that binds Z-DNA in vitro (Schade et al. 1999). Downstream of Zα in the human gene are two duplicated sequences of 49 amino acids. Only one copy of this repeat is found in rodent ADAR1-L. The role in editing of the domains unique to ADAR1-L has largely not been examined.
ADAR1-L is thought to antagonize viral infections because its expression is interferon-inducible and because its cytoplasmic localization would provide it access to viruses replicating in that compartment. Consistent with such a role, vaccinia virus expresses a protein called E3L that has been shown to antagonize ADAR1 editing in vitro (Liu et al. 2001). Moreover, E3L and ADAR1-L share homology and both contain Z-DNA binding and double-stranded RNA binding motifs (Patterson and Samuel 1995). Also consistent with a role for ADAR1-L in an antiviral response, there are several reports of adenosine to guanosine hypermutation observed in cDNA sequences obtained from different RNA viruses (Cattaneo 1994).
It has previously been shown that the two forms of ADAR1 have comparable deaminase activity in vitro (Patterson and Samuel 1995;Liu et al. 1999). However, comparison of the activity of these two forms in vivo, in a site-specific editing assay has not been performed. In the present study, we express either ADAR1-L or ADAR1-S in conjunction with an editing reporter derived from the HDV amber/W site to compare their activity in HEK293 cells. In this reporter, the editing site was retained in the mature message. We report that ADAR1-L was >70-fold more active than ADAR1-S with the HDV amber/W editing reporter in this in vivo assay. Through additional experiments, we showed that the elevated activity of ADAR1-L largely resulted from its cytoplasmic localization, and editing in the cytoplasm can be an extremely efficient process.
RESULTS
ADAR1-L was more active than ADAR1-S in vivo
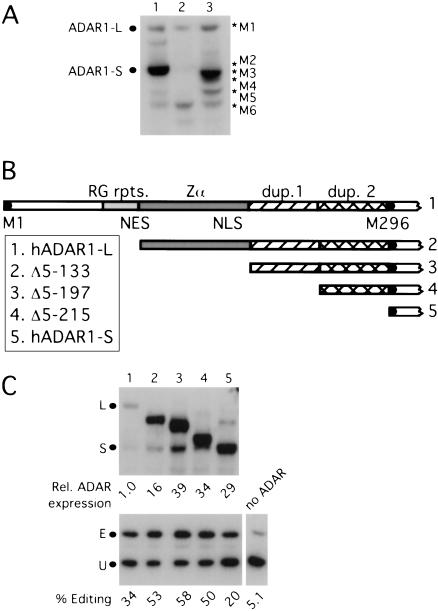
The hADAR1 expression vector used previously (Sato et al. 2001;Wong et al. 2001) contains an HA epitope at the C terminus of the ADAR1 ORF and includes most of the native 5′ UTR from exon 1A (George and Samuel 1999). The HA epitope was used so that protein expressed from the vector could be distinguished from that expressed endogenously in untransfected cells, and we observed that the epitope tag did not alter the activity of the resulting protein. Even though the cDNA in that vector encodes ADAR1-L, we observed much more expression of ADAR1-S than of ADAR1-L (Fig. 1A ▶, lane 1). In an attempt to enhance the expression of ADAR1-L, we mutated the second methionine of ADAR1-L at position 296 to alanine. With this mutant, we observed the disappearance of ADAR1-S but an increase in the expression of faster migrating species (Fig.1A ▶, lane 3). When we mutated the second through fifth methionines to alanines, we observed the expression of a protein, whose mobility was consistent with that predicted after initiation from the sixth methionine (Fig. 1A ▶, lane 2). We concluded that hADAR1-S was translated by internal initiation from a message that encodes hADAR1-L.
FIGURE 1.
Expression of methionine and N-terminal deletion mutants of hADAR1-L. (A) Expression of methionine mutants. The anti-HA Western analysis shows the expression of the two forms of ADAR1 from a C-terminal HA-tagged full-length hADAR1-L expression vector (lane 1). Lane 3 shows protein expression when the second methionine at position 296 of ADAR1-L was mutated to an alanine, and lane 2 shows protein expression when the second through fifth methionines of ADAR1-L were mutated to alanines. M1–M6 refer to the proteins that were predicted to initiate from the first to sixth methionines. (B) Domains present in hADAR1-L but absent in hADAR1-S and the deletion mutants. M1 is the first methionine, and M296 is the second methionine at position 296. RG rpts. indicates arginine/glycine repeats; NES, nuclear export signal, Zα, Z DNA binding domain; NLS, putative nuclear localization signal; and dup. 1 and 2, duplicated sequences. Constructs 2 to 4 have amino acid residues at positions 5–133 (2), 5–197 (3), and 5–215 (4) deleted from ADAR1-L. (C) Expression and editing activity of ADAR1 forms and deletion mutants. The anti-HA Western analysis (top) shows the expression of the C-terminal HA-tagged proteins. The anti-δ antigen (HDAg) Western analysis (bottom) shows the corresponding editing activity with the HDV amber/W reporter (pSS74). L and S mark the positions of ADAR1-L and ADAR1-S, and E and U are the edited and unedited products.
To determine the contribution of the two forms of ADAR1 to editing activity, we next constructed vectors that individually express either ADAR1-L or ADAR1-S. Because mutation of the second through fifth methionines did not increase initiation from the first methionine, we reasoned that the latter is a poor initiator. Consistent with this possibility, we note that there is an out-of-frame AUG upstream of the initiator within the exon 1A UTR that might hinder recognition of the initiator by scanning ribosomal subunits. We next constructed an ADAR1-L expression vector in which the 5′UTR was deleted, and an optimal consensus AUG initiator was inserted at position 1. An ADAR1-S expression vector was constructed by deleting sequences upstream of the M296 initiator. In addition, we constructed N-terminal deletions of ADAR1-L to define the contribution to editing provided by each of the domains unique to that protein (Fig. 1B ▶).
We tested these ADAR1 expression vectors in an in vivo editing assay by using an HDV amber/W editing reporter (Sato et al. 2001;Wong et al. 2001). This reporter is an mRNA that can fold into the antigenomic rod-like structure, thereby creating a functional amber/W site. Prior to editing, the reporter encodes HDAg-S. Editing of the reporter at the amber/W site converts the stop codon of HDAg-S to UIG, allowing for translation of HDAg-L. Thus, the ratio of expression of the large to total δ antigens reflects the extent of editing of this reporter. Figure 1C ▶ shows that even though ADAR1-L was expressed at a 29-fold lower level than that of ADAR1-S, it edited the HDV reporter more efficiently (cf. lanes 1 and 5). Also, note that deletion of amino acids at positions 5–133 of ADAR1-L caused an increase in protein expression (cf. lanes 1 and 2).
To test if the elevated editing activity of hADAR1-L was a unique attribute of the human enzyme, we compared the editing activity of mouse (m) ADAR1-L and mADAR1-S. Even though mADAR1-L was expressed at a lower level, it still edited the HDV reporter much more efficiently than did mADAR1-S (data not shown). To determine if the difference in activity of hADAR1-L in relation to hADAR1-S reflected a unique property of the HDV amber/W site, we compared the ability of the two forms of hADAR1 to edit the GluR-B R/G and +60 sites. In both cases, we observed that hADAR1-L was far more active than was hADAR1-S (data not shown).
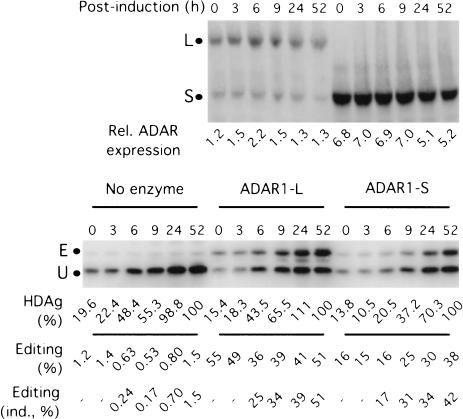
The experiments reported thus far indicated that ADAR1-L exhibited elevated activity in vivo, when assayed after a fairly long period of time (3 d). We next monitored the ability of ADAR1-L to edit transcripts in a much shorter window of time by examining the kinetics of HDV reporter editing by using a tetracycline-inducible construct. Figure 2 ▶ shows the kinetics of editing after induction in the presence of no supplemented ADAR, as well as in the presence of exogenously expressed ADAR1-L and ADAR1-S. In all three cases, some δ antigen (HDAg) expression was observed prior to induction; however, by 24 h after induction, the level of protein expression increased five- to sevenfold. Induced protein synthesis could be detected as early as 6 h after induction in all three cases (Fig. 2 ▶). At that time, in the presence of ADAR1-L, 36% editing was observed, whereas only 16% editing resulted in the presence of ADAR1-S, even though the level of that protein was threefold higher than that of ADAR1-L. In the presence of both enzymes, the percentage of editing continued to increase with time, and comparable kinetics were observed in each case. We concluded that the difference in the editing efficiencies of ADAR1-L and ADAR1-S could not be attributed to differences in editing kinetics and that ADAR1-L efficiently edited transcripts in a rather short (6-h) time frame.
FIGURE 2.
Kinetics of editing by ADAR1-L and ADAR1-S. HEK293 cells were cotransfected with a tetracycline-inducible HDV reporter construct and ADAR1-L, ADAR1-S, or no enzyme. Induction was initiated by the addition of doxycycline 2 d after transfection, and samples were subjected to anti-HA Western analysis to monitor relative ADAR expression (top) and an anti-HDAg Western analysis (bottom) to determine expression and editing of the reporter. HDAg (%) is the HDAg expression, which is given a value of 100 at 52 h after induction, and editing (ind., %) is the % editing of induced HDAg after subtracting the HDAg expression at time 0.
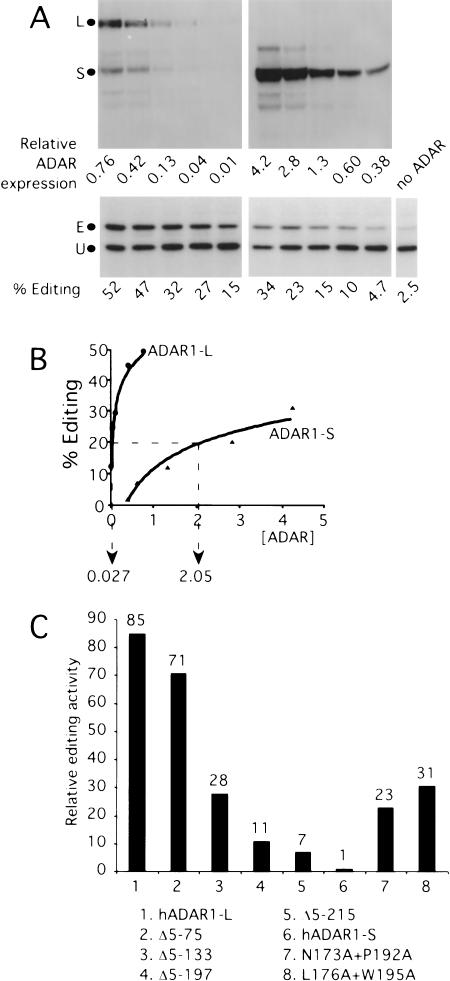
We next set out to accurately quantify the difference in the activity of ADAR1-L versus ADAR1-S, on a per molecule basis, inside the cell. We first tested the ability of our Western assay to accurately quantify ADAR1-L levels over a 32-fold range in concentration by running twofold serial dilutions of the same sample on an SDS-PAGE gel. Analysis of the quantification of the resulting blot yielded a linear plot with a slope of 1.006 and an associated linear fit R value of 0.995 (data not shown). In addition, by using ADAR1-L and ADAR1-S purified from HEK293 cells, we found that the two species were recognized by the Western assay with the same efficiency (data not shown). A more detailed analysis of the difference in the activities of the two forms was then performed by transfecting HEK293 cells with serial dilutions of the hADAR1-L or hADAR1-S expression vectors and assaying the resulting editing activity by using the HDV reporter (Fig. 3A ▶). Figure 3B ▶ is a graphical representation of the data shown in Figure 3A ▶. Compared with ADAR1-L, to achieve 20% editing of the HDV reporter, 76-fold more ADAR1-S was required. In a different experiment, the difference in activity observed between ADAR1-L and ADAR1-S, on a per molecule basis, was 85-fold (Fig. 3C ▶).
FIGURE 3.
Comparison of the in vivo editing activity of ADAR1 forms and mutants. (A) Titration of ADAR1-L and ADAR1-S. HEK293 cells were transiently transfected with serial dilutions of ADAR1 expression vectors and the HDV amber/W editing reporter (pSS74). (Top) Anti-HA Western analysis showing relative enzyme expression levels. (Bottom) Anti-HDAg Western analysis showing the corresponding editing on the HDV amber/W reporter. (B) ADAR1-L was 76-fold more active than was hADAR1-S. The graph represents the quantitation of the above Western analyses. The relative ADAR expression required to edit 20% of the HDV reporter was used to define the comparative activity. (C) The relative editing activity of ADAR1 mutants. Relative editing activity was the activity per molecule of each protein normalized to that of ADAR1-S, for which the activity of ADAR1-S required to edit 20% of the HDV reporter was given a value of one.
No single domain was responsible for the elevated activity of ADAR1-L
Similar titration assays were performed with the N-terminal deletion mutants (Fig. 3C ▶). Deletion of amino acids 5–75 affected neither the activity (Fig. 3C ▶, bar 2) nor expression level of the resulting protein (data not shown). In contrast, the Δ5–133 mutant showed a threefold decrease in activity/molecule (Fig. 3C ▶, bar 3) but showed increased protein expression (Fig. 2 ▶, lanes 1,2). Editing efficiency decreased to 11-fold above that of hADAR1-S when the next domain comprising part of the NES and the Zα domain was deleted (Fig. 3C ▶, bar 4, Δ5–197). Finally, deletion of all of the N terminus except for one of the duplicated sequences resulted in a protein with sevenfold higher activity compared with that of ADAR1-S (Fig. 3C ▶, bar 5, Δ5–215).
Because we did observe approximately a threefold decrease in the editing activity of ADAR1-L when Zα was deleted (Fig. 3C ▶, bars 3,4, cf. Δ5–133 and Δ5–197), we tested whether point mutations in the Zα domain would have a similar effect. Two Zα double-point mutants, N173A + P192A and L176A + W195A, were constructed. The mutations were chosen based on their ability to specifically inhibit Z-DNA binding in vitro (Schade et al. 1999). In addition, three of these residues were found to contact Z-DNA in the Zα crystal structure (Schwartz et al. 1999). Titration assays comparing these mutants to the wild-type enzyme were performed. We observed that the N173A + P192A mutant was 23-fold more active than ADAR1-S, whereas the L176A + W195A mutant was 31-fold more active (Fig. 4C ▶, bars 7,8). Hence, with both double-point mutants, an approximately threefold reduction in activity was observed compared with wild-type ADAR1-L. We conclude that if Z-DNA binding has any role in the editing activity of hADAR1 in vivo, that role is a relatively modest one in the context tested. Consistent with our findings, a threefold decrease in editing efficiency was observed when a Zα-β point mutant was assayed in vivo by using a synthetic 15-bp stem loop substrate (Herbert and Rich 2001).
FIGURE 4.
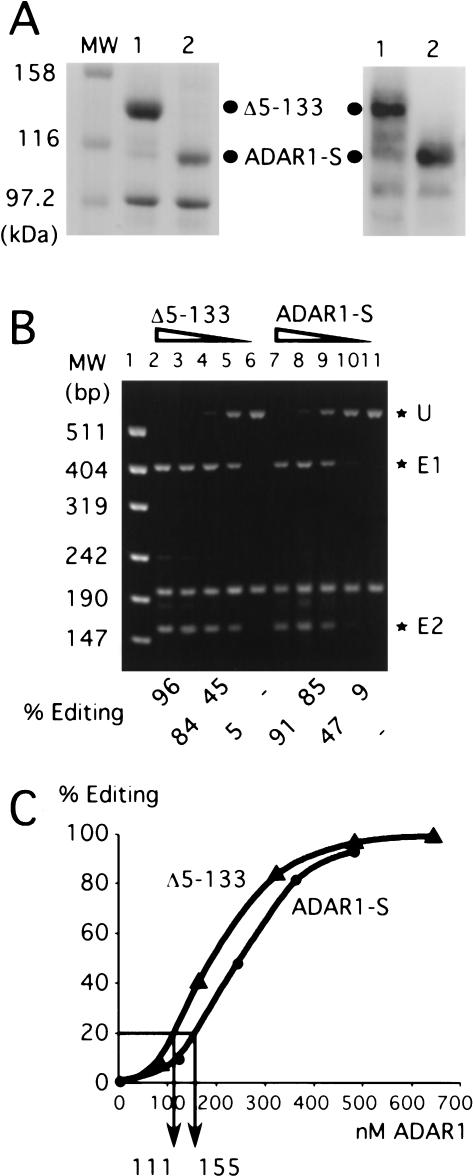
Editing activity per molecule of the N-terminal deletion mutant Δ5–133 and ADAR1-S in vitro. (A) ADAR1-S and Δ5–133 purified from HEK293 cells. Proteins were electrophoresed on a 10% SDS–polyacrylamide gel and stained with Coomassie blue (left) or immunoblotted with anti-HA monoclonal antibody (right). Lane 1 is Δ5–133, and lane 2 is ADAR1-S; the dots mark their positions. (B) In vitro editing assay. Decreasing concentrations of Δ5–133 (lanes 2–6) and ADAR1-S (lanes 7–11) were added to HDV antigenomic RNA. Sty I-digested reverse transcription–PCR products of the in vitro editing reaction were resolved on an agarose gel. *U is the unedited product, and *E1 and *E2 are the two products of editing. MW is the molecular-weight marker (lane 1). The percentage of editing was calculated by E1+E2/U+E1+E2. (C) A graphical representation of the average of two in vitro editing experiments.
ADAR1-S and the Δ 5–133 mutant had similar activity in vitro
Previously, it was reported that the two forms of hADAR1 have comparable specific deamination activity in vitro; however, their in vitro activity in relation to the HDV amber/W site has not been compared. We therefore wished to evaluate the ability of the two forms to edit the HDV amber/W site in vitro, by using protein expressed in and purified from the same cells used in our in vivo assays. To facilitate purification, the high-expressing Nterminal deletion mutant Δ5–133 was used in place of ADAR1-L. In vivo, this mutant was only threefold less active than was ADAR1-L and was 28-fold more active than was ADAR1-S.
HEK293 cells were transfected with cDNAs expressing the Δ5–133 mutant and ADAR1-S that were tagged with two HA repeats and six histidines. These enzymes were purified from cell lysates by using resin chelated with Co2+ (Fig. 4A ▶). The proteins were assayed in vitro by using a T7 RNA polymerase generated transcript that consists of approximately two thirds of the HDV antigenomic rod-like structure, equivalent to that present in the in vivo HDV reporter. Editing of the amber/W site was monitored by digestion of the resulting reverse transcription–PCR product with StyI. Figure 4B ▶ shows the results of an in vitro editing assay using 80–640 nM enzyme and 20 nM substrate. From the quantification of this data, it was clear that the two forms of the enzyme behaved fairly similarly in vitro (Fig. 4C ▶). Although the Δ5–133 mutant was 28fold more active than was ADAR1-S in vivo, only a 1.5fold difference was observed in vitro. Similar results to those obtained with the Δ5–133 mutant were also observed with a less pure preparation of hADAR1-L (data not shown).
ADAR1-L displayed elevated activity in vivo, even in the absence of translation, when assayed with a reporter transcribed by T7 RNA polymerase
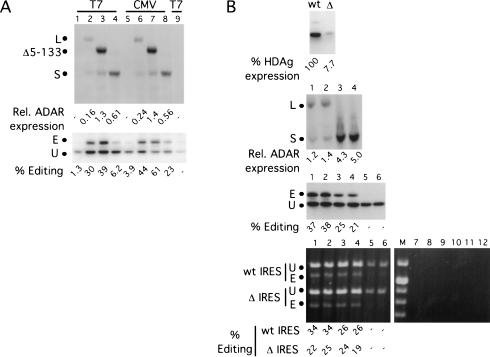
To account for the differences in activity observed in vivo but not in vitro, we postulated that in vivo, ADAR1-L is specifically escorted to nascent transcripts, thereby increasing its effective concentration in relation to the substrate. Perhaps ADAR1-L, but not ADAR1-S, could specifically interact with the transcription machinery of the cell. If this were true, then when assayed on messages transcribed in vivo by a heterologous polymerase such as T7 RNA polymerase, ADAR1-L and ADAR1-S would have similar activities. To address this possibility, we constructed an HDV amber/W reporter driven by a T7 RNA polymerase promoter. To enable translation of this uncapped message, a poliovirus internal ribosomal entry site (IRES) was inserted at the 5′ end, and 100 adenosines were added to the 3′ end. To specify the 3′ end of this reporter, the HDV genomic ribozyme was fused such that cleavage occurs after the final adenosine of the poly A tract. T7 RNA polymerase was expressed from the cytomegalovirus immediate early promoter and contained a SV40 T antigen nuclear localization signal (NLS) fused to its N terminus.
In vivo editing assays were performed by using this reporter. Figure 5A ▶ shows a comparison of editing by ADAR1-L (lanes 2,6), the Δ5–133 mutant (lanes 3,7), and ADAR1-S (lanes 4,8) assayed with reporters transcribed by either T7 RNA polymerase (lanes 1–4) or RNA polymerase II (lanes 5–8). In the control lane, where no T7 RNA polymerase was expressed (lane 9), δ antigen expression was 17% of that obtained with expression of T7 RNA polymerase (lane 1). Hence, the majority of the protein signal observed in lanes 1–4 resulted as a consequence of T7 RNA polymerase-mediated transcription. As shown in Figure 5A ▶, even when assayed by using reporters transcribed by this heterologous polymerase, ADAR1-L and the Δ5–133 mutant were still much more active than was ADAR1-S.
FIGURE 5.
In vivo editing activity of ADAR1 forms when assayed with HDV amber/W editing reporters transcribed by T7 RNA polymerase in the presence or absence of translation. (A) Editing of HDV amber/W editing reporters transcribed by either T7 RNA polymerase or RNA polymerase II. The anti-HA Western analysis (top) shows the enzyme expression levels of ADAR1-L (lanes 2,6), Δ5–133 (lanes 3,7), and ADAR1-S (lanes 4,8). The dots mark the positions of the three proteins. The anti-HDAg Western analysis (bottom) shows the corresponding editing of the HDV reporter with the T7 RNA polymerase promoter (lanes 1–4,9) (pDL730) or the CMV (cytomegalovirus) promoter (lanes 5–8; pSS106). The samples in lanes 1–4 were cotransfected with a T7 RNA polymerase expression vector. Lanes 1 and 5 are no ADAR controls, and lane 9 is the no ADAR and no T7 RNA polymerase control. (B) ADAR1-L is more active than is ADAR1-S, even in the absence of translation. (Top) Anti-HDAg Western analysis showing the expression levels of HDV editing reporters transcribed by T7 RNA polymerase with either the wild-type (wt) or deleted (Δ) IRES. HEK293 cells were cotransfected with a mixture of these two reporter constructs and either ADAR1-L or ADAR1-S expression vectors or no enzyme. The experiment was performed in duplicate, and samples were harvested for Western analysis or RNA purification. The second panel from the top is an anti-HA Western analysis showing the expression levels of ADAR1-L (lanes 1,2) and ADAR1-S (lanes 3,4). The third panel from the top is an anti-HDAg Western analysis showing the corresponding editing of the wild-type HDV reporter. Lanes 5 and 6 are no enzyme controls. (Bottom) The corresponding NcoI digests of the reverse transcription–PCR products from the RNA transcripts of the two reporters. Lanes 7 through 12 are the no reverse-transcription controls of samples 1 through 6, and M is the molecular-weight marker. E and U are the edited and unedited products.
To explore the role of translation in relation to editing by ADAR1-L and ADAR1-S, a derivative of T7 RNA polymerase-based reporter was generated, in which 282 nucleotides of sequence were deleted from the IRES element. The deletion reduced protein expression from the vector by 13-fold (Fig. 5B ▶, first panel). The residual protein expressed from the IRES deletion vector probably resulted from translation of the low level of capped pol II transcripts that are produced from this vector (Fig. 5A ▶, lane 9). We concluded that the IRES deletion effectively prevented translation of the reporter.
We next performed mixed transfections in which equal amounts of the wild-type and deleted IRES constructs were codelivered to the same cells in either the presence or absence of exogenously expressed ADAR1-L and ADAR1-S. Editing of both the wild-type and deleted IRES transcripts was then monitored by NcoI digestion of the products that resulted after reverse transcription–PCR. As shown in Figure 5B ▶, panel 4, with respect to the wild-type IRES construct, higher levels of editing were observed with ADAR1-L than with ADAR1-S, even though the latter was expressed at fourfold higher levels (Fig. 5B ▶, panel 2). The editing levels as determined by the reverse transcription–PCR assay were in good agreement with those obtained from the Western assay (Fig. 5B ▶, cf. panels 3 and 4). In the presence of ADAR1-S, the amount of editing observed with the wild-type and deleted IRES constructs was equivalent (Fig. 5B ▶, panel 4, lanes 3,4), and we concluded that translation did not affect editing by that enzyme. In contrast, in the presence of ADAR1-L, the IRES deletion reporter was edited somewhat less efficiently than was the wild-type reporter (Fig. 5B ▶, panel 4, lanes 1,2), and we concluded that translation stimulated editing by ADAR1-L. Nevertheless, even in the absence of translation, slightly more editing was observed with ADAR1-L than with ADAR1-S, even though the latter was expressed at a fourfold higher level. We concluded that ADAR1-L efficiently edited transcripts even in the absence of translation.
Correlation between activity and localization patterns of wild-type and mutant forms of ADAR1
Because translation stimulated editing by ADAR1-L but not by ADAR1-S, we hypothesized that the former is able to efficiently edit transcripts in the cytoplasm. We next used immunofluorescence to determine if the ability of a particular mutant to localize to the cytoplasm could be correlated to its editing efficiency in vivo.
Consistent with previous reports (Patterson and Samuel 1995;Poulsen et al. 2001), ADAR1-L localized mainly to the cytoplasm, and ADAR1-S to the nucleus (data not shown). The Δ5–133 mutant, similar to ADAR1-L, was also localized mainly to the cytoplasm (data not shown), despite having only one third of the reported NES (Poulsen et al. 2001). We had also mutated five residues (L126A+H129Q+ F130L+L133A+I135A) within this NES in ADAR1-L and found that the resulting protein was located mainly in the cytoplasm (data not shown). Recently, **Yang and colleagues (Yale University, pers. comm.) have identified a second NES in the Zα domain of mouse ADAR1 that resides downstream of the signal identified by Poulsen et al. Perhaps in HEK293 cells, either of the identified NES elements is sufficient to direct ADAR1-L to the cyctoplasm. Of the mutants proteins shown in Figure 3C ▶, only ADAR1-S showed exclusive nuclear staining in all cells, whereas exclusive nuclear staining was seen with most of the cells when the Δ5–197 and Δ5–215 mutants were tested (data not shown). In all other cases, significant cytoplasmic staining was seen and, hence, a correlation was observed between the ability of a mutant to localize to the cytoplasm and its ability to edit with high efficiency.
ADAR1-L was less active than was ADAR1-S when nuclear editing events were assayed
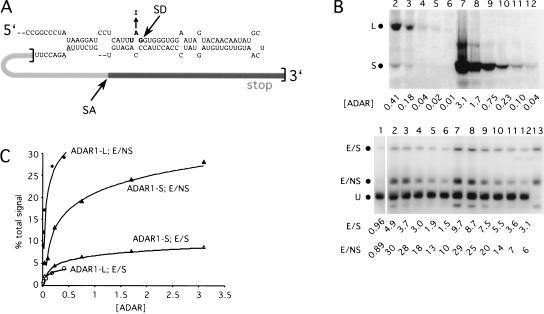
The immunofluorescence data revealed that the more active forms of ADAR1 mainly localized to the cytoplasm. Because the editing reporter could potentially be edited in that compartment, we hypothesized that the high editing activity observed with ADAR1-L in vivo reflected the ability of the protein to efficiently edit transcripts in the cytoplasm. A prediction of this model is that if editing activity were monitored in the nucleus, ADAR1-L would be less active than ADAR1-S. To test this prediction, we constructed a reporter to monitor editing events that can only occur in the nucleus. This reporter has an essential part of its editing site residing within an intron such that editing must occur prior to splicing.
Figure 6A ▶ shows the design of the splicing reporter derived from the amber/W derivative of the GluR-B R/G site (Wong et al. 2001). The GluR-B R/G site has its native splice donor, and the splice acceptor is provided by the rat pre-proinsulin intron. This portion of the intron, together with its adjacent exon and polyadenylation signal, was inserted downstream of the GluR-B R/G sequences. When the resulting reporter mRNA is not edited, translation terminates at the stop codon within amber/W site and a 162 amino acid protein results. If the reporter is edited and then spliced, translation of the message terminates at a stop codon within the next exon and gives rise to a protein of 221 amino acids. The presence of this protein therefore reflects editing that could have only occurred in the nucleus, because the mature spliced message no longer contains a site competent for editing. If the reporter is edited but not spliced, translation terminates at a stop codon within the intron and gives rise to a protein of 185 amino acids. This species reflects editing that could have occurred in either the nucleus or the cytoplasm because, in this case, the editing site is retained in the mature message that is exported to the cytoplasm.
FIGURE 6.
Editing activity of ADAR1-L and ADAR1-S in the nucleus. (A) Design of the GluR-B R/G splicing reporter. The bracketed region denotes the inserted rat pre-proinsulin sequences. SD and SA refer to the splice donor and splice acceptor sites. The stop codon within the intron is underlined. (B) In vivo editing activity of ADAR1-L and ADAR1-S assayed with the GluR-B R/G splicing reporter. (Top) Anti-HA immunoblot showing the relative enzyme expression of serial dilutions of the ADAR1-L (lanes 2–6) or the ADAR1-S (lanes 7–12) expression vector. The dots (L and S) mark the positions of the two forms. (Bottom) Anti-HDAg immunoblot monitoring the corresponding editing on the splicing reporter. E/S and E/NS are the products of the edited/spliced reporter and the edited/not spliced reporter. U is the unedited product. The quantitation of the immunoblot is expressed as a percentage of the total signal. Lane 1 is the no enzyme control, and lane 13 shows the products of the “pre-edited” (UGG) control. (C) Graphical representation of the above titration plotted with a logarithmic curve fit.
The levels of the two forms of ADAR1 were titrated in vivo, and the resulting activity was monitored by using the GluR-B R/G splicing reporter. In the lower panel of Figure 6B ▶, the top band in lanes 1 through 12 of the anti-δ antigen, Western analysis reflects editing that occurred in the nucleus prior to splicing (E/S). The middle band is the product of messages that were edited and not spliced (E/NS). In this case, editing could have occurred in either compartment. The bottom band is the translation product of unedited spliced and unspliced transcripts (U). Note that the E/S product was 4.9% of the total signal when 0.41 units of ADAR1-L were provided, whereas the E/S product represented 5.5% of the total signal when only 0.23 units of ADAR1-S were provided (Fig. 6B ▶, cf. lanes 2 and 10). Hence, ADAR1-S was more active than was ADAR1-L when editing was monitored in the nucleus. In contrast, when editing could occur in the cytoplasm, ADAR1-L was far more active than was ADAR1-S. The E/NS product was 29% of the total signal when 3.1 units of ADAR1-S were provided, whereas the E/NS product represented 28% of the total signal when only 0.18 units of ADAR1-L were provided (Fig. 6B ▶, cf. lanes 7 and 3).
The data from Figure 6B ▶ were fit logarithmically and are depicted graphically in Figure 6C ▶. From this analysis, we concluded that, with respect to the E/S product, the activity of ADAR1-S was threefold higher than that of ADAR1-L, whereas with respect to the E/NS product, it was 13-fold lower. We therefore concluded that much of the elevated activity of ADAR1-L observed with nonsplicing reporters could be attributed to efficient editing that occurred in the cytoplasm.
A substrate that was synthesized in the cytoplasm was efficiently edited by ADAR1-L but not ADAR1-S
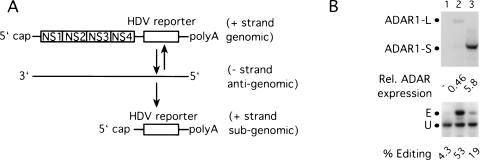
To directly test whether ADAR1-L could efficiently edit transcripts in the cytoplasm, we used a condition in which the reporter would reside only in the cytoplasm and never in the nucleus. Toward this end, a construct that can be transcribed to yield replication-competent Semliki Forest virus (SFV) RNA was modified so that the HDV reporter was inserted to replace the viral structural genes. After transfection of the capped RNA generated in vitro, it should be translated to yield the viral replication proteins NS1-NS4 (Fig. 7A ▶). These proteins cooperate to copy the genomic plus strand RNA into antigenomic minus strand RNA, which in turn serves as template for both genomic and subgenomic positive strand RNA synthesis. The latter species in this case contains the HDV reporter and is the only viral species capable of expressing the δ antigens. Semliki Forest virus is an alphavirus, and as such, all of the events just described are thought to occur solely in the cytoplasm (Strauss and Strauss 1994).
FIGURE 7.
Editing activity of ADAR1-L and ADAR1-S in the cytoplasm. (A) The Simliki Forest virus (SFV) HDV amber/W editing reporter. The replication-competent SFV genomic mRNA contains the HDV reporter sequences in place of the viral structural genes. Replication of the genomic strand gives rise to the antigenomic minus strand RNA, which in turn gives rise to the subgenomic mRNA that encodes the HDV reporter. (B) ADAR1-L is more active than is ADAR1-S in the cytoplasm. HEK293 cells were transfected with the SFV editing reporter mRNA and either ADAR1-L or ADAR1-S expression vectors or no enzyme. (Top) Anti-HA Western analysis shows the expression levels of ADAR1-L (lane 2) and ADAR1-S (lane 3). (Bottom) Anti-HDAg Western analysis shows the corresponding editing of the SFV editing reporter. Lane 1 is the no enzyme control.
Figure 7B ▶ shows the results obtained with the SFV-based editing reporter. Although ADAR1-S was expressed at a 13-fold higher level than was ADAR1-L, it could only weakly stimulate editing of the SFV reporter (15%), whereas ADAR1-L stimulated editing to 49% (Fig. 7B ▶, cf. lanes 2 and 3). We concluded that ADAR1-L could efficiently edit transcripts generated in the cytoplasm.
An ADAR2 mutant that localized to the cytoplasm was more efficient than the wild-type enzyme at editing unspliced reporters
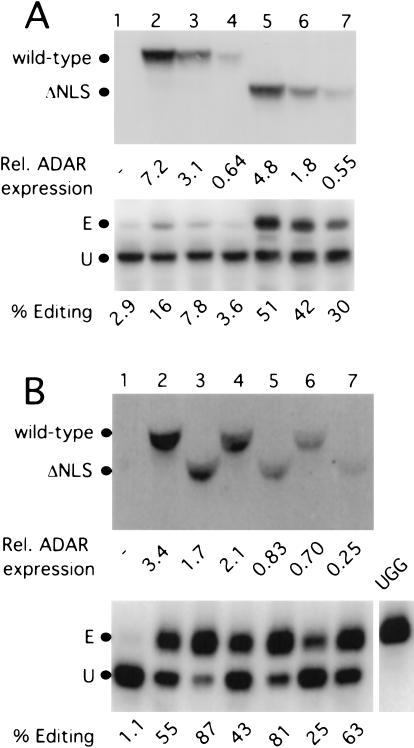
If the cytoplasmic localization of ADAR1-L was largely responsible for its elevated activity observed in vivo with nonsplicing reporters, we reasoned that we would observe a similar effect if ADAR2 were localized to the cytoplasm. We therefore created an N-terminal deletion mutant of ADAR2 in which the putative NLS was removed. The ability of the ADAR2 ΔNLS mutant to edit in the cytoplasm was then compared with that of wild-type ADAR2 by using the Simliki Forest virus editing reporter. As is shown in Figure 8A ▶, although only 13% editing resulted when 7.2 units of the wild-type protein was expressed, 27% editing resulted when only 0.55 units of the ΔNLS mutant was expressed (cf. lanes 2 and 7). Hence, with respect to editing in the cytoplasm, the ΔNLS mutant was at least 26-fold more active than was the wild-type protein. As expected, wild-type ADAR2 was more active than was the ΔNLS mutant when editing in the nucleus was monitored by using the spliced GluR-B R/G reporter (data not shown). We concluded that the putative NLS of ADAR2 indeed does direct ADAR2 to the nucleus, and in its absence, editing in the cytoplasm is dramatically increased.
FIGURE 8.
Deletion of the NLS in ADAR2 increased editing activity in the cytoplasm. (A) The NLS deletion mutant of ADAR2 had increased editing activity with the SFV editing reporter that is synthesized in the cytoplasm. (Top) The anti-HA immunoblot shows the enzyme expression levels of the wild-type (lanes 2–4) and ΔNLS mutant (lanes 5–7) of ADAR2. (Bottom) The anti-HDAg immunoblot monitors the corresponding editing when assayed on the SFV editing reporter. Lane 1 is the no enzyme control. (B) The NLS deletion mutant of ADAR2 had increased editing activity when assayed with a reporter that retains its editing site in the cytoplasm. (Top) The anti-HA immunoblot shows the level of expression of the wild-type (lanes 2,4,6) and ΔNLS mutant (lanes 3,5,7) ADAR2. (Bottom) The anti-HDAg immunoblot shows the corresponding editing when assayed on the nonsplicing GluR-B R/G reporter (pSKW001; Wong et al. 2001). Lane 1 is the no enzymecontrol, and UGG refers to the “pre-edited” control.
We next compared the activity of the ADAR2 ΔNLS mutant (Fig. 8B ▶, lanes 3,5,7) with that of wild-type ADAR2 (Fig. 8B ▶, lanes 2,4,6) by using the nonsplicing GluR-B R/G reporter. This reporter retains its editing site in the mature message and can be edited in either the nucleus or the cytoplasm. Figure 8B ▶ shows that the ΔNLS mutant was much more efficient than was the wild-type enzyme at editing a substrate that can be edited in either the nucleus or the cytoplasm. Note that 3.4 units of the wild-type enzyme were needed to achieve 54% editing, whereas 62% editing was achieved with only 0.25 units of the ADAR2 ΔNLS mutant (cf. lanes 2 and 7). Hence, the activity of the ΔNLS mutant was >15-fold higher than that of wild-type ADAR2 when assayed with a reporter that could be edited in the cytoplasm. We concluded that editing in the cytoplasm is a highly efficient process.1
DISCUSSION
We observed in vivo that hADAR1-L was >70-fold more active than was hADAR1-S at editing an HDV amber/W reporter that retained its editing site in the mature message. The elevated activity of ADAR1-L was also observed with other editing sites, as well as with ADAR1 from another species (mouse). When the activities of ADAR1-L and ADAR1-S were assayed in vitro, essentially no difference was observed. We found that the elevated activity of ADAR1-L observed in vivo could largely be attributed to the protein‘s ability to efficiently edit transcripts in the cytoplasm. When the activities of ADAR1-L and ADAR1-S were compared by using a reporter that could only be edited in the nucleus, ADAR1-L was found to be somewhat less active than ADAR1-S. An SFV-based expression system was used to transcribe the editing reporter in the cytoplasm. Under this condition, editing by ADAR1-L was very efficient. Consistent with the notion that editing in the cytoplasm can be a highly efficient process, deletion of the NLS from ADAR2 resulted in a protein with much higher activity when assayed either with a reporter that maintains its editing site in the cytoplasm or with a reporter that is synthesized in the cytoplasm.
We found that no single domain unique to ADAR1-L was solely responsible for its elevated activity because deletion mutants displayed intermediate phenotypes. Deletion mutants with high activities all showed cytoplasmic staining. However, two mutants, Δ5–197 and Δ5–215, showed predominantly, although not exclusively, nuclear staining (data not shown). These mutants still had 11-fold and sevenfold higher activity, respectively, when compared with that of hADAR1-S. The difference between the two mutants and hADAR1-S is the presence of two copies or one copy of a sequence that was naturally duplicated in the human genome. We do not know how the repeat sequence enhances ADAR1 activity, but our results indicate that the duplication that occurred during evolution may have conferred a slight advantage for ADAR1-L function.
It is also not known whether ADAR1-L and ADAR1-S would have the same or different activities if they could be localized to the same compartment. Consistent with the in vitro data, it is possible that the intrinsic editing capability of the two forms is similar. If the substrate spends a small fraction of its life in the nucleus and most of its life in the cytoplasm, then cytoplasmic ADAR1-L would have much greater access to the substrate and would show elevated activity in our assays. This model would predict that much of the editing observed with ADAR1-L would occur many hours, or even days, after the transcript was first synthesized. However, such a model is not supported by the time-course presented in Figure 2 ▶, in which efficient editing by ADAR1-L was observed at the early (6-h) time point and in which similar kinetics for editing by ADAR1-L and ADAR1-S were observed. Perhaps, compared with cytoplasmic messages, RNAs in the nucleus are more tightly bound by RNA binding proteins (e.g., hnRNP proteins) that might hinder ADAR access to the editing site. Consistent with such a possibility, ADAR2, which presumably has not evolved to function in the cytoplasm, was nevertheless very active when it was localized to that compartment. However, it should be noted that ADAR1-L was more active than was ADAR2 ΔNLS when editing in the cytoplasm was monitored (cf. Fig. 7 ▶, lane 2; Fig. 8A ▶, lane 7). Hence, there is likely to be an additional mechanism operating that enhances the activity of ADAR1-L in the cytoplasm.
RNA transcripts that are edited must contain double-stranded structures, yet such structures would be melted by the ribosome during translation. Furthermore, the many ribosomes on polysomal RNA might sterically interfere with ADAR access to editing sites. Surprisingly, however, translation was found to stimulate rather than inhibit editing by ADAR1-L. It is possible that ADAR1-L is capable of interacting with some translation factor and that this interaction effectively escorts ADAR1-L to its substrate RNA. However, this putative interaction would only stimulate editing by ADAR1-L because editing was still fairly efficient in the absence of translation.
HEK293 cells express both forms of ADAR1 and the endogenous levels of these enzymes can edit reporters at low-to-moderate levels, depending on the specific reporter being tested. Although ADAR1-L represents only ∼5% of the total ADAR1 expressed in these cells, based on its >70-fold higher activity, we would predict that this form is responsible for most of the endogenous editing activity observed with unspliced reporters. Consistent with this prediction, we recently used siRNAs targeted to exon 1A to reduce ADAR1-L levels without affecting ADAR1-S levels and observed a dramatic inhibition of reporter editing (Wong and Lazinski 2002).
Previously, it was established that expression of ADAR1-L is regulated at the level of transcription initiation through the action of an interferon-inducible promoter. Here we provide evidence that ADAR1-L levels may also be controlled during and after translation. We found that the ADAR1-L initiator is poor at directing translation and that frequent internal initiation of the message occurs. Furthermore, we observed that even when an optimal initiator is used, although internal initiation is greatly reduced, ADAR1-L is still poorly expressed. Amino acids 76–133 were found to be responsible for the poor expression. The mechanism by which these residues lower expression is not known, nor is it known whether they have a destabilizing effect under all conditions. Perhaps during interferon induction, these residues are not destabilizing. Given the extraordinarily high activity of ADAR1-L, it is not surprising that the cell uses multiple mechanisms to control its level of expression.
Efficient editing in the cytoplasm is consistent with the postulated antiviral role of ADAR1-L. ADAR activity has been hypothesized to be responsible for biased A-to-G mutations found in the cDNA sequences of several cytoplasmic RNA viruses, including measles virus (Cattaneo 1994), parainfluenza virus (Murphy et al. 1991), and vesicular stomatitis virus (O‘Hara et al. 1984). Because expression of ADAR1-L can be induced by interferon, it is plausible that this enzyme, as part of the cellular immune response, could be responsible for deaminating cytoplasmic viral dsRNAs. However, the role of ADAR1-L with respect to these viruses is still speculative and awaits experimental examination.
We showed here that not only could ADAR1-L edit transcripts in the cytoplasm but also it could do so very efficiently. Most of the known substrates of ADAR1 and ADAR2 are pre-mRNAs that are edited before splicing. Hence, it is thought that editing by ADARs probably occurs cotranscriptionally on nascent premessages. Our experiments show that mature mRNAs can also be edited very efficiently in the cytoplasm. Furthermore, in addition to the thought that viral dsRNAs synthesized in the cytoplasm are targets of ADAR1-L, we found that RNAs synthesized in the nucleus can also serve as substrates.
In addition to the hypothetical antiviral role, our finding that ADAR1-L was highly active at editing mRNAs in the cytoplasm indicates a second potential role for this protein. In an unbiased search for inosine-containing messages in Caenorhabditis elegans and humans, many mRNAs with extensive predicted double-stranded structure in their UTRs were shown to have multiple A-to-I conversions (Morse and Bass 1999,Morse et al. 2002). In these substrates, the editing site is preserved in the mature message, and we propose that ADAR1-L is the form primarily responsible for the observed editing events.
MATERIAL AND METHODS
Protein expression plasmids
All expression vectors were constructed by PCR-assisted cloning and/or mutagenesis, and the regions amplified were sequenced to confirm that they were correct. pDL707 expresses hADAR1-L and was derived from pDL700 (Wong et al. 2001) in which the 5′UTR was deleted, and an optimal Kozak sequence (AGCCACC) was added upstream of the first AUG in the ORF. pDL701 expresses hADAR1-S and was constructed by deleting the 827 bp BglII-BamHI fragment upstream of the second AUG in the ORF of pDL700. pSKW097, pDL722, pDL725, pDL726, and pSKW083 express the Δ5–75, Δ5–133, Δ5–197, Δ5–215, and Δ5–295 deletion mutants respectively. pDL717 and pDL718 express the Zα N173A+P192A and Zα L176A+W195A mutants. pLN007 is derived from pDL700 and has the methionine at position 296 (second methionine) mutated to alanine, whereas in pLN021, the second through fifth methionines were mutated to alanine. mADAR1-L is expressed by pSKW99 and mADAR1-S by pSKW100, and in each case, the C terminus of ADAR1 was fused to sequences encoding two HA epitopes and six histidines. Wild-type ADAR2 was expressed by pMS040 (Wong et al. 2001), and the ΔNLS ADAR2 expression plasmid, pDL754, was constructed by deleting the sequences that encode amino acid residues 4–72. The T7 RNA polymerase expression vector, pKW87, was described previously (Reid and Lazinski 2000).
Editing reporters
The RNA polymerase II-transcribed HDV amber/W reporter was expressed from pSS74 (Sato et al. 2001). The GluR-B R/G nonsplicing reporter was expressed from pSKW001, and pSKW002 (Wong et al. 2001) is the pre-edited (UGG) version. The GluR-B R/G splicing reporter expression vector pDL761 was generated from pSKW001 by inserting four fifths of the rat pre-proinsulin intron sequences together with its adjacent exon and polyadenylation signal sequences (GenBank accession no. V01243, nucleotides 503–1146) six nucleotides after the second stop codon of the ORF. pDL762 expresses the pre-edited (UGG) form of the GluR-B R/G splicing reporter. The HDV amber/W editing reporter transcribed by T7 RNA polymerase was expressed from pDL730 and has the poliovirus IRES sequences, the HDV sequences from pSS106, 100 adenosines, and the HDV ribozyme sequences inserted into an expression vector with a T7 RNA polymerase promoter. The IRES deletion mutant of this reporter was expressed from pDL798, which had 282 nucleotides (PshAI-MscI fragment) of the IRES sequences deleted from pDL730. The tetracycline-inducible HDV amber/W editing reporter was expressed from pDL799, which contained the 1225-bp XbaI-HindIII fragment from pSS99 (Sato et al. 2001) subcloned into the XbaI-HindIII sites of pTRE2 (**Clontech). The Simliki Forest virus HDV amber/W editing reporter was transcribed from pDL800, which contained the 1523-bp Ecl136 II-NotI fragment of pSS74 (Sato et al. 2001) inserted into the SmaI-NotI sites of pSFV4.2 (Pastrana et al. 2001). RNA synthesized from pLN016 was used in in vitro assays. pLN016 was constructed by subcloning the 1164-bp ApaI fragment from the wild-type HDV antigenomic cDNA sequences into an RNA expression vector between the T7 RNA polymerase promoter and terminator.
In vivo editing assays and titrations
In vivo editing assays were performed as described in Wong et al. (2001). Titrations were done with twofold serial dilutions of ADAR cDNAs, which were cotransfected with the RNA polymerase II-transcribed HDV amber/W reporter (pSS74). Each titration had four or five points. Relative ADAR expression was obtained by determination of the I125 signal quantitated by using a phosphoimager, and was compared with that of a HA-tagged hADAR1 standard loaded on the same blot. Equal aliquots of this standard were loaded on all the anti-HA Westerns and were given an arbitrary value of 10. For each set of titrations, the percentage of editing was plotted against relative ADAR expression by using a logarithmic curve fit. The relative ADAR expression required to edit 20% of the HDV reporter was obtained by using the logarithmic curve fit equation. The activity of each enzyme was normalized to that of ADAR1-S, which was given a value of one.
For the reporter induction experiment, HEK293 cells were cotransfected with pDL799, pTet-on (**Clontech), pTet-tTS (**Clontech), and pDL707 (ADAR1-L) or pDL701 (ADAR1-S). Forty-eight hours after transfection, 1 μg/mL of doxycycline was added to the media, and samples were harvested 0, 3, 6, 9, 24, and 52 h after the addition of doxycycline.
For experiments that used T7 RNA polymerase to drive expression, HEK293 cells were cotransfected with equal amounts of the wild-type (pDL730) and deleted IRES (pDL798) expression vectors, pKW87 (T7 RNA polymerase expression vector), and either pDL707 (ADAR1-L) or pDL701 (ADAR1-S). The cells were harvested 3 d after transfection for Western analysis and total RNA purification. Editing of the wild-type and deleted IRES transcripts was monitored by NcoI digestion of the products that resulted after reverse transcription with oligo (dT)20 and PCR with primers oli777 and oli700: Oli777, 5′ CCCAGAGTGT AGCTTAGGTCG ATG 3′; and Oli700, 5′ CCGGGGAACTCGACTTATCGTCCC 3′.
In experiments that used the Semliki Forest virus–based editing reporter, capped mRNA was synthesized from pDL800 by using the mMessage mMachine SP6 Kit (Ambion). HEK 293 cells were cotransfected using lipofectamine 2000 with the capped mRNA synthesized from pDL800 and pDL707 (ADAR1-L), pDL701 (ADAR1-S), pMS040 (ADAR2), or pDL754 (ΔNLS ADAR2). Cells were harvested 2 d after transfection for Western analyses.
Purification of histidine-tagged ADAR1 from HEK293 cells
HEK293 cells were transfected by CaCl2 precipitation; 48 to 72 h after transfection, the cells were trypsinized and washed, and total cell lysate was prepared by a modified Dignam procedure (Dignam et al. 1983). The cell lysate was made up to the same composition as the extraction buffer (40 mM HEPES at pH 7.8, 20% glycerol, 3 mM MgCl2, 600 mM KCl, 5 mM β-mercaptoethanol, 0.5 mM PMSF). Talon cobalt resin (Clontech) was added to the lysate, and purification was performed according to the manufacturer‘s protocol by using imidazole to elute the protein. The protein was concentrated, and the buffer was exchanged with Centricon filters (Millipore) to ∼4 (Δ5–133) and 2 (ADAR1-S) μM in 20 mM HEPES (pH7.8), 10% glycerol, 1.5 mM MgCl, 300 mM KCl, 2.5 mM β-mercaptoethanol, 0.25 mM PMSF, 5 mM EDTA, 1 mM DTT, and 30 mM imidazole.
In vitro editing assay
pLN016 was transcribed by T7 RNA polymerase in vitro, and the resulting RNA was gel-purified. The reaction consisted of 20 nM pLN016 RNA and 80–640 nM ADAR in 20 mM HEPES (pH 7.8), 2% glycerol, 0.3 mM MgCl, 60 mM KCl, 0.5 mM β-mercaptoethanol, 0.05 mM PMSF, 1 mM EDTA, 0.5 mM DTT, 6 mM imidazole and 4 U per μL ribonuclease inhibitor (RNaseOUT by GIBCO) in a total volume of 10 μL. This mixture was incubated for 1 h at 30°C and was heated for 2 min at 95°C to stop the reaction; 2.5 μL of this in vitro editing reaction was reverse-transcribed at 42°C for 1 h with oli577. One-fourth of this reverse-transcription mixture was used in a first-round PCR with oli576 and oli577 (20 cycles). One-fiftieth of this first PCR reaction was used for a second round of PCR with nested primers, oli578 and oli580 (15 cycles). One-tenth of this second PCR reaction was digested with StyI and resolved on an agarose gel. The ethidium bromide–stained DNA was quantitated by using the Kodak Image Station 440CF and Kodak 1D Image analysis software: Oli577, 5′ CAGCGGTGGCAGCAGCCAACTCAGC 3′; Oli576, 5′ GCTTGC ATGCCTGCAGGTCGACTC 3′; Oli578, 5′ GCCGGCCCGCCGG TTGGGGGTG 3′; and Oli580, 5′ AGGGGGAGACCGAAGCGA GGAG 3′.
Acknowledgments
We thank Gordon Carmichael (University of Connecticut) for providing the mouse ADAR1 cDNA, Diana Pastrana and John Schiller (National Institutes of Health) for providing pSV4.2, John Coffin (Tufts University), Claire Moore (Tufts University), and Catherine Squires (Tufts University) for their helpful discussions, and Luciana Nurman for technical assistance. Grant R01-AI40472 from the National Institutes of Health and the Raymond and Beverly Sackler Research Foundation supported this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5160403.
REFERENCES
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., and Emeson, R.B. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308. [DOI] [PubMed] [Google Scholar]
- Casey, J.L. and Gerin, J.L. 1995. Hepatitis D virus RNA editing: Specific modification of adenosine in the antigenomic RNA. J. Virol. 69: 7593–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo, R. 1994. Biased (A→I) hypermutation of animal RNA virus genomes. Curr. Opin. Genet. Dev. 4: 895–900. [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, C.X. and Samuel, C.E. 1999. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. 96: 4621–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert, A. and Rich, A. 2001. The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc. Natl. Acad. Sci. 98: 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert, A., Lowenhaupt, K., Spitzner, J., and Rich, A. 1995. Chicken double-stranded RNA adenosine deaminase has apparent specificity for Z-DNA. Proc. Natl. Acad. Sci. 92: 7550–7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, M., Single, F.N., Kohler, M., Sommer, B., Sprengel, R., and Seeburg, P.H. 1993. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron-exon structure determines position and efficiency. Cell 75: 1361–70. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Maas, S., Single, F.N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R., and Seeburg, P.H. 2000. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81. [DOI] [PubMed] [Google Scholar]
- Kim, U., Wang, Y., Sanford, T., Zeng, Y., and Nishikura, K. 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. 91: 11457–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, F., Drakas, R., and Nishikura, K. 1995. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem. 270: 17098–17105. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Emeson, R.B., and Samuel, C.E. 1999. Serotonin-2C receptor pre-mRNA editing in rat brain and in vitro by splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase ADAR1. J. Biol. Chem. 274: 18351–18358. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Wolff, K.C., Jacobs, B.L., and Samuel, C.E. 2001. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology 289: 378–387. [DOI] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J.R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., and Seeburg, P.H. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266: 1709–1713. [DOI] [PubMed] [Google Scholar]
- Morse, D.P. and Bass, B.L. 1999. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl. Acad. Sci. 96: 6048–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, D.P., Aruscavage, P.J., and Bass, B.L. 2002. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. 99: 7906–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, D.G., Dimock, K., and Kang, C.Y. 1991. Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology 181: 760–763. [DOI] [PubMed] [Google Scholar]
- O‘Connell, M.A., Krause, S., Higuchi, M., Hsuan, J.J., Totty, N.F., Jenny, A., and Keller, W. 1995. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol. Cell. Biol. 15: 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O‘Hara, P.J., Nichol, S.T., Horodyski, F.M., and Holland, J.J. 1984. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell 36: 915–924. [DOI] [PubMed] [Google Scholar]
- Pastrana, D.V., Vass, W.C., Lowy, D.R., and Schiller, J.T. 2001. NHPV16 VLP vaccine induces human antibodies that neutralize divergent variants of HPV16. Virology 279: 361–369. [DOI] [PubMed] [Google Scholar]
- Patterson, J.B. and Samuel, C.E. 1995. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol. 15: 5376–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, J.B., Thomis, D.C., Hans, S.L., and Samuel, C.E. 1995. Mechanism of interferon action: Double-stranded RNA-specific adenosine deaminase from human cells is inducible by α and γ interferons. Virology 210: 508–511. [DOI] [PubMed] [Google Scholar]
- Polson, A.G., Bass, B.L., and Casey, J.L. 1996. RNA editing of hepatitis Δ virus antigenome by dsRNA-adenosine deaminase. Nature 380: 454–456. [DOI] [PubMed] [Google Scholar]
- Poulsen, H., Nilsson, J., Damgaard, C.K., Egebjerg, J., and Kjems, J. 2001. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol. Cell. Biol. 21: 7862–7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, C.E. and Lazinski, D.W. 2000. A host-specific function is required for ligation of a wide variety of ribozyme-processed RNAs. Proc. Natl. Acad. Sci. 97: 424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueter, S.M., T.R. Dawson, T.R., and Emeson, R.B. 1999. Regulation of alternative splicing by RNA editing. Nature 399: 75–80. [DOI] [PubMed] [Google Scholar]
- Sato, S., Wong, S.K., and Lazinski, D.W. 2001. Hepatitis δ virus minimal substrates competent for editing by ADAR1 and ADAR2. J. Virol. 75: 8547–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schade, M., Turner, C.J., Lowenhaupt, K., Rich, A., and Herbert, A. 1999. Structure-function analysis of the Z-DNA-binding domain Zα of dsRNA adenosine deaminase type I reveals similarity to the (α + β) family of helix-turn-helix proteins. EMBO J. 18: 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, T., Rould, M.A., Lowenhaupt, K., Herbert, A., and Rich, A. 1999. Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science 284: 1841–1845. [DOI] [PubMed] [Google Scholar]
- Sommer, B., Kohler, M., Sprengel, R., and Seeburg, P.H. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19. [DOI] [PubMed] [Google Scholar]
- Strauss, J.H. and Strauss, E.G. 1994. The alphaviruses: Gene expression, replication, and evolution. Microbiol Rev. 58: 491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Khillan, J., Gadue, P., and Nishikura, K. 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290: 1765–1768. [DOI] [PubMed] [Google Scholar]
- Wong, S.K. and D.W. Lazinski, D.W. 2002. Replicating hepatitis δ virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. 99: 15118–15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, S.K., Sato, S., and Lazinski, D.W. 2001. Substrate recognition by ADAR1 and ADAR2. RNA 7: 846–58. [DOI] [PMC free article] [PubMed] [Google Scholar]