Abstract
We have examined how splicing affects the expression of a range of human and nonhuman genes in vertebrate cells. Although our data demonstrate that splicing invariably enhances the level of gene expression, this positive effect is generally moderate. However, in the case of the human β-globin gene, splicing is essential for significant protein expression. In the absence of introns, 3′ end processing is inefficient, and this appears to be causally linked to a significant decrease in the level of both nuclear and cytoplasmic 3′ end-processed RNA. In contrast, splicing appears to only modestly enhance nuclear mRNA export. Consistent with this observation, intronless β-globin gene expression was only partially rescued by the insertion of retroviral nuclear mRNA export elements. Surprisingly, in the case of the highly intron dependent β-globin gene, the mRNA that did reach the cytoplasm was also only inefficiently translated if it derived from an intronless expression plasmid. Together, these data argue that splicing can increase gene expression by enhancing mRNA 3′ end processing, and hence, mRNA production. Moreover, in the case of the highly intron-dependent β-globin gene, splicing also significantly enhanced the translational utilization of cytoplasmic β-globin mRNAs.
Keywords: Splicing, polyadenylation, nuclear mRNA export, translation
INTRODUCTION
The life cycle of a eukaryotic mRNA can be subdivided into a number of steps that minimally include transcription, capping, splicing, polyadenylation, nuclear export, and finally, cytoplasmic translation and degradation. Although each of these processes can be studied in isolation, it has also become increasingly clear that each step in the mRNA life cycle is functionally connected to both earlier and later steps (reviewed by Maniatis and Reed 2002). For example, evidence has been presented that demonstrates that recruitment of a range of cellular factors by the carboxy-terminal domain of RNA polymerase II facilitates mRNA capping, splicing, and polyadenylation (Cho et al. 1997; McCracken et al. 1997). Splicing, in turn, is known to increase the efficiency of mRNA polyadenylation (Niwa et al. 1990; Huang and Carmichael 1996; Huang et al. 1999; Vagner et al. 2000), while both splicing and polyadenylation have been proposed to play an important role in the nuclear export of mRNAs (Buchman and Berg 1988; Ecker et al. 1991; Huang and Carmichael 1996; Luo and Reed 1999; Brodsky and Silver 2000; Hilleren et al. 2001; Dower and Rosbash 2002). The recent demonstration that intron removal results in deposition of a large protein complex, termed the exon junction complex (EJC), ∼20 nucleotides (nt) 5′ to the site of intron removal (Kataoka et al. 2000; Le Hir et al. 2000) provides a possible mechanism to explain the effect of splicing on the subsequent fate of an mRNA.
Previous efforts to examine the effect of mRNA splicing on the level of gene expression have demonstrated that a number of intron containing genes, including the genes encoding β-globin, thymidylate synthase, or purine nucleoside phosphorylase, are very poorly expressed when transcribed as cDNAs but effectively expressed if one or more introns are present in cis (Buchman and Berg 1988; Deng et al. 1989; Ryu and Mertz 1989; Jonsson et al. 1992). On the other hand, cDNAs derived from some other intron containing genes have been reported to express at high levels in cultured cells even in the absence of any introns (Pasleau et al. 1987; Brinster et al. 1988; Malim et al. 1988). Finally, a significant number of human genes, including c-jun and several histone and interferon genes, are naturally intronless (Nagata et al. 1980; Hentschel and Birnstiel 1981; Hattori et al. 1988). At least some of these naturally intronless genes, including histone H2A and the Hepatitis B Virus (HBV) genome, contain cis-acting elements that appear to functionally compensate for the absence of introns (Huang and Liang 1993; Huang and Yen 1995; Liu and Mertz 1995; Otero et al. 1998; Huang et al. 1999). In the case of HBV, this element has been termed the post-transcriptional regulatory element (PRE).
Previous efforts to define the step(s) in gene expression that are rendered inefficient in the absence of introns have generally focused on a single, highly intron dependent gene. Ryu and Mertz (1989) showed that the absence of an intron resulted in a significant drop in both the nuclear and cytoplasmic steady-state level of the mRNAs encoded by the SV40 late region, due primarily to a decrease in the nuclear stability of the encoded mRNA. It has also been reported that a lack of introns results in a decrease in the efficiency of both mRNA 3′ end processing and nuclear mRNA export (Ryu and Mertz 1989; Huang and Gorman 1990; Huang et al. 1999). One published study has, in particular, suggested that splicing is critical for rapid and efficient mRNA export in microinjected Xenopus oocytes (Luo and Reed 1999), although this result has not been fully supported by subsequent data from two other groups (Rodrigues et al. 2001; Ohno et al. 2002).
In this article, we have attempted to address the importance of splicing in mediating gene expression in human cells by surveying a number of genes for their intron dependence and by attempting to more clearly define the steps in gene expression that are inefficient in the absence of splicing. Our data show that all genes are expressed less effectively in the absence of splicing, but that this effect is generally moderate. However, the human β-globin gene proved to be highly intron-dependent. Although a lack of introns was found to reduce both the nuclear and cytoplasmic level of expression of the encoded mRNA, most probably by reducing polyadenylation efficiency, splicing only modestly enhanced nuclear mRNA export. Surprisingly, in the case of the highly intron dependent β-globin gene, a lack of splicing also appeared to markedly reduce the translational utilization of the mature mRNAs that do reach the cytoplasm.
RESULTS
Splicing only modestly enhances insulin gene expression
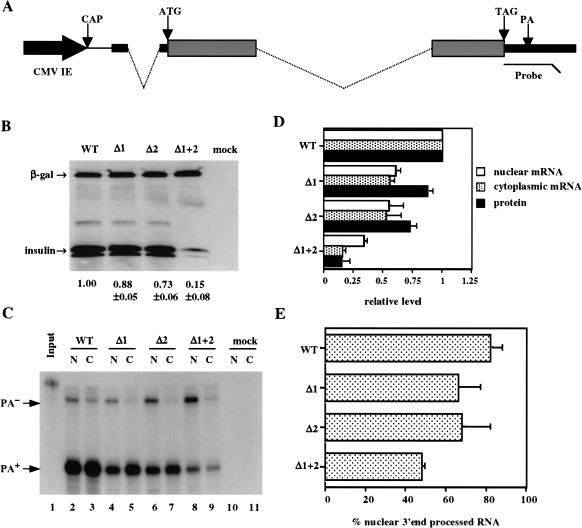
The rat preproinsulin II gene encodes three exons, that are 43, 201, and 199 nt in length, separated by two introns of 119 and 499 nt (Fig. 1A ▶; Lomedico et al. 1979). To test whether these introns enhance the level of preproinsulin mRNA and protein expression, we constructed plasmids designed to express the genomic insulin gene (pCMV/WT/INS), variants precisely lacking the first (pCMV/Δ1/INS) or second (pCMV/Δ2/INS) intron, and finally, a cDNA variant lacking both introns (pCMV/Δ1+2/INS). Importantly, all four plasmids are predicted to encode the identical mature preproinsulin II mRNA. The mRNA sequence encoded by these plasmids differs from the native preproinsulin II mRNA in the 5′ untranslated region (UTR), where the 5′ most 5 nt of the mRNA have been deleted and replaced by 84 nt of noncoding sequence contributed by the cytomegalovirus immediate early (CMV-IE) promoter. In addition, the mRNA has been modified by addition of an influenza hemaglutinin (HA) epitope tag to the carboxy-terminus of the insulin coding sequence. However, these expression plasmids do contain the complete genomic rat preproinsulin II polyadenylation sequence, including 3′ flanking sequences that are excluded from the mature mRNA.
FIGURE 1.
Effect of splicing on insulin gene expression. (A) Schematic representation of pCMV/WT/INS. The black arrow represents the CMV-IE promoter inserted 5′ to the genomic rat preproinsulin II gene. The insulin 5′ and 3′ UTRs are indicated by thick lines, the coding regions by gray boxes, and the introns by dotted lines. The thin line represents 5′ UTR sequences contributed by the CMV-IE promoter. ATG, translation initiation codon; TAG, translation termination codon; CAP, transcription start site; PA, polyadenylation site. The location of the RPA probe used to quantitate insulin RNA expression is also indicated. (B) Western analysis of insulin expression levels. 293T cells were mock transfected or transfected with 400 ng of pCMV/WT/INS, pCMV/δ1/INS, pCMV/δ2/INS, or pCMV/δ1+2/INS, together with 200 ng pCMV/β-gal as an internal control. (C) This RPA measures the expression level of unprocessed (PA−) or 3′ end-processed (PA+) insulin RNA encoded by pCMV/WT/INS, pCMV/δ1/INS, pCMV/δ2/INS, or pCMV/δ1+2/INS, in the nucleus [N] or cytoplasm [C] of 293T cells either mock transfected or transfected with 400 ng of the above expression plasmids. The RPA probe used is shown in lane 1 (1% of input). (D) The level of expression of nuclear and cytoplasmic 3′ end-processed insulin RNA or insulin protein is shown normalized to the level seen in 293T cells transfected with pCMV/WT/GLB, which is arbitrarily set at 1. Average of three experiments with SD indicated. (E) The level of nuclear 3′ end-processed insulin RNA in 293T cells transfected with the indicated expression plasmids is given as a percentage of total nuclear RNA, as quantified by RPA.
To examine the importance of these two introns in mediating preproinsulin II gene expression, we transfected human 293T cells with the above expression plasmids and then assayed insulin expression levels by Western analysis (Fig. 1B ▶) and RNase protection analysis (RPA; Fig. 1C ▶). As can be seen in Figure 1B ▶, precise deletion of either the first or the second intron had little effect on the level of insulin protein expression. However, deletion of both introns reduced insulin expression to ∼15% of the level seen with the parental genomic expression plasmid. Steady-state insulin RNA expression levels, in either the nucleus or cytoplasm of the transfected cells, were quantified using an RPA probe that can detect transcripts that have undergone proper 3′ end processing or that have not undergone appropriate processing (Fig. 1A ▶). This analysis revealed that removal of either intron decreased the level of both nuclear and cytoplasmic 3′ end-processed mRNA by ∼twofold (Fig. 1C,D ▶). Removal of both introns reduced the level of both nuclear and cytoplasmic 3′ end-processed RNA by four- to sixfold, that is, to about the level seen for insulin protein expression (Fig. 1D ▶). Therefore, in the case of the rat preproinsulin II gene, the introns present in the genomic copy of the gene appear to enhance protein expression, and cytoplasmic and nuclear 3′ end-processed RNA expression, by approximately the same ∼fivefold increment. Of interest, the level of insulin RNA that was appropriately 3′ end processed decreased from ∼82% to ∼48% of total nuclear insulin RNA when both introns were removed (Fig. 1E ▶). These data are consistent with earlier reports suggesting that the presence of a functional 3′ splice site in cis significantly enhances the utilization of a genomic polyadenylation signal (Huang and Gorman 1990; Huang et al. 1999).
β-globin gene expression is unusually intron dependent
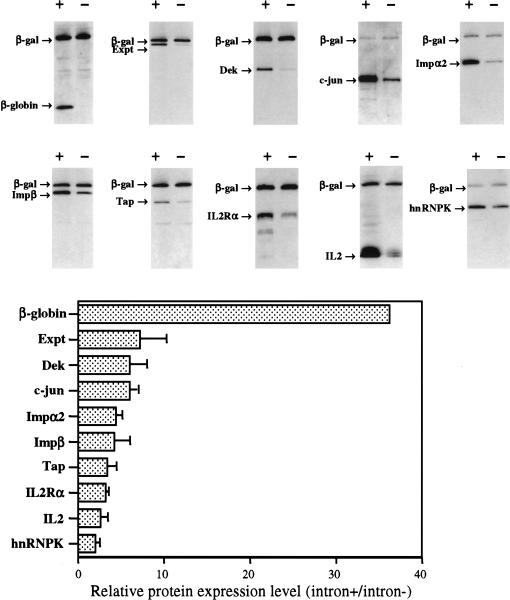
Although the enhancing effect of introns of rat preproinsulin II expression was readily detectable, it was modest compared to several previous reports showing that certain genes, such as β-globin, require introns for their successful expression from viruses or plasmids (Bushman and Berg 1988; Huang et al. 1999). We therefore wondered if insulin might be unusual in showing a limited dependence on introns. To test this hypothesis, we precisely substituted the insulin open reading frame in either pCMV/δ2/INS or in pCMV/δ1+2/INS with 1 of 10 open reading frames encoding a range of human proteins of different sizes and functions. These two matched expression plasmids would, therefore, in each case, express the same mature mRNA, containing 5′ and 3′ UTRs derived from the rat preproinsulin II gene (Fig. 1A ▶). However, in one case the pre-mRNA would contain an intron in the 5′ UTR, while in the second this intron would have been precisely removed prior to transcription. The human genes chosen included β-globin, which, as noted above, has been reported to be highly intron dependent, and c-jun, which is naturally intronless (Hattori et al. 1988). In all cases, we modified the encoded open reading frame by addition of either an N-terminal or C-terminal HA epitope tag. Each expression plasmid was then cotransfected into 293T cells, together with a plasmid encoding a HA-tagged form of β-galactosidase (β-gal) as an internal control, and protein expression quantified by Western analysis using an anti-HA monoclonal antibody. Efficient excision of the 5′ UTR intron was confirmed by RPA (data not shown).
As shown in Figure 2 ▶, all 10 tested genes were expressed at higher levels when they encoded an intron in their 5′ UTR. As expected from previous work, β-globin showed a high degree of intron dependence, giving an ∼35-fold higher level of protein expression when an intron was present in cis. In contrast, the other open reading frames were less strongly intron dependent, with the enhancement ranging from ∼twofold in the case of hnRNPK to ∼eightfold in the case of exportin t (Exp t). Curiously, c-jun, which is naturally intronless, was at least as responsive to the inclusion of an intron as were most intron-containing genes (Fig. 2 ▶). Together, these data suggested that the effect of an intron on the level of expression of the rat preproinsulin II gene in 293T cells was actually fairly typical, while β-globin expression appeared unusually sensitive to the absence of introns in cis.
FIGURE 2.
Effect of intron inclusion on the level of expression of a range of human genes. Western analysis of the level of protein expression for each of 10 different genes in 293T cells transfected with 400 ng each of a matched intron+ or intron− expression plasmid, together with pCMV/β-gal as an internal control. The relative increase in the level of expression of each protein that is seen upon intron inclusion is indicated at the bottom after normalization to the internal control. An average of three independent experiments with standard deviation is indicated. (The intronless β-globin construct gave rise to a level of protein expression that was 2.8 ± 1.8% of the level seen when an intron was present in cis.)
The relative intron dependence of genes is conserved in different cell types
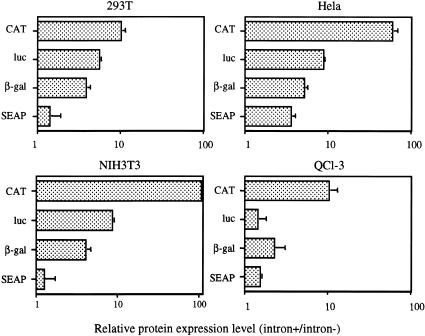
All of the genes tested in Figure 2 ▶ are of mammalian origin, and it could, therefore, be argued that they might contain sequences designed to either enhance or inhibit their expression in the absence of introns. To address this hypothesis, and also to determine whether 293T cells are unusually permissive for intronless gene expression, we constructed additional expression plasmids, again derived by precise substitution of the insulin open reading frame in either pCMV/δ2/INS or pCMV/δ1+2/INS. However, in this case we introduced two indicator genes of bacterial origin, that is, chloramphenicol acetyl transferase (CAT) and β-gal, one gene of insect origin, that is, luciferase (luc), and finally one additional gene of human origin, that is, secreted alkaline phosphase (SEAP). These plasmids were then transfected into human 293T or HeLa cells, into the murine cell line NIH3T3 or into the quail cell line QCl-3 (Fig. 3 ▶).
FIGURE 3.
Effect of intron inclusion on the expression of indicator genes in four cell lines. 293T, HeLa, NIH3T3, and quail QCl-3 cells were transfected with 200 ng each of a matched intron+ or intron− plasmid encoding CAT, β-gal, luc, or SEAP. Cells were also transfected with pCMV/β-gal or pCMV/CAT as an internal control. The fold increase seen upon intron inclusion is indicated, after correction for the internal control. An average of three experiments with SD is indicated.
In 293T cells, these four indicator genes behaved essentially as observed for the majority of human genes analyzed in Figure 2 ▶. Thus, CAT expression was enhanced by ∼10-fold by addition of an intron in cis, while β-gal expression was increased by ∼fivefold. In contrast, the SEAP indicator gene gave rise to only a minimal, ∼twofold expression increase upon inclusion of the identical intron (Fig. 3 ▶). Interestingly, the qualitative pattern seen in 293T cells, with CAT showing strong intron dependence, SEAP showing little effect and luc and β-gal being intermediate, was reproduced in the three other cell lines tested. However, although the data obtained in QCl-3 cells was quantitatively quite similar to the results seen in 293T cells, both HeLa and NIH3T3 generally showed a significantly greater increase in gene expression upon intron introduction, with CAT expression increasing by almost 100-fold in both cell lines (Fig. 3 ▶). Therefore, although these data suggest that different cell types can vary significantly in the absolute level of enhancement seen upon introduction of an intron into a specific gene expression plasmid, they also suggest that the higher dependence of certain genes on the presence of an intron is both intrinsic and reproducible in different cell and species contexts, that is, that the degree of intron dependence is encoded within the cDNA sequence of the gene. As the β-globin gene is clearly unusually dependent on introns for effective gene expression in 293T cells, even compared to genes of prokaryotic origin (Figs. 2, 3 ▶ ▶), these data imply that currently undefined sequences present in the β-globin gene act to inhibit β-globin expression when no introns are present in cis. In fact, Liu and Mertz (1996) have previously reported that sequences located in the 3′ terminal exon of human β-globin can inhibit appropriate mRNA processing.
Splicing has distinct effects on β-globin and insulin gene expression
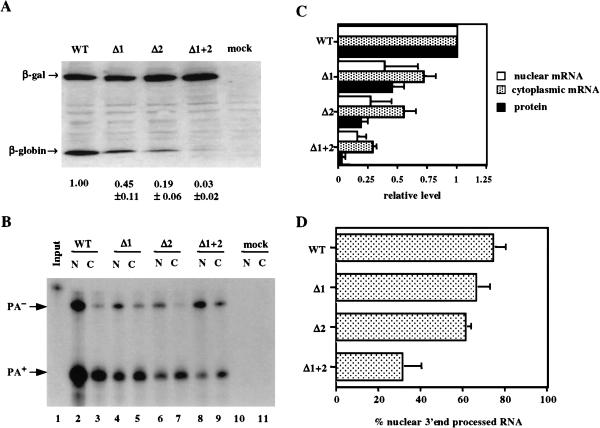
The structure of the genomic β-globin gene is in some ways quite similar to the preproinsulin II gene in that both genes consist of three exons, separated by two small introns (Lomedico et al. 1979; Buchman and Berg 1988). Both genes are also small, and are expressed at a very high level but in a developmentally regulated manner. We therefore wished to more completely analyze the effect of introns on β-globin gene expression when compared to their effect on preproinsulin expression (Fig. 1 ▶). For this purpose, we introduced the genomic β-globin coding sequence, including both introns, into pCMV/δ1+2/INS in place of the insulin open reading frame to give pCMV/WT/GLB. We then prepared derivatives precisely lacking the 5′ intron (pCMV/δ1/GLB), the 3′ intron (pCMV/δ2/GLB), or both introns (pCMV/δ1+2/GLB), and analyzed β-globin protein and RNA production. It is important to note that these β-globin expression constructs contain the same promoter, 5′ UTR, 3′ UTR, and genomic polyadenylation sequence present in the insulin expression plasmids analyzed in Figure 1 ▶.
As expected, removal of both introns resulted in a dramatic, ∼33-fold drop in the level of β-globin protein expression (Fig. 4A ▶). Removal of only the 5′ intron reduced β-globin protein expression by ∼twofold, while removal of the 3′ intron reduced protein expression by ∼fivefold. Remarkably, analysis of the nuclear and cytoplasmic level of 3′ end-processed mRNA revealed a significantly more modest effect at this level. Thus, removal of the second intron only reduced cytoplasmic 3′ end-processed mRNA by ∼twofold while removal of both introns reduced cytoplasmic 3′ end-processed mRNA by a mere ∼fourfold (Fig. 4B,C ▶). Therefore, although removal of both introns from the genomic β-globin gene has a similar effect on the level of cytoplasmic β-globin mRNA expression as does removal of both introns from the rat preproinsulin II gene (cf. Fig. 1C,D ▶ and Fig. 4B,C ▶), β-globin protein expression is clearly more severely impacted than is insulin protein expression.
FIGURE 4.
Effect of introns on β-globin gene expression. (A) Western analysis of β-globin expression levels. 293T cells were either mock transfected or transfected with 400 ng of pCMV/WT/GLB, pCMV/δ1/GLB, pCMV/δ2/GLB, or pCMV/δ1+2/GLB together with 200 ng pCMV/β-gal as an internal control. (B) This RPA was performed as described in Figure 1C ▶. (C) This quantitation was performed as described in Figure 1D ▶. (D) The level of nuclear 3′ end-processed β-globin RNA in 293T cells transfected with the indicated expression plasmids is given as a percentage of total nuclear β-globin RNA, as determined by RPA.
If the absence of introns reduces the efficiency of nuclear mRNA export, then one would predict that there would be a significant drop in the cytoplasmic level of that mRNA while nuclear mRNA levels might be largely unaffected, as has indeed been previously reported (Chang and Sharp 1989; Malim et al. 1989). However, intron removal, in fact, resulted in a comparable decline in the level of nuclear and cytoplasmic poly(A)+ mRNA in the case of both the insulin (Fig. 1C,D ▶) and β-globin gene (Fig. 4B,C ▶), thus suggesting that splicing may not greatly enhance nuclear mRNA export.
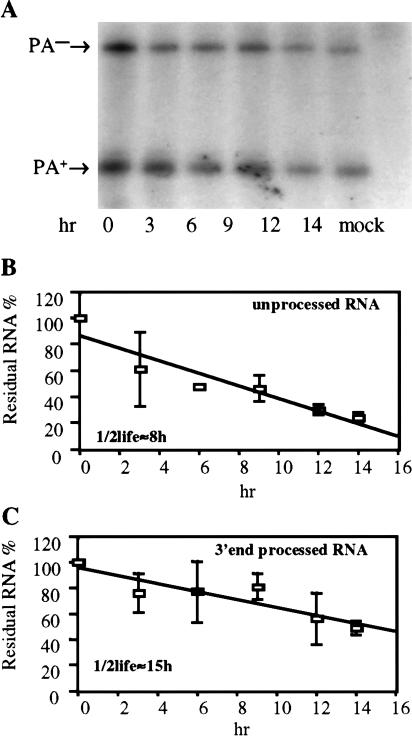
As previously noted for the rat preproinsulin II gene (Fig. 1E ▶), removal of both introns from the β-globin gene reduced the efficiency of 3′ end processing at the flanking genomic rat preproinsulin II polyadenylation sequence. This effect reduced the proportion of the total nuclear globin RNA that was correctly 3′ end processed from ∼75% to ∼30% at steady state (Fig. 4D ▶). However, if the half-life of the unprocessed β-globin RNA is significantly less than the half-life of the 3′ end-processed RNA, then this steady-state measurement might actually significantly underestimate the indirect effect of inefficient 3′ end formation on β-globin mRNA expression. To address this question, we measured the half-life of 3′ end-processed and unprocessed β-globin RNA in 293T cells transfected with β-globin expression plasmids that do or do not contain an intron, after treatment of cells with actinomycin D. These data (Fig. 5 ▶; data not shown) demonstrate that the half-life of 3′ end-processed β-globin RNA, derived from an intron containing β-globin expression plasmid, was 21 ± 7 h, while the half-life of the unprocessed RNA was a substantially shorter 9 ± 1 h. Similarly, the half-life of 3′ end-processed β-globin RNA derived from the intronless pCMV/δ1+2/GLB plasmid was 15 ± 3 h, while the unprocessed RNA had a half-life of 8 ± 1 h. Although the minor differences in the observed half-lives of the in principle identical mRNAs obtained from these two different β-globin expression plasmids are of uncertain significance, it is clear that the unprocessed β-globin RNA is substantially less stable than the 3′ end-processed RNA. These data therefore imply that the steady-state ratio of processed versus unprocessed globin RNA expression measured in Figure 4 ▶ may significantly underestimate the inhibitory effect of intron removal on polyadenylation efficiency. As unprocessed globin RNA is largely retained in the nucleus and then degraded (Fig. 4B ▶), inefficient polyadenylation appears to be sufficient to explain most of the ∼fourfold drop in the cytoplasmic level of globin mRNA noted in cells transfected with the intronless pCMV/δ1+2/GLB plasmid (Fig. 4C ▶).
FIGURE 5.
Half-life of β-globin RNAs. (A) This RPA measures the relative level of total 3′ end-processed (PA+) and unprocessed (PA−) RNA in 293T cells transfected with the intronless pCMV/δ1+2/GLB plasmid after incubation with the transcription inhibitor actinomycin D for the indicated number of hours. (B) This figure shows the rate of decay of the unprocessed β-globin RNA, determined as shown in (A). An average of three independent experiments with SD is indicated. (C) Same as (B), except that the rate of decay of the 3′ end-processed RNA encoded by pCMV/δ1+2/GLB is measured. Analogous experiments were also performed using 293T cells transfected with an intron containing β-globin expression plasmid.
The HBV PRE, unlike retroviral RNA export elements, effectively rescues intronless β-globin gene expression
As an alternative approach to addressing whether intron removal compromises mRNA expression by inhibiting nuclear mRNA export, we next asked if expression of an intronless β-globin gene would be enhanced by the introduction of known nuclear mRNA export elements into the 3′ UTR. The inserted elements included the Mason Pfizer Monkey Virus (MPMV) constitutive transport element (CTE), which is able to directly recruit the cellular Tap nuclear mRNA export factor (Grüter et al. 1998; Kang and Cullen 1999). Importantly, Tap is believed to normally mediate the nuclear export of cellular mRNAs, and has been reported to be recruited by the EJC (Kataoka et al. 2001; Le Hir et al. 2001b). In addition, we also inserted the human immunodeficiency virus type-1 (HIV-1) Rev Response Element (RRE) which, in the presence of the HIV-1 Rev protein, can efficiently recruit the cellular Crm1 nuclear RNA export factor (Fornerod et al. 1997; Malim et al. 1989). Finally, we also introduced the HBV PRE, an RNA element whose mechanism of action is unknown but that has been shown to post-transcriptionally enhance the expression of intronless HBV mRNAs (Huang and Liang 1993; Huang and Yen 1995; Otero et al. 1998).
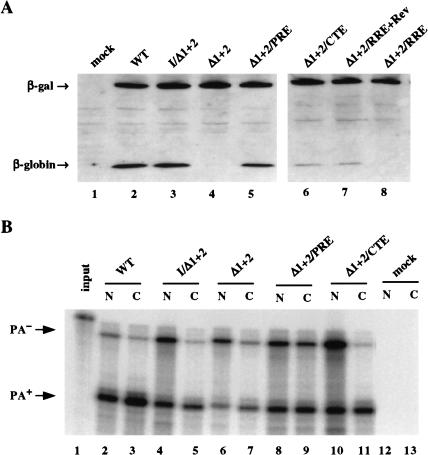
As shown in Figure 6A ▶, and quantitated in Table 1 ▶, efficient β-globin protein expression is sustained by either the two genomic β-globin introns or by insertion of the 5′ UTR intron from the preproinsulin II gene, while little β-globin protein is expressed when no intron is present (Fig. 6A ▶, lanes 2,4). Insertion of either the MPMV CTE or the HIV-1 RRE into the 3′ UTR of pCMV/δ1+2/GLB enhanced β-globin protein expression by ∼fivefold, that is, to ∼20% of the level seen when an intron is present (Fig. 6A ▶, lanes 6,7; Table 1 ▶). In contrast, the HBV PRE was able to essentially completely rescue protein expression from this intronless β-globin gene (Fig. 6A ▶, lane 5; Table 1 ▶).
FIGURE 6.
Effect of various viral RNA elements on intronless β-globin gene expression. (A) Western analysis of the β-globin expression level in 293T cells either mock transfected or transfected with 400 ng of the indicated expression plasmid, together with 200 ng pCMV/β-gal as an internal control. (B) This representative RPA was performed as described in Figure 1C ▶.
TABLE 1.
Comparison of β-globin protein and 3′ end-processed β-globin RNA expression
| Introduced β-globin expression plasmid | Relative protein expression | Relative level of nuclear 3′ end-processed RNA | Relative level of cytoplasmic 3′ end-processed RNA |
| pCMV/WT/GLB | 1.00 | 1.00 | 1.00 |
| pCMV/I/δ1+2/GLB | 1.04 ± 0.11 | 0.66 ± 0.13 | 0.41 ± 0.07 |
| pCMV/δ1+2/GLB | 0.04 ± 0.02 | 0.17 ± 0.09 | 0.23 ± 0.12 |
| pCMV/δ1+2/PRE/GLB | 0.77 ± 0.19 | 0.49 ± 0.27 | 0.50 ± 0.03 |
| pCMV/δ1+2/CTE/GLB | 0.23 ± 0.07 | 0.59 ± 0.30 | 0.43 ± 0.17 |
| pCMV/δ1+2/RRE/GLB+pcRev | 0.17 ± 0.03 | ND | ND |
| pCMV/δ1+2/RRE/GLB | 0.02 ± 0.03 | ND | ND |
This table shows the relative level of β-globin protein, as well as nuclear and cytoplasmic 3′ end-processed mRNA, detected upon transfection of 293T cells with the indicated expression plasmids. These data are normalized to the level seen with the pCMV/WT/GLB genomic β-globin expression plasmid and represent the average of three independent experiments, with standard deviation indicated. No RNA analysis was performed with the cultures transfected with the pCMV/δ1+2/RRE/GLB plasmid because the pcRev expression plasmid contains the same polyadenylation site. ND, not determined.
We next performed an RPA to quantify the level of 3′ end-processed and unprocessed RNA expression in the nucleus and cytoplasm of cells transfected with the plasmids listed in Figure 6A ▶ (Fig. 6B ▶; Table 1 ▶). The most important point to emerge from this analysis was that introduction of the insulin-derived 5′ UTR intron into pCMV/δ1+2/GLB, or insertion of the PRE into the 3′ UTR, both increased the level of cytoplasmic 3′ end-processed mRNA by only two- to threefold (Fig. 6B ▶, cf. lanes 5,7,9) yet increased β-globin protein expression by up to 25-fold (Fig. 6A ▶; Table 1 ▶). In contrast, while the MPMV CTE also enhanced the cytoplasmic level of intronless unprocessed β-globin mRNA expression by ∼twofold (Fig. 6B ▶, lane 11; Table 1 ▶), this did not result in an equivalently dramatic increase in β-globin protein expression (Fig. 6A ▶, lane 6; Table 1 ▶). Therefore, both splicing and the PRE appear to significantly enhance the cytoplasmic translational utilization of β-globin mRNAs. However, the insertion of an intron into the 5′ UTR of the intronless β-globin gene, or insertion of the PRE into the 3′ UTR, appeared to only modestly enhance the efficiency of 3′ end processing at the genomic preproinsulin polyadenylation site (Fig. 6B ▶).
As noted above, the MPMV CTE functions by binding to the Tap nuclear mRNA export factor while Rev recruits the cellular Crm1 RNA export factor to the RRE (Fornerod et al. 1997; Grüter et al. 1998; Kang and Cullen 1999). In contrast, the mechanism of action of the HBV PRE remains unclear. To address whether the HBV PRE indeed functions as an effective nuclear mRNA export sequence, we asked if the HBV PRE could rescue the cytoplasmic expression of the late, structural proteins encoded by a Rev− HIV-1 proviral clone when inserted in cis. As shown in Table 2 ▶, this Rev− HIV-1 provirus, as expected (Malim et al. 1989; Yi et al. 2002), failed to produce detectable levels of the p24 Gag protein when transfected into 293T cells. However, p24 Gag production could be effectively rescued by either cotransfection of a Rev expression plasmid or by insertion of the MPMV CTE into a unique XhoI site present in the proviral clone. In contrast, insertion into the same XhoI site of the HBV PRE fragment that could efficiently rescue β-globin protein expression (Fig. 6A ▶) had no effect on p24 Gag production (Table 2 ▶), although cotransfection of a Rev expression plasmid again fully rescued p24 Gag protein production (data not shown). Therefore, at least in this rigorous assay for nuclear mRNA export ability, the HBV PRE differs from the MPMV CTE or the HIV-1 Rev/RRE combination in being inactive.
TABLE 2.
The HBV PRE cannot rescue Gag production by a Rev− HIV-1 Proviral Clone
| HIV-1 proviral clone | p24 Gag production (ng/mL) |
| pNL4-3 Rev− | 0.2 |
| pNL4-3 Rev− + pcRev | 92.3 |
| pNL4-3 Rev−/CTE | 74.7 |
| pNL4-3 Rev−/PRE | 0.6 |
293T cells were transfected with 500 ng of the indicated proviral expression plasmid and with 1000 ng of pcRev, or of the pBC12/CMV plasmid as a negative control. At 72 h after transfection, secreted p24 Gag production was measured by ELISA.
DISCUSSION
Two recent discoveries have prompted us to reexamine the importance of splicing in modulating the fate of mRNA transcripts. The most significant of these is the observation that splicing results in the deposition of the EJC proteins 20 to 22 nt 5′ to each exon junction site (Kataoka et al. 2000; Le Hir et al. 2000). The EJC includes the protein Aly/Ref, which has been proposed to recruit the Tap nuclear mRNA export factor (Zhou et al. 2000; Le Hir et al. 2001b; Rodrigues et al. 2001), as well as the proteins Y14, Magoh and RNPS1, which have been proposed to remain at least transiently bound to mRNAs after their export to the cytoplasm (Kataoka et al. 2001; Kim et al. 2001; Lykke-Andersen et al. 2001). These EJC proteins therefore have the potential to regulate the cytoplasmic fate of an mRNA and, in fact, EJC components are now thought to be involved in regulating mRNA subcellular localization and in activating nonsense-mediated mRNA decay (Kataoka et al. 2001; Le Hir et al. 2001a, 2001b; Lykke-Andersen et al. 2001). It has also been reported that intronless mRNAs are very inefficiently exported from the nucleus of microinjected Xenopus oocytes when compared to identical mRNAs that have undergone splicing after injection (Luo and Reed 1999). Although this result has been controversial (Rodrigues et al. 2001; Ohno et al. 2002), these observations, nevertheless, suggest that mRNA nuclear export might be strongly enhanced by the EJC-mediated recruitment of Tap to spliced mRNA molecules.
Previous efforts to address the importance of splicing in mRNA expression have identified several intron containing genes that are very poorly expressed when they are transcribed as cDNAs (Buchman and Berg 1988; Deng et al. 1989; Ryu and Mertz 1989; Jonsson et al. 1992). However, other intron-containing genes have been reported to be expressed efficiently in the absence of introns, while a number of human genes are known to be naturally devoid of introns (Nagata et al. 1980; Hentschel and Birnstiel 1981; Pasleau et al. 1987; Brinster et al. 1988; Hattori et al. 1988; Malim et al. 1988). Attempts to identify the steps in the mRNA life cycle that are affected by splicing in highly intron-dependent genes have suggested that a lack of introns (1) significantly reduces both the nuclear and cytoplasmic mRNA level, (2) reduces the efficiency of transcript polyadenylation, and (3) can reduce the efficiency of nuclear mRNA export (Ryu and Mertz 1989; Huang and Gorman 1990; Huang et al. 1999). Why other genes can be effectively expressed in the absence of introns has not previously been addressed in detail, although it has been proposed that at least some genes that naturally lack introns may contain cis-acting RNA elements that compensate for this deficiency (Huang and Liang 1993; Huang et al. 1999).
Many genes are not highly intron-dependent
In this article, we have examined a range of human and nonhuman genes for their intron dependence in vertebrate cells and have observed that highly intron-dependent genes, such as the β-globin gene, appear to be unusual (Fig. 2 ▶). Although all genes tested were clearly expressed more efficiently when an intron was present in cis, the observed activation, while significant, tended to be relatively modest, at least in 293T cells. Interestingly, the relative intron dependence of genes, but not the quantitative effect of introns on the level of gene expression, proved to be conserved when measured in different cell lines derived from different species (Fig. 3 ▶). This argues that genes, either fortuitously or by evolutionary design, contain sequence information that allows them to be more or less effectively expressed as cDNA transcripts. As the two prokaryotic genes tested displayed the same moderate intron dependence typical of most human genes (Figs. 2, 3 ▶ ▶), it appears that highly intron-dependent genes such as β-globin probably contain sequences that inhibit intron-independent gene expression (Liu and Mertz 1996).
Splicing enhances 3′ end processing and thereby increases nuclear and cytoplasmic mRNA levels
A comparison of the highly intron-dependent β-globin gene (Fig. 4 ▶) with the moderately intron-dependent preproinsulin II gene (Fig. 1 ▶) demonstrated that a lack of introns markedly reduced the efficiency of 3′ end processing at the genomic preproinsulin II poly(A) addition site present in both expression plasmids (Figs. 1E , 4D ▶ ▶). Largely as a result, a lack of introns also led to a readily detectable drop in the level of nuclear 3′ end-processed mRNA, although total nuclear RNA levels were less affected. However, as the unprocessed RNA is both significantly more labile than the analogous 3′ end-processed mRNA (Fig. 5 ▶), and largely unable to reach the cytoplasm (Figs. 1C, 4B ▶ ▶), this defect in polyadenylation appeared to largely explain the four- to fivefold drop in cytoplasmic mRNA observed in cultures transfected with intronless expression plasmids (Figs. 1D, 4C ▶ ▶). It remains possible, as suggested by others, that splicing may also enhance gene transcription (Furger et al. 2002). However, this effect would have to be fairly modest in the experiments reported here, possibly due to the use of the highly active CMV-IE promoter.
Splicing only modestly enhances nuclear mRNA export
If splicing is critical for effective nuclear mRNA export, then one would predict that a lack of introns would selectively reduce the cytoplasmic level of the encoded mRNA transcript. In fact, mutation of the 5′ or 3′ splice site of the second intron present in β-globin mRNA has previously been shown to prevent the nuclear export of the unspliced β-globin mRNA and instead induces its nuclear accumulation (Chang and Sharp 1989). In contrast, we did not observe a disproportionate effect of splicing on the cytoplasmic level of either insulin or β-globin 3′ end-processed mRNA (Figs. 1C,D, 4B,C ▶ ▶). It is important to note that an experimental strategy that only measured total RNA levels in the cell nucleus and cytoplasm (e.g., Ryu and Mertz 1989) would have detected a significantly greater inhibitory effect of an absence of introns on the cytoplasmic level of mRNA expression. However, this effect would, in fact, have been primarily due to the presence in the nucleus of a substantial pool of improperly 3′ end-processed RNA that is not effectively exported (e.g., Fig. 1C ▶). The observed inability of this nonpolyadenylated RNA to exit the human nucleus is interesting, given recent data suggesting a strong functional link between mRNA 3′ end formation and nuclear export (Custodio et al. 1999; Brodsky and Silver 2000; Hilleren et al. 2001; Dower and Rosbash 2002).
The MPMV CTE has been shown to avidly bind to the Tap nuclear mRNA export factor (Grüter et al. 1998; Kang and Cullen 1999), and Tap has also been reported to interact with the EJC (Le Hir et al. 2001b). We, therefore, asked if the MPMV CTE would be able to functionally substitute for introns in mediating β-globin gene expression. In fact, both the MPMV CTE, as well as the functionally analogous HIV-1 Rev/RRE combination, were able to partially rescue β-globin protein expression induced by the intronless β-globin gene (Fig. 6 ▶; Table 1 ▶). Although this result suggests that the stimulatory effect of splicing on mRNA expression may, in part, be due to enhanced nuclear mRNA export, it is also clear that splicing is not essential for the nuclear export and translation of most mRNA species (Fig. 2 ▶). The recent demonstration that EJC proteins differ from bona fide nuclear export factors such as Tap, p15, and UAP56 in being largely dispensable for nuclear mRNA export in metazoan cells (Gatfield and Izaurralde 2002) is consistent with this hypothesis. Importantly, recent data have demonstrated that recruitment of mRNA export factors occurs not only during splicing but also cotranscriptionally, and these two recruitment processes may therefore be partly redundant (Lei et al. 2001; Sträβer et al. 2002; Zenklusen et al. 2002).
Splicing can affect mRNA translation efficiency
A striking observation to emerge from these studies is that splicing can enhance mRNA translation, at least in the case of the highly intron-dependent β-globin gene. The first hint of this phenomenon came from a comparison of the effect of splicing on the level of insulin and β-globin 3′ end-processed mRNA and protein expression. As shown in Figure 1D ▶, a lack of introns reduced the cytoplasmic level of insulin mRNA and protein expression by the same ∼sixfold. In contrast, while intron removal reduced cytoplasmic β-globin mRNA levels by four- to fivefold, the drop in protein expression was a more dramatic ∼33-fold (Fig. 4C ▶).
More evidence in favor of an effect of splicing on mRNA utilization came from an analysis of the effect of various cis-acting RNA elements on β-globin protein and mRNA expression (Fig. 6 ▶; Table 1 ▶). Specifically, we observed that insertion of an intron into the 5′ UTR of pCMV/δ1+2/GLB, which has no effect on the encoded mature mRNA, fully restored β-globin protein expression (an ∼25-fold increase) yet increased cytoplasmic mRNA levels by less than threefold (Fig. 6 ▶; Table 1 ▶). A similar effect was noted with the HBV PRE, which increased β-globin protein expression by ∼20-fold yet enhanced cytoplasmic mRNA levels by only two- to threefold. In contrast, the MPMV CTE nuclear RNA export signal also increased cytoplasmic mRNA levels by ∼twofold yet increased β-globin protein expression by only ∼fivefold. Therefore, these data suggest that both splicing and the HBV PRE enhance not only the cytoplasmic level of 3′ end-processed β-globin mRNA but also the utilization of that mRNA. In contrast, the MPMV CTE enhanced protein expression by almost the same increment as mRNA expression.
The HBV PRE can act at a level distinct from nuclear mRNA export
An interesting observation reported in this manuscript is that the HBV PRE differs from the retroviral MPMV CTE and Rev/RRE nuclear mRNA export signals in two important ways. First, the HBV PRE is essentially fully competent to rescue intronless β-globin gene expression when introduced in cis, while the MPMV CTE and the HIV-1 Rev/RRE have only a moderate enhancing effect (Fig. 6A ▶). Second, the HBV PRE was found to differ from both the Rev/RRE and the MPMV CTE in being unable to support the nuclear export and expression of the unspliced, late mRNAs encoded by a Rev− HIV-1 provirus (Table 2 ▶). These data suggest that the mechanism of action of the HBV PRE, which remains poorly understood, may be quite different from the mechanism of action of the MPMV CTE and the HIV-1 Rev/RRE, both of which act exclusively to export intron-containing retroviral RNA transcripts (Malim et al. 1989; Grüter et al. 1998; Kang and Cullen 1999; Yi et al. 2002). Although the HBV PRE may contribute to the nuclear export of intronless nuclear mRNAs (Huang and Liang 1993; Huang and Yen 1995; Otero et al. 1998), these data suggest that the HBV PRE may also reproduce the positive effect of splicing on other stages in the mRNA life cycle, including enhanced cytoplasmic utilization.
In conclusion, our data agree with earlier reports indicating that splicing enhances mRNA 3′ end formation and, at least in part as a result, enhances both the nuclear and cytoplasmic level of mRNA expression (Ryu and Mertz 1989; Huang and Gorman 1990; Huang et al. 1999). Our data do not support a critical role for splicing in mediating mRNA nuclear export but do demonstrate, at least in the case of the highly intron-dependent β-globin gene, that splicing can markedly enhance the cytoplasmic translation of an mRNA. These results agree with data reported previously by Matsumoto et al. (1998), suggesting that splicing can markedly enhance mRNA translational utilization in microinjected Xenopus oocytes. Our observations also closely agree with results recently obtained by Nott et al. (2003) who also observed that splicing can significantly enhance mRNA translational utilization. In total, these observations suggest that the EJC proteins may not only regulate the cytoplasmic localization and stability of spliced mRNAs but also their translation.
MATERIALS AND METHODS
Construction of molecular clones
All expression plasmids utilized in this work are based on pBC12/CMV. The following expression plasmids have been described previously: pBC12/CMV, pBC12/CMV/β-gal, pcRev, and pNL4-3Rev− (Malim et al. 1988; Yi et al. 2002). pCMV/WT/INS was generated by insertion of the entire genomic rat preproinsulin II gene (Lomedico et al. 1979) into pBC12/CMV. pCMV/δ1/INS, pCMV/δ2/INS, and pCMV/δ1+2/INS are variants of pCMV/WT/INS respectively lacking the first, the second, or both introns. pCMV/WT/GLB was generated by insertion of the genomic form of the complete human β-globin open reading frame into a modified form of pCMV/δ1+2/INS bearing a unique NcoI site coincident with the translation initiation codon and a unique XhoI site immediately 3′ to the translation termination codon. pCMV/δ1/GLB, pCMV/δ2/GLB, and pCMV/δ1+2/GLB are intron deletion variants generated from pCMV/WT/GLB using recombinant PCR. pCMV/I/δ1+2/GLB was generated by insertion of the insulin 5′ UTR intron into pCMV/δ1+2/GLB.
pCMV/CAT, pCMV/β-gal, pCMV/luc, pCMV/SEAP, pCMV/TAP, pCMV/Impα2, pCMV/Impβ, pCMV/DEK, pCMV/Xpot, pCMV/c-jun, pCMV/IL2, and pCMV/IL2Rα were generated by insertion of the relevant PCR amplified gene coding sequence between the NcoI and XhoI sites present in the modified pCMV/δ1+2/INS plasmid. Each equivalent intron containing plasmid was generated by insertion of the genomic insulin 5′ UTR.
pCMV/δ1+2/GLB/PRE, pCMV/δ1+2/GLB/CTE, and pCMV/δ1+2/GLB/RRE were generated by insertion of the PCR amplified, full-length HBV PRE, MPMV CTE, or HIV-1 RRE (Malim et al. 1989; Grüter et al. 1998; Otero et al. 1998; Kang and Cullen 1999) into the unique XhoI site present in pCMV/δ1+2/GLB. All expression plasmids used for Western blot analysis have either an amino terminal or carboxyl terminal HA tag added in frame. The integrity of the DNA sequences introduced into expression plasmids was confirmed by DNA sequencing.
The pNL4-3Rev−/PRE and pNL4-3Rev−/CTE HIV-1 proviral expression plasmids were generated by insertion of the HBV PRE or MPMV CTE sequence into a unique XhoI site present in the dispensable nef gene of pNL4-3Rev− (Yi et al. 2002).
Cell culture, transfection, and reporter assay
Human 293T cells and Hela cells, mouse NIH3T3 cells, and quail QCl-3 cells were maintained as described previously (Kang and Cullen 1999), and were transfected using either Fugene-6 (Roche Molecular Biochemicals) or DEAE-dextran. CAT, β-gal, SEAP, and luc enzyme activities were determined ∼48 h after transfection. The observed reporter activities were adjusted for minor differences in transfection efficiency or sample recovery, as revealed by cotransfected internal control plasmids.
RNA isolation and ribonuclease protection assay
Nuclear and cytoplasmic RNA were isolated using an RNeasy Mini kit (Qiagen) and RPA performed using a Hyspeed RPA kit (Ambion) at ∼48 h after transfection of 293T cells, as described previously (Yi et al. 2002). The RNA probes used in the RPA were generated by in vitro transcription of PCR products containing a T7 promoter sequence, using a Riboprobe kit (Promega). The RNA probe that traverses the preproinsulin genomic polyadenylation site is 320 nt in length, while probe fragments rescued by polyadenylated and nonpolyadenylated mRNA transcripts are predicted to be 166 and 290 nt, respectively. Protected probe fragments were separated by gel electrophoresis, visualized by autoradiography, and individual bands then quantified using a Phosphorimager.
Half-lives of poly(A)+ and poly(A)− β-globin RNAs were measured by addition of Actinomycin D (100 μg/mL final) to the culture medium at ∼40 h after transfection of 293T cells. Cells were harvested at 0, 3, 6, 9, 12, or 14 h after drug addition and total RNA isolated using the RNeasy Mini Kit (Qiagen) followed by RPA as described above.
Western blot analyses
Western blot assays and quantitation of reactive protein levels were performed using 293T cells at ∼48 h after transfection. A murine monoclonal anti-HA antibody (Covance), followed by treatment with horseradish peroxidase-conjugated rabbit antimouse antiserum (Amersham) was used. Reactive proteins were visualized by enhanced chemiluminescence followed by autoradiography and quantified by NIH Image 1.62 software after scanning the film.
Virus replication assay
293T cells were cotransfected with 500 ng of the pNL4-3Rev−, pNL4-3Rev−/PRE, or pNL4-3Rev−/CTE HIV-1 proviral expression plasmid and 1000 ng of pBC12/CMV or pcRev. At ∼72 h after transfection, supernatant media were collected, and p24 Gag antigen production quantified using an enzyme-linked immunosorbent assay (ELISA) kit (NEN Life Science), as described previously (Yi et al. 2002).
Acknowledgments
The authors thank Heather Wiegand and Rui Yi for help with parts of this research project. We also thank Tom Hope, Iain Mattaj, and Gideon Dreyfuss for reagents used in this work and Elisa Izaurralde and Melissa Moore for communication of their results prior to publication.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5260303.
REFERENCES
- Brinster, R.L., Allen, J.M., Behringer, R.R., Gelinas, R.E., and Palmiter, R.D. 1988. Introns increase transcriptional efficiency in transgenic mice. Proc. Natl. Acad. Sci. 85: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, A.S. and Silver, P.A. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6: 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman, A.R. and Berg, P. 1988. Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 8: 4395–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D.D. and Sharp, P.A. 1989. Regulation by HIV Rev depends upon recognition of splice sites. Cell 59: 789–795. [DOI] [PubMed] [Google Scholar]
- Cho, E.J., Takagi, T., Moore, C.R., and Buratowski, S. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 11: 3319–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio, N., Carmo-Fonseca, M., Geraghty, F., Pereira, H.S., Grosveld, F., and Antonion, M. 1999. Inefficient processing impairs release of RNA from the site of transcription. EMBO J. 18: 2855–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, T., Li, Y., and Johnson, L.F. 1989. Thymidylate synthase gene expression is stimulated by some (but not all) introns. Nucleic Acids Res. 17: 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower, K. and Rosbash, M. 2002. T7 RNA polymerase-directed transcripts are processed in yeast and link 3′ end formation to mRNA export. RNA 8: 686–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner, R., Ellmeier, W., and Birnstiel, M.L. 1991. Mature messenger RNA 3; End formation stimulates RNA export from the nucleus. EMBO J. 10: 3513–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod, M., Ohno, M., Yoshida, M., and Mattaj, I.W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060. [DOI] [PubMed] [Google Scholar]
- Furger, A., O’Sullivan, J.M., Binnie, A., Lee, B.A., and Proudfoot, N.J. 2002. Promoter proximal splice sites enhance transcription. Genes & Dev. 16: 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield, D. and Izaurralde, E. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüter, P., Tabernero, C., von Kobbe, C., Schmitt, C., Saavedra, C., Bachi, A., Wilm, M., Felber, B.K., and Izaurralde, E. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1: 649–659. [DOI] [PubMed] [Google Scholar]
- Hattori, K., Angel, P., Beau, M.M.L., and Karin, M. 1988. Structure and chromosomal localization of the functional intronless human JUN protooncogene. Proc. Natl. Acad. Sci. 85: 9148–9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel, C.C. and Birnstiel, M.X. 1981. The organization and expression of histone gene families. Cell 25: 301–313. [DOI] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R,. and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542. [DOI] [PubMed] [Google Scholar]
- Huang, J. and Liang, T.J. 1993. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol. Cell. Biol. 13: 7476–7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, M.T.-F. and Gorman, C.M. 1990. Intervening sequences increase efficiency and accumulation of cytoplasmic RNA. Nucleic Acids Res. 18: 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. and Carmichael, G.G. 1996. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 16: 1534–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y., Wimler, K.M., and Carmichael, G.G. 1999. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 18: 1642–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Z.-M. and Yen, T.S.B. 1995. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol. Cell. Biol. 5: 3864–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson, J.J., Goresman, M.D., Wilson, N., and McIvor, R.S. 1992. Intron requirement for expression of the human purine nucleoside phosphorylase gene. Nucleic Acids Res. 20: 3191–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y. and Cullen, B.R. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes & Dev. 13: 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, N., Diem, M.D., Kim, V.N., Yong, J., and Dreyfuss, G. 2001. Magoh, a human homolog of Drosophila magno nashi protein, is a component of the splicing-dependent exon–exon junction complex. EMBO J. 20: 6424–6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, N., Yong, J., Kim, V.N., Velazquez, F., Perkinson, R.A., Wang, F., and Dreyfuss, G. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6: 673–682. [DOI] [PubMed] [Google Scholar]
- Kim, V.N., Yong, J., Kataoka, N., Abel, L., Diem, M.D., and Dreyfuss, G. 2001. The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J. 20: 2062–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Braun, I.C., Forler, D., and Izaurralde, E. 2001a. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 21: 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001b. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, E.P., Krebber, H., and Silver, P.A. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes & Dev. 15: 1771–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. and Mertz, J. 1995. HnRNP L binds a cis-acting RNA sequence element that enables intron-independent gene expression. Genes & Dev. 9: 1766–1780. [DOI] [PubMed] [Google Scholar]
- Liu, X. and Mertz, J. 1996. Sequence of the polypyrimidine tract of the 3′-terminal 3′ splicing signal can affect intron-dependent pre-mRNA processing in vivo. Nucleic Acids Res. 24: 1765–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomedico, P., Rosenthal, N., Efstratiadis, A., Gilbert, W., Kolodner, R., and Tizard, R. 1979. The structure and evolution of the two nonallelic rat preproinsulin genes. Cell 18: 545–558. [DOI] [PubMed] [Google Scholar]
- Luo, M.-j., and Reed, R. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. 96: 14937–14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.-D., and Steitz, J.A. 2001. Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836–1839. [DOI] [PubMed] [Google Scholar]
- Malim, M.H., Hauber, J., Fenrick, R., and Cullen, B.R. 1988. Immunodeficiency virus Rev trans-activator modulates the expression of the viral regulatory genes. Nature 335: 181–183. [DOI] [PubMed] [Google Scholar]
- Malim, M.H., Hauber, J., Le, S.-Y., Maizel, J.V., and Cullen, B.R. 1989. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338: 254–257. [DOI] [PubMed] [Google Scholar]
- Maniatis, T. and Reed, R. 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506. [DOI] [PubMed] [Google Scholar]
- Matsumoto, K., Wassarman, K.M., and Wolffe, A.P. 1998. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 17: 2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S., Fong, N., Yankulov, K., Ballantyne, S., Pan, G., Greenblatt, J., Patterson, S.D., Wickens, M., and Bentley, D.L. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361. [DOI] [PubMed] [Google Scholar]
- Nagata, S., Mantei, N., and Weissmann, C. 1980. The structure of one of the eight or more distinct chromosomal genes for human interferon-α. Nature 287: 401–408. [DOI] [PubMed] [Google Scholar]
- Niwa, M., Rose, S.D., and Berget, S.M. 1990. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes & Dev. 4: 1552–1559. [DOI] [PubMed] [Google Scholar]
- Nott, A., Meislin, S.H., and Moore, M.J. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA (this issue). [DOI] [PMC free article] [PubMed]
- Ohno, M., Segref, A., Kuersten, S., and Mattaj, I. W. 2002. Identity elements used in export of mRNAs. Mol. Cell 9: 659–671. [DOI] [PubMed] [Google Scholar]
- Otero, G.C., Harris, M.E., Donello, J.E., and Hope, T.J. 1998. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus Rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J. Virol. 72: 7593–7597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasleau, F., Leung, F., and Kopchick, J.J. 1987. A comparison of bovine growth hormone expression directed by bGH genomic or intronless DNA in transiently transfected eukaryotic cells. Gene 57: 47–52. [DOI] [PubMed] [Google Scholar]
- Rodrigues, J.P., Rode, M., Gatfield, D., Blencowe, B.J., Carmo-Fonseca, M., and Izaurralde, E. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. 98: 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, W.-S. and Mertz, J.E. 1989. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J. Virol. 63: 4386–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sträβer, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzl, M., Rodriguez-Navarro, S., Rondón, A.G., Agullera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–307. [DOI] [PubMed] [Google Scholar]
- Vagner, S., Vagner, C., and Mattaj, I.W. 2000. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes & Dev. 14: 403–413. [PMC free article] [PubMed] [Google Scholar]
- Yi, R., Bogerd, H.P., and Cullen, B.R. 2002. Recruitment of the Crm1 nuclear export factor is sufficient to induce cytoplasmic expression of incompletely spliced human immunodeficiency virus mRNAs. J. Virol. 76: 2036–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen, D., Vinciguerra, P., Wyss, J.-C., and Stutz, F. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yralp and Sub2p by Hprlp. Mol. Cell. Biol. 22: 8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Luo, J.-j., Straesser, K., Katahira, J., Hurt, E., and Reed, R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405. [DOI] [PubMed] [Google Scholar]