Abstract
Genetic control by metabolite-binding mRNAs is widespread in prokaryotes. These riboswitches are typically located in noncoding regions of mRNA, where they selectively bind their target compound and subsequently modulate gene expression. We have identified mRNA elements in fungi and in plants that match the consensus sequence and structure of thiamine pyrophosphate-binding domains of prokaryotes. In Arabidopsis, the consensus motif resides in the 3′-UTR of a thiamine biosynthetic gene, and the isolated RNA domain binds the corresponding coenzyme in vitro. These results suggest that metabolite-binding mRNAs are possibly involved in eukaryotic gene regulation and that some riboswitches might be representatives of an ancient form of genetic control.
Keywords: Allosteric RNA, aptamer, genetic control, riboswitch, thiamine pyrophosphate
INTRODUCTION
Riboswitches are genetic control elements that can be found in the 5′-untranslated region of certain messenger RNAs of prokaryotes (Mironov et al. 2002; Nahvi et al. 2002; Winkler et al. 2002a,b). These genetic switches exhibit two surprising properties. First, the mRNA is able to form a highly selective binding site for the target metabolite without the aid of proteins. Second, metabolite binding brings about an allosteric reorganization of RNA structure that leads to alterations in genetic expression. Unlike many other genetic control systems, riboswitches do not require metabolite-binding proteins to serve as sensors, and thus offer a direct link between the genetic information that is encoded by an mRNA and its chemical surroundings.
A number of distinct types of riboswitches have been confirmed by biochemical and genetic analyses. For example, a coenzyme B12-binding RNA has been shown (Nahvi et al. 2002) to control expression of the Escherichia coli btuB gene, which encodes a cobalamin transport protein. Riboswitches triggered by thiamine pyrophosphate (TPP) have been shown to control operons in E. coli (Winkler et al. 2002a) and Bacillus subtilis (Mironov et al. 2002) that are responsible for biosynthesis of this coenzyme. In addition, the RFN element, which frequently is found in the 5′-untranslated region of genes responsible for the biosynthesis or import of riboflavin and FMN, serves as the receptor portion of FMN-dependent riboswitches in B. subtilis (Mironov et al. 2002; Winkler et al. 2002b). Recently, we have determined that certain S-box motifs that are located in the 5′-UTRs of numerous genes in B. subtilis bind the coenzyme S-adenosylmethionine (SAM) with high affinity and precision (W.C. Winkler, A. Nahvi, N. Sudarsan, J.E. Barrick, and R.R. Breaker, in prep.). These findings indicate that riboswitches are used to recognize a diverse collection of metabolites, and that direct sensing of small molecules by mRNAs is an important form of genetic control for certain organisms. Herein, we provide evidence that metabolite-binding domains are embedded in certain mRNAs of eukaryotes, indicating that higher organisms might also exploit riboswitches for genetic control.
RESULTS AND DISCUSSION
Several groups have reported that the conserved cores of TPP riboswitches (Miranda-Rios et al. 2001; Stormo and Ji 2001; Rodionov et al. 2002) and FMN riboswitches (Gelfand et al. 1999; Vitreschak et al. 2002) are widely distributed across the phylogenetic landscape of bacteria and archaea. Likewise, we have identified dozens of RNA elements in prokaryotes that exhibit sequence similarity to the B12- and SAM-dependent riboswitches (W.C. Winkler, A. Nahvi, N. Sudarsan, J.E. Barrick, and R.R. Breaker, in prep.; A. Nahvi, J.E. Barrick, and R.R. Breaker, unpubl.). Given the relatively large size and sequence complexity of these RNA motifs, it is unlikely that numerous evolutionary reinventions of the same elements would have occurred. Furthermore, the metabolite triggers of these genetic switches are predicted to have been present in a time before the emergence of proteins (White III 1976; Benner et al. 1989;Jeffares et al. 1998). Therefore, we speculate that the known classes of metabolite-sensing RNAs might have originated in the ancient RNA world, which is believed to be a time before the emergence of proteins, when metabolism was guided entirely by RNA (Joyce 2002).
If the present-day riboswitches are of ancient origin, then eukaryotes might possess RNA genetic switches that are descendent from the last common ancestor of modern cells. We find that several eukaryotes carry RNA domains that conform to the consensus sequence and structure of the metabolite-binding domain of the TPP riboswitch class (Fig. 1A ▶). For example, a putative thiamine biosynthesis gene1 of Arabidopsis thaliana carries an RNA element (Fig. 1B ▶) in its 3′-UTR that conforms to the consensus TPP-binding domain. Similar RNA elements are found in rice (Oriza sativa) and bluegrass (Poa secunda). RNA elements that conform to the TPP-binding sequence and structure are also present in fungi such as Neurospora crassa (Fig. 1C ▶) and Fusarium oxysporum. As with plants, the riboswitch homologs in fungi are located in genes that have been implicated in the biosynthesis of thiamine,1 suggesting that in each case their role is to maintain required coenzyme levels by modulating expression of the appropriate biosynthetic genes. A sequence alignment of the homologous domains found in eukaryotes compared with that of the gram negative bacterium E. coli (thiC and thiM) and the gram-positive bacterium Clostridium acetobutylicum (thiC) is depicted in Figure 2 ▶.
FIGURE 1.
Putative eukaryote riboswitches. (A) Consensus TPP-binding domain based on 100 bacteria and archaea RNAs. Nucleotides in red are most conserved (>90%). Open circles represent nucleotide positions and domains that vary in sequence and length are designated var. The consensus model is similar to that reported recently (Rodionov et al. 2002). (B) The TPP-binding domain of A. thaliana. Variations in O. sativa (orange) and P. secunda (green) are shown. (C) Putative TPP-binding domain in the intron of N. crassa.
FIGURE 2.
Sequence alignments of eukaryotic domains related to bacterial TPP-dependent riboswitches. Base-paired stems are shaded in black and labeled as defined previously (Winkler et al. 2002a). The P3 sequences, which in eukaryotes are significantly expanded in length and number of base pairs, are represented as a stem-loop structure. The highly conserved nucleotide positions in bacteria that were used to search for eukaryotic domains are shaded gray. For each identified (ID) sequence, the position of the conserved CUGAGA sequence within the given GenBank entry is given along with the accession identification, sequence name, and gene identification. Additional protein annotations based on sequence similarity are shown in brackets. (Methods) Riboswitch-like domains were initially identified by sequence similarity to bacterial sequences (Eco2 and Cac) by a BLASTN search of GenBank using default parameters. These hits were verified and expanded by searching for degenerate matches to the pattern (CTGAGA [200] ACYTGA [5] <<< GNTNNNNC >>> [5] CGNRGGRA) using the program SequenceSniffer (unpublished algorithm). Angle brackets indicate base pairing, and bracketed numbers are variable gaps with constrained maximum lengths. All of the eukaryotic sequences have one or zero mismatches to this pattern except for one (Aor), which initially had three mismatches due to a single A insertion in the final search element. The presence of this mutation results in an inactive aptamer, whereas removal permits TPP binding (data not shown). Comparison of mRNA (M33643.1) and genomic (AB033416.1) sequences demonstrated that the F. oxysporum element is in an intron in the 5′ UTR of the sti35 gene. Other fungal sequences (Ncr, Aor, and Fso) are flanked by consensus splicing sequences.
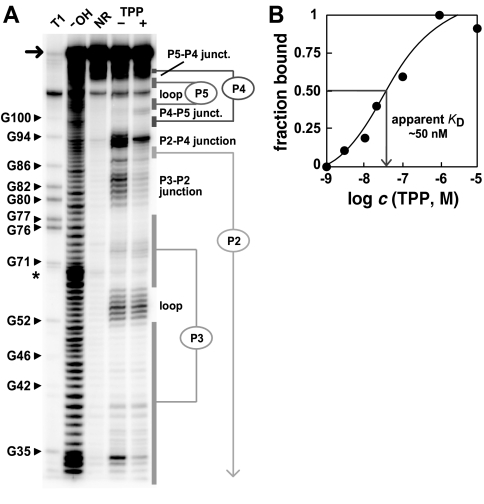
The RNA element corresponding to the consensus TPP-binding domain of A. thaliana (Fig. 1A ▶) was generated by in vitro transcription of a synthetic DNA template and the RNA was subjected to in-line probing (Fig. 3A ▶). This method relies on the spontaneous breakdown of RNA phosphodiester linkages, whose pattern of cleavage can be used to reveal the structural and functional features of ligand-binding RNAs (Nahvi et al. 2002; Winkler et al. 2002a,b). The riboswitch-like element exhibits TPP-dependent structural modulation and has a fragmentation pattern that is consistent with the predicted secondary structure of TPP riboswitches from bacteria (Winkler et al. 2002a). In addition, we have used this structure-probing method to establish that the RNA binds TPP with an apparent dissociation constant (KD) of ∼50 nM (Fig. 3B ▶), which is similar to that determined previously for an E. coli riboswitch variant. Similarly, we have demonstrated that the sequence elements of fungi that correspond to the TPP riboswitch consensus also bind TPP with high affinity (data not shown).
FIGURE 3.
Structural probing of the putative TPP-riboswitch from Arabidopsis. (A) Fragmentation pattern of the 128-nucleotide RNA (arrow) of A. thaliana (Fig. 1B ▶), which was generated by incubation in the absence (−) or presence (+) of 100 μM TPP. T1, OH, and NR identify RNAs that were partially digested with RNase T1 (cleaves 3′ to G residues), alkali, or were not reacted, respectively. Reactions were conducted as described previously (Winkler et al. 2002a). (B) Apparent KD for TPP binding by the A. thaliana RNA. Fraction bound was determined by in-line probing as described previously (Nahvi et al. 2002; Winkler et al. 2002a,b).
Sequestering of the ribosome-binding site and transcription termination are demonstrated mechanisms for TPP riboswitches in E. coli (Fig. 4 ▶). Because the TPP-binding element in plants is located immediately upstream from the poly(A) tail, it is possible that metabolite binding might regulate mRNA processing and stability. Alternatively, a consensus TPP-binding sequence (Fig. 1C ▶) identified in the fungal genome of N. crassa resides in an intron, suggesting that RNA splicing might also be guided by metabolite-binding pre-mRNAs. In prokaryotes, ligand binding typically brings about allosteric changes in the Watson–Crick base-pairing arrangements near gene control elements such as transcription terminators and ribosome-binding sites. Likewise, secondary structure rearrangements by metabolite-binding riboswitches could be used to modulate a greater variety of RNA processing, transport and expression pathways in eukaryotes.
FIGURE 4.
Genetic structures, thiamine biosynthetic genes, and possible mechanisms of riboswitch control. The location and mechanism of the E. coli and B. subtilis riboswitches are detailed elsewhere (Mironov et al. 2002; Winkler et al 2002a). The putative TPP riboswitch from P. secunda resides immediately upstream from the poly(A) tail in the cDNA clone of the thiC gene. The putative TPP riboswitch domain in F. oxysporum is located in a 5′-UTR intron of the sti35 gene according to the genomic sequence, but is absent in the cDNA clone.
Although we favor the hypothesis that these TPP-binding domains and those for coenzyme B12, FMN, and SAM are of ancient origin, it is possible that other examples of metabolite-binding mRNAs have emerged more recently in evolution. These newer riboswitches would be more narrowly distributed across the phylogenetic landscape, therefore, efforts to search for new riboswitches that are triggered by compounds that are not ancient and universally distributed will be difficult. Regardless of the scope of riboswitch use in modern organisms, both natural and engineered riboswitches could have significant utility. Given the central role that known riboswitches serve in modulating the concentration of key coenzymes, these RNAs might serve as new targets for drug discovery efforts. Moreover, previous efforts to engineer RNAs that perform as ligand-dependent molecular switches have proven that RNA has an enormous untapped potential for molecular sensing (Seetharaman et al. 2001; Hesselberth et al. 2003). Therefore, reverse engineering of natural riboswitches could be used to establish a conceptual basis for creating designer riboswitches for the purposeful control of eukaryotic genes.
Acknowledgments
We thank members of the Breaker laboratory for comments on the manuscript. This work was supported by the NIH (GM 559343), by the NSF (EIA-0129939), and by a fellowship to R.R.B. from the David and Lucile Packard Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5090103.
Footnotes
The mRNAs that carry the TPP-binding domains encode for a protein that is homologous to the thiC protein of E. coli. This protein enzyme catalyzes the conversion of 5-aminoimidazole ribotide (AIR) to hydroxymethyl pyrimidine phosphate (HMP-P), which is a key biosynthetic step in the synthesis of thiamine and ultimately TPP (Vander Horn et al. 1993; Begley et al. 1999).
REFERENCES
- Begley, T.P., Downs, D.M., Ealick, S.E., McLafferty, F.W., Van Loon, A.P., Tayla, S., Campobasso, N., Chiu, H.J., Kinsland, C., Reddick, J.J., et al. 1999. Thiamin biosynthesis in prokaryotes. Arch. Microbiol. 171: 293–300. [DOI] [PubMed] [Google Scholar]
- Benner, S.A., Ellington, A.D., and Tauer, A. 1989. Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. 86: 7054–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand, M.S., Mironov, A.A., Jomantas, J., Kozlov, Y.I., and Perumov, D.A. 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 15: 439–442. [DOI] [PubMed] [Google Scholar]
- Hesselberth, J.R., Robertson, M.P., Knudsen, S.M., and Ellington, A.D. 2003. Simultaneous detection of diverse analytes with aptazyme ligase array. Anal. Biochem. 312: 106–112. [DOI] [PubMed] [Google Scholar]
- Jeffares, D.C., Poole, A.M., and Penny, D. 1998. Relics from the RNA world. J. Mol. Evol. 46: 18–36. [DOI] [PubMed] [Google Scholar]
- Joyce, G.F. 2002. The antiquity of RNA-based evolution. Nature 418: 214–221. [DOI] [PubMed] [Google Scholar]
- Miranda-Rios, J., Navarro, M., and Soberón, M. 2001. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. 98: 9736–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov, A.S., Gusarov, I., Rafikov, R., Lopez, L.E., Shatalin, K., Kreneva, R.A., Perumov, D.A., and Nudler, E. 2002. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111: 747–756. [DOI] [PubMed] [Google Scholar]
- Nahvi, A., Sudarsan, N., Ebert, M.S., Zou, X., Brown, K.L., and Breaker, R.R. 2002. Genetic control by a metabolite binding RNA. Chem. Biol. 9: 1043–1049. [DOI] [PubMed] [Google Scholar]
- Rodionov, D.A., Vitreschak, A.G., Mironov, A.A., and Gelfand, M.S. 2002. Comparative genomics of thiamin biosynthesis in prokaryotes. J. Biol. Chem. 277: 48949–48959. [DOI] [PubMed] [Google Scholar]
- Seetharaman, S., Zivartz, M., Sudarsan, N., and Breaker, R.R. 2001. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat. Biotechnol. 19: 336–341. [DOI] [PubMed] [Google Scholar]
- Stormo, G.D and Ji, Y. 2001. Do mRNAs act as direct sensors of small molecules to control their expression? Proc. Natl. Acad. Sci. 98: 9465–9467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Horn, P.B., Backstrom, A.D., Stewart, V., and Begley, T.P. 1993. Structural genes for thiamine biosynthetic enzymes (thiCEFGH) in Escherichia coli K-12. J. Bacteriol. 175: 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak, A.G., Rodionov, D.A., Mironov, A.A., and Gelfand, M.S. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30: 3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White III, H.B. 1976. Coenzymes as fossils of an earlier metabolic state. J. Mol. Evol. 7: 101–104. [DOI] [PubMed] [Google Scholar]
- Winkler, W., Nahvi, A., and Breaker, R.R. 2002a. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Cohen-Chalamish, S., and Breaker, R.R. 2002b. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. 99: 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]