Abstract
In humans, recognition of a stop codon by protein release factor eRF1 leads to release of the nascent peptide from the ribosome. Although efficient eRF1 activity is usually desirable, numerous pathologies result from eRF1 recognition of premature stop mutations in essential genes. In these cases, decreased eRF1 activity could increase readthrough of the premature stop codon, thereby making full-length protein. To broaden the means available to beneficially decrease eRF1 activity, we have targeted eRF1 mRNA using siRNAs and antisense oligonucleotides. We show that both eRF1-targeted siRNA and antisense oligonucleotides decrease eRF1 mRNA and eRF1 protein concentrations, and increase UAG readthrough in cultured human cells.
Keywords: Human, release factor, stop codon, siRNA, antisense
INTRODUCTION
Although termination codons are only about 4.7% of all codons, mutations that lead to premature termination usually have a noteworthy phenotype. Typically, the resulting truncated protein is nonfunctional, leading to insufficiency and in some cases to disease. Therefore, of the numerous human clinical conditions caused by point mutations, about 20% result from a premature termination codon in the open reading frame of an otherwise normal mRNA (Cooper et al. 1998; Krawczak et al. 1998). In principle, reducing the efficiency of translation termination could reduce disease pathology by allowing readthrough past the premature stop codon, leading to the production of a low level of functional protein.
In eukaryotes, stop codons are decoded on the ribosome by eukaryotic release factors eRF1 and eRF3 (Frolova et al. 1994; Stansfield et al. 1995; Zhouravleva et al. 1995). eRF1 is the sequence-specific element, recognizing all three stop codons and stimulating hydrolysis of the peptide–tRNA bond. eRF3 assists eRF1 function in a GTP-dependent manner (Zavialov et al. 2001; Kisselev et al. 2003). Because eRF1 is the sole release factor to decode all stop codons, its activity is critical for translation termination. As would be predicted, in yeast, eRF1 levels have been shown to directly correlate with termination efficiency. Depletion of eRF1 protein decreases termination efficiency (Stansfield et al. 1996), and overexpression of eRF1 increases termination efficiency (Le Goff et al. 1997). Thus high eRF1 activity prevents readthrough of premature stop codons, reducing erroneous insertion of an amino acid.
Previous techniques directed at increasing translational readthrough of premature stop codons targeted either the ribosome or eRF1. These approaches include the use of suppressor tRNAs, aminoglycosides, and eRF1-binding RNAs. Suppressor tRNAs, complexed with translation elongation factors, compete with eRF1 for stop codon recognition by virtue of an anticodon loop complementary to a particular stop codon (Murgola 1995). Suppressor tRNAs promote incorporation of an amino acid at a stop codon, and have been shown to induce readthrough in vivo (Buvoli et al. 2000). Aminoglycosides have also been used to induce readthrough of premature stop codons as a particular application of their general ability to decrease the fidelity of translation (Stansfield et al. 1998). The loss of fidelity leads to global mistranslation, and many codons are misread during translation. Among the misread codons are UAA, UAG, and UGA, which are mistranslated by a noncognate amino acid–tRNA. Although aminoglycosides can be quite effective in inducing readthrough (Barton-Davis et al. 1999; Keeling et al. 2001; Keeling and Bedwell 2002), the induction of errors throughout translation complicates this approach. Nevertheless, aminoglycosides are being tested in clinical trials to treat diseases caused by premature stop codons (Wagner et al. 2001). Another approach to promote readthrough uses eRF1-binding RNAs, called aptamers, which have been shown to inhibit eRF1 function and affect suppression during translation in vitro (Carnes et al. 2000). The efficacy of these eRF1 aptamers, however, has not yet been demonstrated in living cells.
In all three approaches that induce readthrough, eRF1 competes with the desired activity. Decreasing cellular levels of eRF1 therefore seemed an alternative and complementary approach to elevate readthrough of a stop codon in human cells. To test this procedure, we designed siRNAs and antisense oligonucleotides that targeted eRF1 mRNA. Both techniques specify mRNA for degradation by sequence complementarity (Agrawal and Zhao 1998; Elbashir et al. 2001; Nishikura 2001). In this work, we demonstrate that transfection with these two types of oligonucleotides decreases eRF1 mRNA concentration, causes eRF1 protein to decline, and increases readthrough of a premature stop codon in human cells in culture.
RESULTS AND DISCUSSION
Both siRNAs and antisense oligonucleotides promote readthrough of a stop codon in mammalian cells
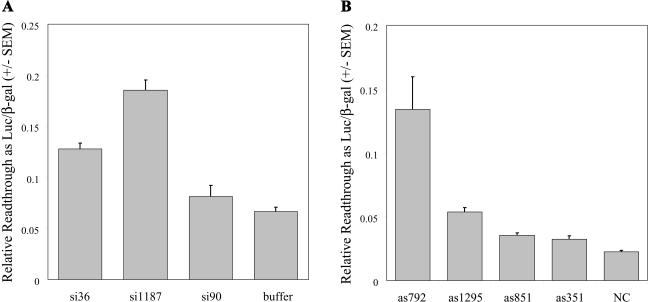
Three siRNAs and four antisense oligonucleotides targeting sequences predicted to be accessible in the human eRF1 mRNA were synthesized to determine if decreased eRF1 protein could promote stop codon readthrough (Fig. 1 ▶). To assay readthrough in cells, we stably transfected human embryonic kidney (HEK) 293 cells with the pAC-TMV readthrough reporter plasmid (Stahl et al. 1995) to create cell line Q24. Q24 cells express a transcriptional fusion of β-galactosidase to luciferase with these sequences separated by a leaky viral UAG stop codon. Although β-galactosidase protein is constitutively produced from this construct (and therefore serves as an internal control), production of luciferase protein requires readthrough of a stop codon within a sequence from Tobacco Mosaic Virus (TMV; Skuzeski et al. 1991). Ratios of luciferase activity to β-galactosidase activity (proportional to readthrough) are shown in Figure 2A ▶ for siRNA treatment and Figure 2B ▶ for antisense treatment. Both si1187 and as792 significantly increased readthrough of the UAG stop codon in comparison to their respective negative controls, buffer alone and noncomplementary (NC) oligonucleotide. The ability of these oligonucleotides to increase readthrough supports the idea that targeting eRF1 mRNA holds potential as a therapeutic strategy.
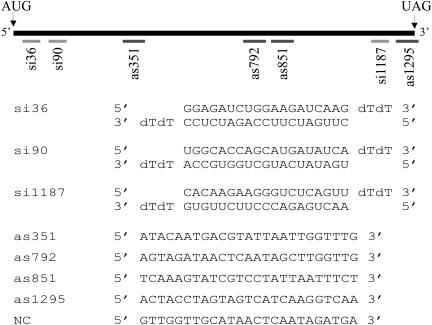
FIGURE 1.
Schematic of eRF1 mRNA coding region showing relative locations of siRNA (light gray bars) and antisense oligonucleotide (dark gray bars) complementary regions, with both siRNA and antisense sequences shown below.
FIGURE 2.
Readthrough of a UAG stop codon after treatment with siRNAs (A) or antisense (B). Luciferase activity is normalized to β-galactosidase activity. Bars represent an average of eight measurements per sample; error bars are ±1 standard error of the mean. Results consistent with Figure 2 ▶ were repeated in at least three independent experiments.
Although both si36 and as1295 also increased the amount of reported readthrough compared to their respective negative controls, the extent was less dramatic, close to twofold above background. This result suggests that oligonucleotides that target different sites in the same mRNA have varying effectiveness. Indeed, oligonucleotides si90, as351, or as851 had no significant effect on readthrough levels.
Effect of siRNAs and antisense oligonucleotides on eRF1 mRNA concentration
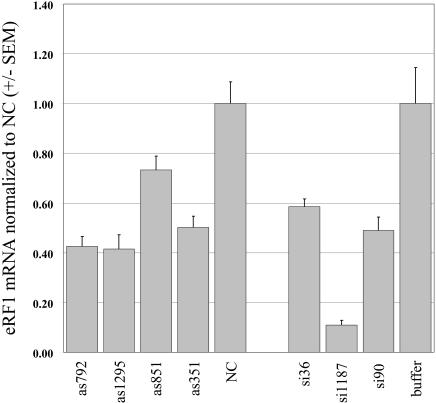
To determine if the observed increase in readthrough was due to the predicted reduction in eRF1, we measured the levels of eRF1 mRNA in siRNA- and antisense-treated cells. Quantitative PCR analysis indicated that eRF1 mRNA concentration was decreased by both siRNA and antisense treatments with respect to negative controls (Fig. 3 ▶). A similar decrease in eRF1 mRNA after treatment with siRNAs was independently confirmed by RNase protection (data not shown). As expected, the oligonucleotides that produced the most significant increase in readthrough (si1187 and as792) were also very effective in decreasing eRF1 mRNA levels. In fact, all of the oligonucleotides complementary to eRF1 decreased eRF1 mRNA to some extent.
FIGURE 3.
Taqman analysis of eRF1 mRNA concentration after treatment with antisense or siRNA oligonucleotides. Bars denote amounts of eRF1 mRNA relative to negative control oligonucleotide NC. Bars represent an average of three measurements per sample; error bars are ±1 standard error of the mean.
Surprisingly, some oligonucleotides that effectively decreased eRF1 mRNA had little or no effect in the readthrough assay (Fig. 2 ▶). For example, although as792 and as1295 decreased eRF1 mRNA similarly, as792 was clearly more effective in the readthrough assay. A possible explanation is that some oligonucleotides not only affect eRF1 mRNA degradation, but also alter eRF1 mRNA translation. Certain peptide nucleic acid (PNA) antisense oligonucleotides can inhibit translation elongation on target mRNAs (Dias et al. 2002). Transfection with as792 might impede eRF1 mRNA translation in addition to promoting eRF1 mRNA degradation, whereas as1295 may not slow overall translation significantly. In any case, measuring translational readthrough is a more direct assay for eRF1 activity than measuring eRF1 mRNA concentration, and is a more apt criterion for comparing different suppression strategies.
Effect of siRNAs and antisense oligonucleotides on eRF1 protein concentration
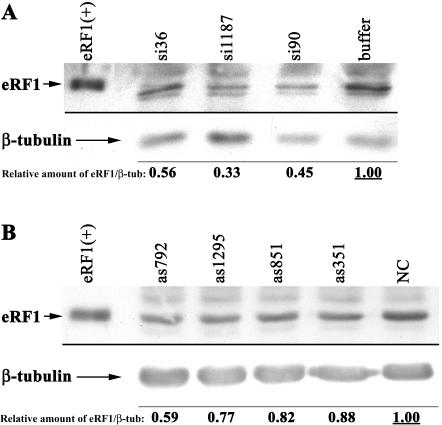
To determine if siRNAs and antisense oligonucleotides decrease eRF1 protein concentration as expected, samples from transfected Q24 cells were analyzed by Western blotting. The amount of eRF1 in equivalent cell lysates after treatment with siRNAs or antisense is shown in Figure 4A and B ▶, respectively. Quantitation of the Western blots was less reproducible from experiment to experiment than either eRF1 mRNA or readthrough assays. We have presented quantitation of eRF1 relative to β-tubulin, normalized to the ratio in control cells. The normalized decrease in eRF1 observed on Western blots is moderate. However, comparing these data with eRF1 mRNA and readthrough assays, the same oligonucleotides clearly tend to be effective.
FIGURE 4.
(A) Western blot of eRF1 protein concentration after treatment with siRNAs, with Western blot of β-tubulin serving as a loading control. (B) Similar Western analysis showing eRF1 protein concentration after antisense treatment. Lane 1 shows purified eRF1 protein as a positive control.
Experimental variability could explain the sometimes inexact correlation between decreased eRF1 levels and increased measured readthrough levels. However, there is a general relation between mRNA and protein decreases, and, notably, treatment with the most effective oligonucleotides si1187 or as792 led to the largest decrease in eRF1 protein.
Accordingly, these experiments demonstrate a new route to suppression of a stop codon in human cells, via eRF1 mRNA levels. Because eRF1-targeted oligonucleotides reduce the amount of eRF1 mRNA and consequently the protein present in the cell, their use potentially complements other strategies to decrease termination at premature stop codons. In addition, because eRF1 decodes all three stop codons, the siRNA- and antisense-mediated readthrough observed at a UAG stop codon should also occur at UAA and UGA stop codons (Drugeon et al. 1997).
The effects of stop codon context may well alter the degree to which readthrough is observed at other stop codons (Martin 1994; Cassan and Rousset 2001). Experiments with both suppressor tRNAs and aminoglycosides indicate that bases surrounding a stop codon significantly influence readthrough (Martin et al. 1993; Howard et al. 2000). Stop codon context could also potentially prevent effective readthrough at authentic stop codons when the concentration of eRF1 is decreased, thereby minimizing possible toxic effects. Western blots probed for β-tubulin did not reveal any larger molecular weight band. Readthrough of the β-tubulin natural stop TAA would add ∼40 amino acids to the protein, an addition that would have been detectable. Therefore, readthrough of this authentic stop codon was probably infrequent.
The extension of this work to expression of siRNAs in vivo holds potential as a therapeutic treatment for human pathologies tracing to premature stop codons. Recent development of methods to express siRNAs in mammalian tissue culture (Brummelkamp et al. 2002; Lee et al. 2002; Miyagishi & Taira 2002; Paul et al. 2002; Yu et al. 2002) obviate the need for expensive chemical synthesis of siRNA oligonucleotides. A significant step toward realizing the potential of siRNAs as therapeutics is the report of their application in mammals. Transfection of siRNAs in vivo has successfully downregulated gene expression in mice, with expression of a chimeric Hepatitis C virus NS5B protein fused to luciferase reduced by 75% (McCaffrey et al. 2002). An inherent barrier to such therapeutic approaches in humans remains the difficulties in delivery to target cells in vivo. In addition, although siRNA-mediated inhibition can persist for many cell divisions in tissue culture (Elbashir et al. 2001), the duration of siRNA-mediated effect in vivo remains undetermined. Experiments that address these issues of delivery and persistence in cells may manifest new possibilities for treatment of diseases caused by premature stop codons. We have now shown that, given delivery and persistence, therapeutic oligonucleotide suppression could rely on aptamers (Carnes et al. 2000), antisense sequences or RNA interference (this work), or perhaps a more effective combination thereof.
MATERIALS AND METHODS
Stable cell line
Human embryonic kidney 293 cells were cotransfected with a 10:1 molar ratio of pAC-TMV (Stahl et al. 1995) plasmid to pQBI-25 (Quantum Biotechnologies) plasmid. Clones were selected in DMEM (Invitrogen) containing 2 mM L-glutamine (Invitrogen), 10% calf serum (HyClone), and 200 μg/mL G418 (Mediatech). Clones were screened for β-galactosidase activity, and positive clones were subsequently tested for luciferase activity. A clone designated as Q24 was subsequently used to test the effectiveness of the various oligonucleotides.
siRNA and AS oligonucleotide sequences
Three siRNA sequences and four antisense sequences were tested: si36, si90, si1187, as351, as792, as851, and as1295 (Fig. 1 ▶). Oligonucleotide numbers denote distance from the A of the translation start codon in eRF1 mRNA. siRNAs were obtained from Dharmacon Research as desalted and deprotected single-strand RNAs, and were annealed per manufacturer instructions in annealing buffer (100 mM potassium acetate, 30 mM HEPES-KOH at pH 7.4, 2 mM magnesium acetate) prior to transfection. Chimeric antisense oligonucleotides were obtained from Sequitur, Inc.
Transfection protocols
The day prior to transfection, cells were plated at a density of 250,000 cells/mL, with 100 μL/well in 96-well plates and 600 μL/well in 24-well plates. For siRNA transfections, siRNAs were diluted in OptiMEM (Invitrogen) media to a final concentration of 700 nM. LipofectAMINE 2000 (Invitrogen) was diluted 1:25 in OptiMEM, and equal volumes of diluted LipofectAMINE 2000 and siRNA were combined and gently mixed by inversion. After a 10-min incubation at room temperature, a volume of DMEM containing 2 mM L-glutamine and 10% calf serum (equivalent to 2.5 times the volume of the LipofectAMINE 2000:siRNA mixture) was added. An equal volume of this transfection mixture was then added to either 96-well (100 μL) or 24-well (600 μL) wells, for a final volume of 200 μL in 96-well and 1.2 mL in 24-well wells. For antisense transfections, 20 μM stocks of oligonucleotide were diluted 1:10 in OptiMEM containing 10% calf serum, and LipofectAMINE 2000 was diluted 1:25 in OptiMEM containing 10% calf serum. Equal volumes of diluted oligonucleotide and LipofectAMINE 2000 were combined and incubated at room temperature for 10 min. Twenty-six μL of this transfection mixture were added to each well in a 96-well plate, or 156 μL to each well in a 24-well plate.
Readthrough assay
Forty-eight hours post-transfection, Q24 293 cells were resuspended in 40 μL of Galacto-Star Lysis buffer (Tropix) for 24-well plates, or 30 μL of Lysis buffer for 96-well plates. Cell lysates were either assayed immediately or frozen on dry ice and stored at −80°C. Cell lysates were centrifuged for 2 min at 16,000 g, and 20 μL removed for luciferase assay. The remaining cell lysate was heated to 48°C for 1 h to inactivate endogenous β-galactosidase activity. Both β-galactosidase and luciferase activities were measured using a Turner Designs 20/20 luminometer. For the luciferase assay, 50 μL of luciferase buffer containing luciferin (Promega) were added to 20 μL of cell lysate and the activity measured as relative light units. For β-galactosidase assay, 100 μL of Galacto-Star Diluent buffer containing Galacton-Star Substrate were added to 15 μL of heat-inactivated cell lysate, incubated at room temperature for 50 min, and the activity measured as relative light units.
Taqman analysis
RNA for quantitative PCR analysis was isolated from Q24 cells 24 h post-transfection using the RNeasy Kit with QIAshredder (Qiagen) as described by the manufacturer. Total RNA concentrations were assayed in triplicate in 96-well plates on a Perkin-Elmer HTS 7000 fluorescence plate reader using the RiboGreen RNA Quantitation Kit (Molecular Probes). The amount of eRF1 mRNA was measured by analyzing equivalent amounts of total RNA. Amplification primers for eRF1 mRNA quantitation were 5′-GAGCTAC GTTGGAAATTGTCAC-3′ and 5′-ATACCTCCAATTCCACCAA ATC-3′ (IDTDNA). A dual-labeled detection probe 5′-AAATCAC AAGAAGGGTCTCAGTTTGTGAAAG-3′ contained 5′ 6-FAM and 3′ BHQ-1 modifications (IDTDNA). Taqman quantitation was performed on an ABI Prism 5700 using Applied Biosystems Taqman One-Step RT-PCR Master Mix Reagents (Applied Biosystems) following the manufacturer’s protocol. Standard curves were used to ensure that all PCR results fell within the linear range of the assay.
Western blot
Q24 293 cells from 24-well plates were harvested 48 h post-transfection by resuspending cells in 40 μL of Galacto-Star Lysis buffer (Tropix). Cell lysates were either assayed immediately or frozen on dry ice and stored at −80°C. Cell lysates were centrifuged for 2 min at 16,000 g, and 2 μL were added to 400 μL of Bio-Rad Protein Assay (Bio-Rad) to determine total protein concentration by absorbance at 595 nm. Based on total protein concentration, sample volumes were adjusted for equivalent loading. Then, Laemmli loading buffer (Bio-Rad) containing β-mercaptoethanol was added, and the samples were boiled for 3 min. Samples were loaded on 7.5% polyacrylamide gel and subjected to electrophoresis. The gel was subsequently electrotransferred onto Hybond-P PVDF membrane (Amersham Pharmacia Biotech), and the membrane blocked overnight at 4°C in 1× phosphate-buffered saline (PBS) containing 3% milk, 2% Normal Goat Serum, and 0.1% Tween-20. Blots were probed with anti-eRF1 serum (a generous gift from Michel Philippe (Université de Rennes I); Tassan et al. 1993) in blocking solution at room temperature for 60 min. Blots were washed three times for 15 min in 30 mL of 1× PBS containing 0.1% Tween-20. The blot was then probed with goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (Jackson Labs) at a 1:10,000 dilution in blocking solution. Incubation with antibody and subsequent washes were as before. Blots were then visualized by chemiluminescence (NEN) and exposure to x-ray film. After visualization, blots were stripped at 55°C in 62.5 mM Tris (pH 6.8), 2% SDS, 0.7% β-mercaptoethanol for 30 min, washed four times for 15 min in 30 mL of 1× PBS, and blocked overnight as before. Blots were then reprobed (following protocol above) with an anti-β-tubulin monoclonal antibody (Amersham Pharmacia Biotech) at 1:10,000 dilution and goat anti-mouse secondary antibody conjugated to horseradish peroxidase (Jackson Labs) at 1:2000 dilution. X-ray films were subsequently scanned on a UMAX PowerLook III and quantified using ImageQuant 5.1 (Molecular Dynamics).
Acknowledgments
We thank James D. Thompson for consultations concerning antisense design, Jean-Pierre Rousset for plasmid pAC-TMV, Michel Philippe for eRF1 antibody, Dhinakar Kompala for use of fluorescence plate reader, Somalogic, Inc. for use of ABI 5700, Anastasia Khvorova for technical advice, and Ludmilla Frolova and Lev Kisselev for eRF1 expressing plasmid (Merkulova et al. 1999).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5280103.
REFERENCES
- Agrawal, S. and Zhao, Q. 1998. Antisense therapeutics. Curr. Opin. Chem. Biol. 2: 519–528. [DOI] [PubMed] [Google Scholar]
- Barton-Davis, E.R., Cordier, L., Shoturma, D.I., Leland, S.E., and Sweeney, H.L. 1999. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J. Clin. Invest. 104: 375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553. [DOI] [PubMed] [Google Scholar]
- Buvoli, M., Buvoli, A., and Leinwand, L.A. 2000. Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Mol. Cell. Biol. 20: 3116–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes, J., Frolova, L., Zinnen, S., Drugeon, G., Phillippe, M., Justesen, J., Haenni, A.L., Leinwand, L., Kisselev, L.L., and Yarus, M. 2000. Suppression of eukaryotic translation termination by selected RNAs. RNA 6: 1468–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassan, M. and Rousset, J.P. 2001. UAG readthrough in mammalian cells: Effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol. Biol. 2: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, D.N., Ball, E.V., and Krawczak, M. 1998. The human gene mutation database. Nucleic Acids Res 26: 285–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias, N., Senamaud-Beaufort, C., Forestier, El, E., Auvin, C., Helene, C., Ester Saison-Behmoaras, T. 2002. RNA hairpin invasion and ribosome elongation arrest by mixed base PNA oligomer. J. Mol. Biol. 320: 489–501. [DOI] [PubMed] [Google Scholar]
- Drugeon, G., Jean-Jean, O., Frolova, L., Le Goff, X., Philippe, M., Kisselev, L., and Haenni, A.L. 1997. Eukaryotic release factor 1 (eRF1) abolishes readthrough and competes with suppressor tRNAs at all three termination codons in messenger RNA. Nucleic Acids Res. 25: 2254–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Frolova, L., Le Goff, X., Rasmussen, H.H., Cheperegin, S., Drugeon, G., Kress, M., Arman, I., Haenni, A.L., Celis, J.E., Philippe, M., et al. 1994. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372: 701–703. [DOI] [PubMed] [Google Scholar]
- Howard, M.T., Shirts, B.H., Petros, L.M., Flanigan, K.M., Gesteland, R.F., and Atkins, J.F. 2000. Sequence specificity of aminoglycoside-induced stop condon readthrough: Potential implications for treatment of Duchenne muscular dystrophy. Ann. Neurol. 48: 164–169. [PubMed] [Google Scholar]
- Keeling, K.M. and Bedwell, D.M. 2002. Clinically relevant aminoglycosides can suppress disease-associated premature stop mutations in the IDUA and P53 cDNAs in a mammalian translation system. J. Mol. Med. 80: 367–376. [DOI] [PubMed] [Google Scholar]
- Keeling, K.M., Brooks, D.A., Hopwood, J.J., Li, P., Thompson, J.N., and Bedwell, D.M. 2001. Gentamicin-mediated suppression of Hurler syndrome stop mutations restores a low level of α-L-iduronidase activity and reduces lysosomal glycosaminoglycan accumulation. Hum. Mol. Genet. 10: 291–219. [DOI] [PubMed] [Google Scholar]
- Kisselev, L., Ehrenberg, M., and Frolova, L. 2003. New EMBO member’s review: Termination of translation: Interplay of mRNA, rRNAs and release factors? EMBO J. 22: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak, M., Ball, E.V., and Cooper, D.N. 1998. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet. 63: 474–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff, X., Philippe, M., and Jean-Jean, O. 1997. Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol. Cell. Biol. 17: 3164–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, N.S., Dohjima, T., Bauer, G., Li, H., Li, M.J., Ehsani, A., Salvaterra, P., and Rossi, J. 2002. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 20: 500–505. [DOI] [PubMed] [Google Scholar]
- Martin, R. 1994. On the relationship between preferred termination codon contexts and nonsense suppression in human cells. Nucleic Acids Res. 22: 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, R., Phillips-Jones, M.K., Watson, F.J., and Hill, L.S. 1993. Codon context effects on nonsense suppression in human cells. Biochem. Soc. Trans. 21: 846–851. [DOI] [PubMed] [Google Scholar]
- McCaffrey, A.P., Meuse, L., Pham, T.T., Conklin, D.S., Hannon, G.J., and Kay, M.A. 2002. Gene expression: RNA interference in adult mice. Nature 418: 38–39. [DOI] [PubMed] [Google Scholar]
- Merkulova, T.I., Frolova, L.Y., Lazar, M., Camonis, J., and Kisselev, L.L. 1999. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 443: 41–47. [DOI] [PubMed] [Google Scholar]
- Miyagishi, M. and Taira, K. 2002. U6 promoter driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 20: 497–500. [DOI] [PubMed] [Google Scholar]
- Murgola, E.J. 1995. Translational suppression: When two wrongs do make one right. American Society for Microbiology, Washington, DC.
- Nishikura, K. 2001. A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107: 415–418. [DOI] [PubMed] [Google Scholar]
- Paul, C.P., Good, P.D., Winer, I., and Engelke, D.R. 2002. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 20: 505–508. [DOI] [PubMed] [Google Scholar]
- Skuzeski, J.M., Nichols, L.M., Gesteland, R.F., and Atkins, J.F. 1991. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol. 218: 365–373. [DOI] [PubMed] [Google Scholar]
- Stahl, G., Bidou, L., Rousset, J.P., and Cassan, M. 1995. Versatile vectors to study recoding: Conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 23: 1557–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield, I., Eurwilaichitr, L., Akhmaloka, Tuite MF. 1996. Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol. Microbiol. 20: 1135–1143. [DOI] [PubMed] [Google Scholar]
- Stansfield, I., Jones, K.M., Kushnirov, V.V., Dagkesamanskaya, A.R., Poznyakovski, A.I., Paushkin, S.V., Nierras, C.R., Cox, B.S., Ter-Avanesyan, M.D., and Tuite, M.F. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14: 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansfield, I., Jones, K.M., Herbert, P., Lewendon, A., Shaw, W.V., and Tuite, M.F. 1998. Missense translation errors in Saccharomyces cerevisiae. J. Mol. Biol. 282: 13–24. [DOI] [PubMed] [Google Scholar]
- Tassan, J.P., Le Guellec, K., Kress, M., Faure, M., Camonis, J., Jacquet, M., and Philippe, M. 1993. In Xenopus laevis, the product of a developmentally regulated mRNA is structurally and functionally homologous to a Saccharomyces cerevisiae protein involved in translation fidelity. Mol. Cell. Biol. 13: 2815–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, K.R., Hamed, S., Hadley, D.W., Gropman, A.L., Burstein, A.H., Escolar, D.M., Hoffman, E.P., and Fischbeck, K.H. 2001. Gentamicin treatment of Duchenne and Becker muscular dystrophy due to nonsense mutations. Ann. Neurol. 49: 706–711. [PubMed] [Google Scholar]
- Yu, J.Y., DeRuiter, S.L., and Turner, D.L. 2002. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl. Acad. Sci. 99: 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov, A.V., Buckingham, R.H., and Ehrenberg, M. 2001. A post-termination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107: 115–124. [DOI] [PubMed] [Google Scholar]
- Zhouravleva, G., Frolova, L., Le Goff, X., Le Guellec, R., Inge-Vechtomov, S., Kisselev, L., and Philippe, M. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14: 4065–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]