Abstract
In eukaryotes, the m7GpppN cap structure is added to all nascent RNA polymerase II transcripts, and serves important functions at multiple steps of RNA metabolism. The predominantly nuclear cap-binding complex (CBC) binds to the cap during RNA synthesis. The predominantly cytoplasmic eukaryotic initiation factor 4F (eIF4F) is thought to replace CBC after export of mature mRNA to the cytoplasm, and mediates the bulk of cellular translation. Yeast as well as mammalian CBC interacts in vitro with eIF4G, a subunit of eIF4F. In this work, we investigate a potential role of this interaction during translation in yeast. We identify a mutation (DR548/9AA) in Tif4631p, one of two isoforms of yeast eIF4G, that abolishes its binding to CBC. Cells expressing this mutant protein as the sole source of eIF4G grow at wild-type rates, and bulk cellular translation, as assessed by metabolic labeling and polysome profile analysis, is unchanged. Importantly, we find that the DR548/9AA mutation neither diminishes nor delays the translation of newly induced reporter mRNA. Finally, microarray analysis reveals marked transcriptome alterations in CBC subunit deletion strains, whereas eIF4G point mutants have essentially a wild-type transcriptome composition. Collectively, these data suggest that in yeast, the phenotypic consequences of CBC deletions are separable from its interaction with eIF4G, and that the CBC–eIF4G interaction is dispensable for a potential “pioneering round” of translation in yeast.
Keywords: Cap-binding protein, CBC, eIF4E, “pioneer round” of translation
INTRODUCTION
The m7GpppN cap structure at the 5′ end of eukaryotic mRNAs influences many aspects of their metabolism including splicing (Izaurralde et al. 1994; Lewis et al. 1996), 3′ end formation (Flaherty et al. 1997), export from the nucleus (Görlich et al. 1996; Visa et al. 1996; Shen et al. 2000), stability (Furuichi et al. 1977), and translation (Shatkin 1985). Principally, the roles of the cap structure are mediated through the binding and dynamic interchange of specific protein complexes (Lewis and Izaurralde 1997; Mitchell and Tollervey 2001; Preiss 2002). Two well-characterized complexes of this kind are the predominantly nuclear CBC and the predominantly cytoplasmic eIF4F. CBC is a heterodimer consisting of cap-binding protein (CBP) 20 (or MUD13 in yeast) and CBP80 (STO1 or GCR3 in yeast; Ohno et al. 1990; Colot et al. 1996). The smaller subunit CBP20 contains a common RNA-binding motif, the RNP domain (Izaurralde et al. 1994) and is highly conserved from yeast to human. CBP80 is far less conserved and comprises three tandem domains resembling the middle domain of eIF4G (MIF4G, see below; Aravind and Koonin 2000; Ponting 2000). CBP20 recognizes capped RNA in conjunction with CBP80 (Mazza et al. 2002). Furthermore, CBC exits the nucleus to the cytoplasm together with the mRNA (Görlich et al. 1996; Visa et al. 1996; Shen et al. 2000) where it is thought to be replaced by the eIF4F complex.
The eIF4F complex plays a central role in cap-dependent initiation of translation (for review, see Gingras et al. 1999). eIF4F consists of the cap-binding subunit eIF4E, the modular protein eIF4G, and the RNA-helicase eIF4A (Dominguez et al. 1999; Gingras et al. 1999; Neff and Sachs 1999). There are two isoforms of eIF4G in yeast (eIF4G1 and 2), which are encoded by the TIF4631 and TIF4632 genes (Goyer et al. 1993). Like CBP80, eIF4G is a member of the MIF4G domain family of proteins, which comprises several proteins involved in RNA metabolism (Aravind and Koonin 2000; Ponting 2000). In addition to eIF4E and eIF4A, eIF4G also interacts with the poly(A)-binding protein PABP (Pab1p in yeast; Tarun and Sachs 1996). Based on observations in higher eukaryotes (Lamphear et al. 1995), it is also thought to contact the small ribosomal subunit-associated multimeric factor eIF3, thus recruiting translation preinitiation complexes to the mRNA. The interaction between PABP and eIF4G facilitates the functional association of the 3′ end of an mRNA with its 5′ end to promote translation (Tarun et al. 1997; Preiss 2002), whereas the association of eIF4G with eIF4E markedly enhances the binding of the latter to the cap structure (Haghighat and Sonenberg 1997).
Interestingly, eIF4G interacts with CBC in yeast as well as in human cells (Fortes et al. 2000; McKendrick et al. 2001). In yeast, the interaction site for CBC on eIF4G is located between the eIF4E-binding motif and the MIF4G domain (Fortes et al. 2000). eIF4E and CBC can simultaneously bind to eIF4G, but adversely affect each others affinity for eIF4G. Moreover, CBC addition to yeast in vitro translation reactions inhibited cap-dependent translation in wild-type extracts but stimulated the expression of capped mRNA in extracts containing a mutant form of eIF4G that is deficient for eIF4E and Pab1 binding (Fortes et al. 2000). These findings are consistent with a role for the CBC-eIF4G interaction in the exchange of CBC for eIF4F and/or a direct recruitment of nascent mRNA for translation. Based on conventional thinking, these events might be expected to take place once the mRNA reaches the cytoplasm or concomitantly with its nucleocytoplasmic transport. However, because CBC (Görlich et al. 1996) and many components of the translation machinery (Iborra et al. 2001; Dahlberg et al. 2003; Nathanson et al. 2003) are present in both cellular compartments, it cannot be excluded that the eIF4G–CBC interaction functions in the nucleus. Indeed, the interaction between the mammalian factors was revealed in nuclear preparations (McKendrick et al. 2001). Further support for a direct role of CBC in translation comes from recent work in human cell lines suggesting that both nuclear and cytoplasmic nonsense mediated decay (NMD) occurs on CBC-associated rather than eIF4E-associated mRNA (Ishigaki et al. 2001; Lejeune et al. 2002). NMD is a surveillance mechanism comprising the recognition and subsequent degradation of mRNAs bearing a premature termination codon and, importantly, is translation dependent (Hentze and Kulozik 1999; Maquat and Carmichael 2001; Schell et al. 2002; Wilkinson and Shyu 2002). Thus, it has been proposed that NMD takes place during a “pioneering round” of translation that is stimulated by CBC—perhaps through its interaction with eIF4G.
In this study, we investigate a potential role of CBC in translation in the yeast Saccharomyces cerevisiae. For this purpose, we examine the CBC-binding site of eIF4G1, identify a point mutation (DR548/9AA) that abolishes its interaction with CBC, and study the phenotypic consequences of this mutation in vivo. In contrast to deletion of either CBC subunit in vivo, we find that cells expressing eIF4G1 with the (DR548/9AA) substitution as the sole source of eIF4G (termed eIF4G1–548 protein and 4G1–548 strain in the following) show no detectable phenotype at the level of cell growth or transcriptome composition. Unlike eIF4G1–459, a mutant protein with reduced binding to eIF4E, eIF4G1–548 shows no defect in bulk cellular translation, and we find no detectable delay in the translation of newly made mRNA molecules. Taken together, these data suggest that the eIF4G–CBC interaction is dispensable in yeast. Moreover, the indistinguishable kinetics of translation of an induced reporter mRNA in eIF4G1–548 and wild-type cells reveal that the first round of mRNA translation does not require the wild-type eIF4G–CBC interaction.
RESULTS
A point mutant of eIF4G1 defective in CBC binding
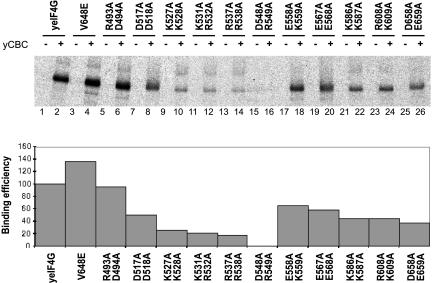
The minimal CBC-interacting region of the yeast eIF4G1 protein was previously shown to reside between amino acid residues 490 and 592, with a further contribution by amino acids 593–656 (Fortes et al. 2000). To isolate point mutants with reduced CBC binding for further functional analyses, we assayed the binding of a suitable wild-type eIF4G1 fragment (eIF4G1486–656) as well as a series of amino acid substitution mutants to CBC in vitro (Fig. 1 ▶). Antibody-immobilized CBC was incubated with in vitro translated, [35S]-labeled eIF4G1486–656 proteins, as previously described (Fortes et al. 2000). Quantitative analysis of labeled proteins bound to CBC after extensive washes revealed strongly reduced binding with mutants in the region from amino acids 527 to 549 (lanes 9–16). The mutant protein eIF4G1–548, corresponding to alanine substitutions of an aspartate and an arginine residue in positions 548 and 549, lacked any detectable CBC binding (lanes 15–16).
FIGURE 1.
Mutation of amino acids 548 and 549 of eIF4G1 to alanine impairs binding to CBC in vitro. [35S]-labeled wild-type fragment of eIF4G1 protein (amino acids 486–656) or various point mutants as indicated above the panel were translated in rabbit reticulocyte lysate. All reactions yielded similar amounts of labeled protein as confirmed by SDS-PAGE (not shown). Equal amounts of each eIF4G1 protein were then incubated with a column with antibody-immobilized CBC (+) or a control column, prepared with preimmune serum (−), as previously described (Fortes et al. 2000). After incubation and washing, the bound fractions were analyzed by 8% SDS-PAGE (top) and binding was quantified by fluorimetry. The relative binding efficiencies of the mutants in comparison to the wild-type eIF4G1 are shown (bottom). The integrity and amount of unbound material was also analyzed on a separate gel confirming that there was no selective loss of [35S]-labeled eIF4G1 in any of the binding reactions (not shown).
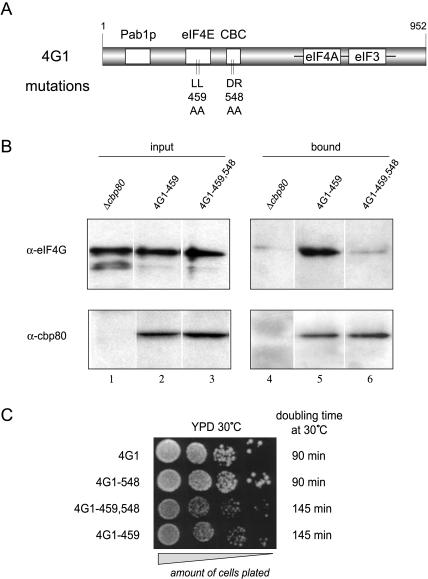
To study the functional consequences of the eIF4G interaction with CBC in vivo, we chose to employ a series of yeast strains expressing the eIF4G1 protein from a centromeric plasmid as a sole source of eIF4G (Table 1 ▶). Wild-type eIF4G1 protein was used as a reference (strain 4G1). Additionally, we introduced versions of eIF4G1 carrying the (DR548/9AA) substitution to affect binding to CBC (strain 4G1–548) or the previously described (LL459/60AA) substitution affecting the interaction of eIF4G with eIF4E (strain 4G1–459; Tarun and Sachs 1997), alone or in combination (strain 4G1–459,548; Fig. 2A ▶). To ascertain the efficacy of the (DR548/9AA) substitution in vivo, we performed immunoprecipitation experiments (Fig. 2B ▶) using antibodies raised against CBP80 protein (Görlich et al. 1996). Because the level of complex formation between CBC and wild-type eIF4G1 is below detection limits in this type of approach (Fortes et al. 2000), we chose to compare strains 4G1–459 and 4G1–459,548. Through an elimination of negative cooperative effects between eIF4E and CBC, the –459 mutation increases the amount of CBC-eIF4G complex present at steady state to detectable levels (see Fig. 2B ▶, lane 5, and Fortes et al. 2000). In this context, the –548 mutation strongly reduces the eIF4G1 signal in the CBC-bound fraction (lane 6). The residual level of CBC with strain 4G1–548 represents nonspecific background binding to the antibody resin, as shown by performing a control experiment with a Δcbp80 yeast strain. Even in this context, a small amount of eIF4Gs signal is still detectable in the bead-bound material (lane 4). These results show that the –548 mutation also impairs the eIF4G-CBC interaction in vivo.
TABLE 1.
Yeast strains used in this study
| Systematic name | Name used in this study | Disrupted interaction | Plasmids |
| YAS 2632 (α) | 4G1 | none | TRP1-CEN-pTIF4631 |
| YAS 2757 (α) | 4G1-548 | CBC | TRP1-CEN-ptif4631-548 |
| YAS 2074 (β) | 4G1-459 | eIF4E | TRP1-CEN-ptif4631-459 |
| YAS 2758 (α) | 4G1-459,548 | eIF4E, CBC | TRP1-CEN-ptif4631-459,548 |
| YBJ 220 (δ) | none | TRP1-CEN-pTIF4631 URA-CEN-pGal-Luc | |
| YBJ 221 (δ) | CBC | TRP1-CEN-ptif4631-548 URA-CEN-pGal-Luc | |
| Name | Genotype | ||
| Δcbp80 | MATa ade2 ade3 his3 leu2-3,112 trp1 ura3 ycbp80/gcr3::TRP1 | ||
| Δcbp20 | MATa ade2 ade3 his3 leu2-3,112 trp1 ura3 ycbp20/mud13::HIS3 | ||
α: These strains have the following genotype: MATa ade2-1 his3-11,15 leu2-3,111 trp1-1 ura3-1 pep4::HIS3 tif4631::leu2 tif4632::ura3.
β: This strain is described by Tarun and Sachs (1997); it has the following genotype: MATa ade2-1 his3-11,15 leu2-3,111 trp1-1 ura3-1 pep4::HIS3 tif4631::LEU2.
δ: YBJ 220 and YBJ 221 derived respectively from the strains YAS 2632 and YAS 2757.
FIGURE 2.
Phenotypic characterization of the eIF4G1–548 mutation. (A) Schematic representation of the eIF4G1 protein. Boxed regions represent binding sites for the proteins indicated above. Vertical bars indicate the nature and position of the mutations described in this study. (B) Immunoprecipitation using rabbit polyclonal α-CBP80 antibodies and extracts from indicated yeast strains. A Western blot analysis is shown revealing the amount of eIF4G (upper panel) and CBP80 (lower panel) proteins present in the input (left panels, 1% of the input is loaded), and bound fractions (right panels, 100% of the eluate) of the immunoprecipitation. A residual level of eIF4G binding to the antibody resin is seen even in the absence of CPB80 protein (lane 4). Variation of several parameters of the immunoprecipitation protocol failed to reduce this background binding (data not shown); however, robust binding of eIF4G1–459 protein and a lack of detectable binding of eIF4G1–459,548 protein to the CBC resin was consistently seen in three independent repeat experiments. (C) Serial dilutions (10-fold) of the indicated yeast strains growing exponentially were plated on YPD and incubated for 48 h at 30°C. Doubling times listed on the right were measured in liquid YPD medium over 8 h of incubation at exponential growth at 30°C.
In agreement with earlier reports (Tarun and Sachs 1997), the strain 4G1–459 has a marked growth phenotype in complete medium at 30°C when compared to the reference strain 4G1 (145 versus 90 min doubling time, respectively, in liquid culture; Fig. 2C ▶). By contrast, the strain 4G1–548 grows normally (90 min doubling time), and the combination of both mutations in strain 4G1–459,548 causes no additional growth retardation (145 min doubling time). Analysis of the strains on galactose-based medium and various temperatures (16°C, 23°C, 30°C, and 37°C) revealed no contribution to the growth phenotype by the –548 mutation (data not shown). These results argue that the eIF4G-CBC interaction is dispensable in vivo, even when the binding between eIF4G and eIF4E is compromised.
The eIF4G-CBC interaction is dispensable for cellular translation
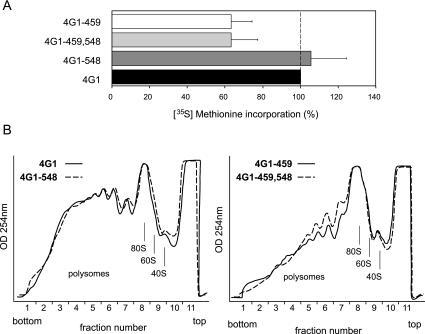
Next, we characterized the consequences of disrupting the eIF4G-CBC interaction for cellular translation. To this end, we measured the overall rate of protein synthesis in exponentially growing cells by metabolic labeling with [35S] methionine. Figure 3A ▶ displays the results of these measurements with the wild-type and mutant strains as used for Figure 2C ▶. As expected (Winstall et al. 2000), the 4G1–459 cells show a significantly reduced rate of protein synthesis (approximately 60% of the wild-type reference), which correlates with its slow growth phenotype. By contrast, the –548 mutation displays no significant effect on the protein synthesis rate when measured in the context of an otherwise wild-type eIF4G1 protein (compare strain 4G1–548 with 4G1), or in the context of the –459 mutation (compare strain 4G1–459 with 4G1–459,548). These findings are substantiated by polysome profile analyses of extracts from strains expressing the different eIF4G mutants on linear sucrose density gradients (Fig. 3B ▶). The 4G1–459 strain displays a drastic decline in polysomal complexes and a concomitant increase in monosome complexes (Fig. 3B ▶, compare the absorbance profiles at 254 nm from strain 4G1–459, right panel, and the 4G1 strain, left panel), typical of a defect in translation initiation. By contrast, the –548 mutation induces no appreciable profile shifts in either context (compare profiles within each panel). These results demonstrate that the –548 mutation has no significant effect on bulk translation in exponentially growing cells.
FIGURE 3.
Effects of eIF4G mutants on general translation activity. (A) The amount of newly synthesized proteins in different strains was measured by pulse labeling with [35S]-methionine. The data are expressed as a percentage of methionine incorporation measured with each mutant strain relative to the 4G1 wild-type control. Shown are averaged results from 10 to 16 measurements corresponding to 3 to 5 independent yeast cultures with standard deviations. (B) Polysome profile analysis. Yeast extracts from the indicated strains were analyzed by sucrose density gradient centrifugation. Absorbance profiles at 254 nm were recorded after centrifugation. The profiles are representative of several biological repeat experiments.
The eIF4G-CBC interaction is dispensable for efficient translation of newly made mRNAs
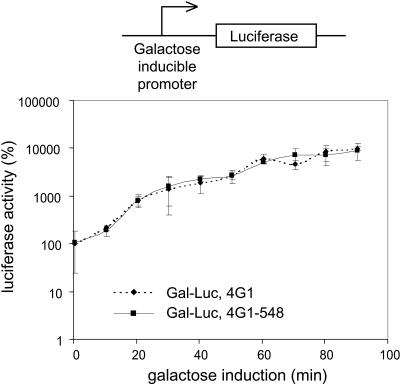
In mammalian cells, the first (“pioneer”) round of translation has been suggested to be mediated by CBC rather than eIF4E (Lejeune et al. 2002). In this context, CBC might require its interaction with eIF4G (Ishigaki et al. 2001; Wilkinson and Shyu 2002). Because only a small proportion of total cellular protein synthesis would derive from the first round of translation at steady state, we devised an experiment to measure the accumulation of protein product from newly made mRNA molecules in cells harboring different forms of eIF4G1 protein. For this purpose, we constructed the yeast strains YBJ 220 and YBJ 221 (Table 1 ▶), which are derivatives of the strains 4G1 and 4G1–548, respectively, and bear an additional plasmid encoding a luciferase reporter mRNA under the control of a strong, galactose-inducible promoter. Both strains were shifted to galactose-based media and aliquots of cells collected for luciferase activity assays at multiple time points ranging from 0 to 90 min (Fig. 4 ▶). Given the strong and rapid induction of luciferase gene transcription, it is a reasonable assumption that the accumulation of luciferase activity over time directly reflects the initial recruitment of newly made luciferase mRNA for translation. We measured steep increases of luciferase activity over time (∼50-fold within 60 min) that were, however, completely indistinguishable between the two yeast strains analyzed, even at the earliest time points measured (that is, 10 min). These results argue against the eIF4G-CBC interaction as a requirement for a pioneering round of translation in yeast.
FIGURE 4.
Translation of newly made mRNA is independent of the eIF4G-CBC interaction. The yeast strains 4G1 and 4G1–548 were transformed with a plasmid expressing the luciferase reporter mRNA under the control of a galactose-inducible promoter. The resulting strains YBJ 220 and YBJ 221, respectively, were used to monitor the accumulation of luciferase activity over time after galactose induction as described in Materials and Methods. The luciferase activity was normalized against the number of cells per sample. The graph represents the average of three independent experiments.
Effects of disrupting CBC function on the composition of the cellular transcriptome
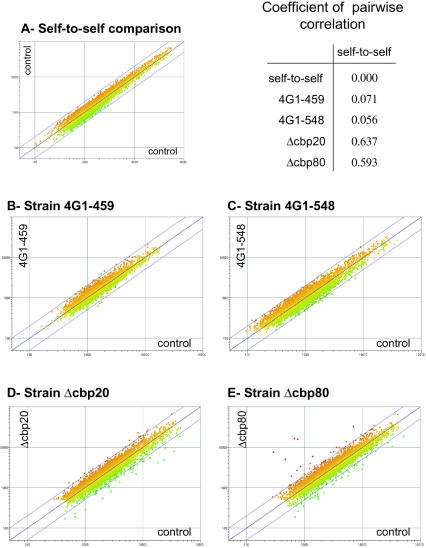
Analysis of transcriptome changes using DNA microarrays is an alternative way to uncover the cellular consequences of compromised CBC function. Given the tight coupling between multiple steps in mRNA metabolism, including translation and mRNA decay, it is conceivable that a selective impairment of a given step for specific mRNAs might ultimately be reflected in an altered steady-state level of that mRNA. Thus, we chose to perform transcriptome profiling experiments for a selected panel of yeast strains, including 4G1–459, 4G1–548, and the wild-type reference strain 4G1, as well as strains lacking either subunit of CBC (strains Δcbp80 and Δcbp20) and a suitable wild-type control. Total RNA was isolated and subjected to microarray analysis using cDNA microarrays comprising approximately 95% of all annotated ORFs in the yeast genome as detailed in Materials and Methods. Two separate array experiments were performed for each mutant to wild-type comparison with fluorescent dye swap (Fig. 5 ▶). Using a twofold threshold for changes in expression level, both CBP deletion strains revealed relevant transcriptome alterations (Fig. 5D ▶, E ▶). Strain Δcbp20 displayed increased levels for 18 and decreases for 34 different mRNAs. In strain Δcbp80, 33 different mRNAs scored as increased and 41 as decreased in levels relative to wild-type cells. A proportion of these mRNAs showed consistent changes in both deletion strains: 15 mRNAs had a decreased abundance, and 5 genes an increased abundance (see Table 2 ▶ for a list of these mRNAs). By contrast, the analysis of the strains 4G1–548 and 4G1–459 reveal no significant specific alterations in transcriptome composition (Fig. 5B ▶,C ▶). Calculating a coefficient of pairwise correlation (as described in the legend of Fig. 5 ▶) as a measure to compare the array data sets with a self-to-self control experiment (Fig. 5A ▶) confirmed this notion: although both CBC deletion strains scored as significantly different from the self-to-self control (0.637 for the strain Δcbp20 and 0.593 for the Δcbp80), both eIF4G point mutants are virtually indistinguishable from the wild-type control (0.071 for 4G1–548 and 0.056 for 4G1–459). Remarkably, we do not register any significant transcriptome alterations in strain 4G1–459, despite its marked translational and growth phenotype. Note that the microarray data normalization would “mask” any uniform changes in the amount of all mRNA present in cells to focus on mRNAs that deviate from any general trend (the normalization strategy is described in Materials and Methods). These data indicate that alterations of the transcriptome composition due to deletions of either CBC subunit are not primarily due to a disruption of the CBC–eIF4G complex.
FIGURE 5.
Transcriptome composition analysis. Scatter plot representation of the transcriptome microarray analysis. Each dot represents the averaged measurements for a single gene. On the vertical and horizontal axes, the intensities measured for the indicated probes corresponding to two fluorescent channels are plotted. Each panel represents the average of two or three array experiments (including dye-swap). (A) A self-to-self experiment using identical RNA samples to prepare the Cy3- and C5-labeled probes. Comparisons of probes derived from strains 4G1–459 (B) and 4G1–548 (C) to control strain 4G1, and from the deletion strains Δcbp20 (D) and Δcbp80 (E) to corresponding isogenic wild-type cells are also shown. The coefficient of pairwise correlation is a measure of the distance between a control-to-control and mutant-to-control comparison: a coefficient of 0 corresponds to the minimal distance between two identical arrays and a coefficient of 1 represents the maximal distance for two divergent experiments.
TABLE 2.
mRNAs whose expression is affected in both, Δcbp20 and Δcbp80, strains
| Expression ratio | |||
| ORF name | Δcbp20 | Δcbp80 | Description |
| mRNAs with increased abundance | |||
| YNL338W | 2.151 | 2.409 | |
| YGL209W | 2.082 | 2.257 | MIG2 (protein containing zinc fingers very similar to zinc fingers in Mig1p) |
| YDL021W | 2.118 | 2.235 | GPM2 (phosphoglycerate mutase, involved in glycolysis) |
| YHR183W | 2.503 | 2.010 | GND1 (phosphogluconate dehydrogenase, decarboxylating) |
| YHL050C | 2.108 | 2.184 | |
| mRNAs with decreased abundance | |||
| YMR162C | 0.274 | 0.271 | |
| YLR012C | 0.462 | 0.429 | |
| YBL044W | 0.445 | 0.428 | |
| YFL017W-A | 0.487 | 0.416 | SMX2 (snRNP G protein, homolog of Human Sm-G) |
| YJL088W | 0.394 | 0.354 | ARG3 (ornithine carbamoyltransferase) |
| YFR007W | 0.446 | 0.372 | |
| YLR446W | 0.366 | 0.341 | |
| YLR181C | 0.440 | 0.407 | |
| YOR122C | 0.440 | 0.398 | PFY1 (profiling, actin-binding protein) |
| YLR062C | 0.411 | 0.409 | BUD28 |
| YMR120C | 0.226 | 0.434 | ADE17 (5-aminoimidazole-4-carboxamide ribonucleotide transformylase) |
| YKL152C | 0.209 | 0.286 | GPM1 (phosphoglycerate mutase) |
| YKL153W | 0.387 | 0.454 | |
| YGR254W | 0.223 | 0.317 | ENO1 (enolase) |
| YHR174W | 0.423 | 0.358 | ENO2 (enolase) |
DISCUSSION
The study presented here extends our previous investigations on the interaction between CBC and eIF4G in yeast. Using a series of amino acid substitution mutants in eIF4G1 protein, we identify critical amino acids for CBC binding that lie approximately in the middle of the region that was previously mapped for CBC binding (Fortes et al. 2000). In particular, we identify one mutation (DR548/9AA) that abolishes binding to CBC in vitro and in vivo. Surprisingly, our experiments reveal no detectable phenotypic consequences of the disrupted CBC–eIF4G complex formation at various levels of analysis. Cells expressing the eIF4G1–548 protein grow normally under laboratory conditions, and display wild-type levels of bulk protein synthesis. Translation of a newly made reporter mRNA is unaffected and shows no kinetic delay associated with a lack of interaction between CBC and eIF4G. Furthermore, we find no phenotypic differences between a strain carrying a doubly mutated eIF4G protein with disrupted binding to both eIF4E and CBC (4G1–459,548) and a strain just compromised in eIF4Ebinding (4G1–459). This may suggest that the previously observed synthetic lethality between the eIF4G1–459 protein and a frameshift mutation in CBP80 (Fortes et al. 1999, 2000) is not simply a consequence of a disrupted binding between the two proteins, but implies a further impairment of CBP80 function.
Alternatively, disruption of the interaction between CBC and eIF4G may affect the translation of a smaller subset of mRNAs that do not contribute appreciably to bulk cellular translation. The levels of the translation products of these mRNAs may not be sufficiently altered to affect cellular growth under the conditions we tested here. Although we have not exhaustively investigated this scenario, our preliminary analysis of changes in polysome distribution of mRNAs using microarray-based methods did not reveal such a subset of mRNAs (J. Baron-Benhamou, data not shown). On balance, our observations argue against a direct role of the CBC-eIF4G in translation initiation and favor a nonessential contribution, if any, to the remodeling of mRNPs from a nuclear to a cytoplasmic configuration in yeast.
Deletion of either subunit of CBC alone, or in combination, is not lethal, but it incurs a growth impairment (Colot et al. 1996; Fortes et al. 1999; Das et al. 2000). We present evidence here that deletions of the CBP20 or CBP80 genes selectively affect the steady-state levels of a small subset of mRNAs. These transcriptome alterations do not persist in the 4G1–548 strain, suggesting that CBC’s effect on target mRNAs is not mediated through its interaction with eIF4G. CBC is known to affect multiple steps in mRNA metabolism, including a recently identified nuclear pathway for mRNA degradation (Das et al. 2000). Further experiments are required to delineate the step(s) in target mRNA metabolism that is affected by the absence of CBC subunits.
Collectively, our data send a cautionary message with regard to current models of a pioneering round of translation and the molecular mechanism that mediates it (Wilkinson and Shyu 2002). It is noticeable that, unlike CBP20, CBP80 is evolutionarily not a highly conserved protein. Thus, it is possible that our findings in yeast are reflective of an evolutionary divergence between the yeast and human proteins. More detailed analyses will be required to understand the putative role of CBC in an mRNP remodeling process that prepares nascent mRNA for the initial recognition by the translation machinery.
MATERIALS AND METHODS
Plasmids constructs and yeast strains
PCR mutagenesis was used to introduce the alanine codons into the recombinant eIF4G1 gene on a TRP1CEN plasmid as previously described (Hershey et al. 1999). The mutated plasmids were used to produce templates for coupled in vitro transcription–translation reaction to express [35S]-labeled eIF4G1 fragments. The centromeric plasmid URA-CEN-pGal-Luc was obtained by insertion of a HindIII–XhoI fragment from pGEM-Luc (Promega) into the HindIII–XhoI sites of p416Gal1 kindly provided by Michael Knop (EMBL, Heidelberg, Germany). The strains of yeast S. cerevisiae used in this study are described in Table 1 ▶. Standard yeast growth conditions and manipulations were used (Sherman 2002).
CBC column preparation and in vitro binding assays
CBC column preparation and in vitro binding assays were performed as previously described (Fortes et al. 1999).
Immunoprecipitation and Western blot analysis
Binding buffer (10× concentrated: 100 mM Tris-HCl at pH 8.0, 1 M NaCl, 1% NP40) was added to equal amounts (650 μL) of yeast extracts prepared as for gradient separation (see below). Polyclonal antibodies raised against the yeast CBP80 protein (Görlich et al. 1996) were chemically cross-linked to protein A sepharose beads (Amersham) as described by Harlow and Lane (1988). The extracts were then incubated with 15 μL of bead-cross-linked antibodies for 1 h 30 min at 4°C under agitation. The unbound fraction was collected and the beads were extensively washed (five times with binding buffer at 4°C). The bound fraction was eluted from the beads using 40 μL elution buffer (10 mM Tris-HCl at pH 8.0, 1 M NaCl, 0.2% SDS). One percent of the input extracts and 100% of the bound fractions were loaded on a SDS-PAGE gel. Western blot was performed as described by De Gregorio et al. (2001). Rabbit polyclonal antisera against the N-terminal region of the yeast eIF4G1 protein were raised against the synthetic peptide SEAENTKRLFLEQVRLRKAAMERKKNG (Genosys) coupled to maleimide-activated keyhole limpet hemocyanin (KLH; Pierce). α-eIF4G antibodies (dilution 1/1000) or α-CBP80 antibodies (dilution 1/1000; Görlich et al. 1996) were used for Western analysis, followed by anti-rabbit IgG coupled to horseradish peroxidase (dilution 1/5000; Amersham).
Pulse labeling
Yeast cells were grown exponentially at 30°C in minimal media lacking methionine. An equal amount of cells (0.2 OD600 nm units) was collected for each strain and incubated at 30°C with 3.5 μL of a mix containing 0.44 μCi/μL [35S]-methionine (Amersham) and 6 ng/μL cold methionine. After 5 min, the reaction was stopped by addition of an equal volume of 20% trichloroacetic acid. Samples were boiled for 20 min at 90°C and applied to glass microfibre Whatman filters (Ø = 25 mm). The amount of radioactivity, retained on the filter after extensive washes, was measured using a scintillation counter.
Yeast extract and sucrose gradient separation
Yeast cells were grown exponentially at 30°C in complete media (YPD), and 50 μg/mL cycloheximide was added to the cultures 1 min before harvesting the cells by centrifugation. Cells were washed once with breaking buffer (20 mM Tris-HCl at pH 7.4, 100 mM KCl, 2 mM MgCl2, 200 μg/mL heparin, 100 μg/mL cycloheximide, 2 mM DTT, 0.5 mM PMSF) and repelleted. One and a half times the wet cell weight of breaking buffer and five times wet cell weight of glass beads (425–600 μm) were added, and cells were broken by five times vortexing for 1 min with intermittent cooling on ice. After two consecutive centrifugations at 12,000g, supernatants were collected and the equivalent of 25 OD260 nm units was then layered onto linear 17.5 to 50% sucrose density gradients in gradient buffer (50 mM Tris-HCl at pH 7.4, 50 mM NH4Cl, 4 mM MgCl2, 2 mM DTT). Extracts were then separated by ultracentrifugation at 202,000g for 2.5 h. The gradients were then fractionated starting from the bottom and the absorbance profiles at 254 nm were recorded.
Total RNA extraction and microarray analysis
Yeast extracts prepared as for gradient separation were used to prepare total RNA by phenol-chloroform-isoamyl alcohol extraction. Twenty micrograms of total RNA were used to prepare Cy 3- and Cy5-labeled cDNA for hybridization to glass DNA microarrays comprising approximately 95% of all annotated ORF sequences in the S. cerevisiae genome as described by T. Preiss, J. Baron-Benhamou, W. Ansorge, and M.W. Hentze (in prep.). Microarray data were analyzed using Gene Pix 4.0 and Gene Spring 5.0 (Silicon Genetics) software. Only spots classified as “present” and satisfying a minimal intensity requirement above background were included in the analysis (representing approximately 90% of all yeast ORFs). Intensity ratios were normalized to the 50th percentile for each chip.
Luciferase assay
Yeast strains were grown exponentially at 30°C in selective minimal medium complemented with 2% raffinose. At time 0, galactose was added to a final concentration of 2%. At each time point 1-mL aliquots of each culture were collected, cells were washed with H2O and resuspended in 100 μL lysis buffer (50 mM Tris-HCl at pH 7.4, 100 mM NaCl, 1 mM PMSF). One hundred microliters of glass beads (425–600 μm) were added and cells were lysed by vortexing. Sixty percent of the supernatant was used to measure luciferase activity using a luciferase assay system (Promega), following the manufacturer’s instructions.
Acknowledgments
The authors thank Iain Mattaj and Alan Sachs for many helpful discussions and their generous support of this work. We thank Dirk Görlich for antibodies against yeast CBP80 protein. This work was supported by grants from the Deutsche Forschungsgemeinschaft (PR616/1–1 and 2) to T.P. and M.W.H. (Gottfried Wilhelm Leibniz Prize), as well as the Human Frontiers in Science Program (RG0038/1999-M) to M.W.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5100903.
REFERENCES
- Aravind, L. and Koonin, E.V. 2000. Eukaryote-specific domains in translation initiation factors: Implications for translation regulation and evolution of the translation system. Genome Res. 10: 1172–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot, H.V., Stutz, F., and Rosbash, M. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes & Dev. 10: 1699–1708. [DOI] [PubMed] [Google Scholar]
- Dahlberg, J.E., Lund, E., and Goodwin, E.B. 2003. Nuclear translation: What is the evidence? RNA 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, B., Guo, Z., Russo, P., Chartrand, P., and Sherman, F. 2000. The role of nuclear cap binding protein Cbc1p of yeast in mRNA termination and degradation. Mol. Cell. Biol. 20: 2827–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio, E., Baron, J., Preiss, T., and Hentze, M.W. 2001. Tethered-function analysis reveals that elF4E can recruit ribosomes independent of its binding to the cap structure. RNA 7: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, D., Altmann, M., Benz, J., Baumann, U., and Trachsel, H. 1999. Interaction of translation initiation factor eIF4G with eIF4A in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274: 26720–26726. [DOI] [PubMed] [Google Scholar]
- Flaherty, S.M., Fortes, P., Izaurralde, E., Mattaj, I.W., and Gilmartin, G.M. 1997. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. 94: 11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes, P., Kufel, J., Fornerod, M., Polycarpou-Schwarz, M., Lafontaine, D., Tollervey, D., and Mattaj, I.W. 1999. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell. Biol. 19: 6543–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes, P., Inada, T., Preiss, T., Hentze, M.W., Mattaj, I.W., and Sachs, A.B. 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6: 191–196. [PubMed] [Google Scholar]
- Furuichi, Y., LaFiandra, A., and Shatkin, A.J. 1977. 5′-Terminal structure and mRNA stability. Nature 266: 235–239. [DOI] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Görlich, D., Kraft, R., Kostka, S., Vogel, F., Hartmann, E., Laskey, R.A., Mattaj, I.W., and Izaurralde, E. 1996. Importin provides a link between nuclear protein import and U snRNA export. Cell 87: 21–32. [DOI] [PubMed] [Google Scholar]
- Goyer, C., Altmann, M., Lee, H.S., Blanc, A., Deshmukh, M., Woolford Jr., J.L., Trachsel, H., and Sonenberg, N. 1993. TIF4631 and TIF4632: Two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol. 13: 4860–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighat, A. and Sonenberg, N. 1997. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272: 21677–21680. [DOI] [PubMed] [Google Scholar]
- Harlow, E. and Lane, D. 1988. Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Hentze, M.W. and Kulozik, A.E. 1999. A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96: 307–310. [DOI] [PubMed] [Google Scholar]
- Hershey, P.E., McWhirter, S.M., Gross, J.D., Wagner, G., Alber, T., and Sachs, A.B. 1999. The cap-binding protein eIF4E promotes folding of a functional domain of yeast translation initiation factor eIF4G1. J. Biol. Chem. 274: 21297–21304. [DOI] [PubMed] [Google Scholar]
- Iborra, F.J., Jackson, D.A., and Cook, P.R. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293: 1139–1142. [DOI] [PubMed] [Google Scholar]
- Ishigaki, Y., Li, X., Serin, G., and Maquat, L.E. 2001. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106: 607–617. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E., Lewis, J., McGuigan, C., Jankowska, M., Darzynkiewicz, E., and Mattaj, I.W. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668. [DOI] [PubMed] [Google Scholar]
- Lamphear, B.J., Kirchweger, R., Skern, T., and Rhoads, R.E. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270: 21975–21983. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., Ishigaki, Y., Li, X., and Maquat, L.E. 2002. The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: Dynamics of mRNP remodeling. EMBO J. 21: 3536–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.D. and Izaurralde, E. 1997. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 247: 461–469. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D., Görlich, D., and Mattaj, I.W. 1996. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 24: 3332–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. and Carmichael, G.G. 2001. Quality control of mRNA function. Cell 104: 173–176. [DOI] [PubMed] [Google Scholar]
- Mazza, C., Segref, A., Mattaj, I.W., and Cusack, S. 2002. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21: 5548–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKendrick, L., Thompson, E., Ferreira, J., Morley, S.J., and Lewis, J.D. 2001. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 21: 3632–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2001. mRNA turnover. Curr. Opin. Cell. Biol. 13: 320–325. [DOI] [PubMed] [Google Scholar]
- Nathanson, L., Xia, T., and Deutscher, M.P. 2003. Nuclear protein synthesis: A reevaluation. RNA 9: 9–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, C.L. and Sachs, A.B. 1999. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol. Cell. Biol. 19: 5557–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, M., Kataoka, N., and Shimura, Y. 1990. A nuclear cap binding protein from HeLa cells. Nucleic Acids Res. 18: 6989–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting, C.P. 2000. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem. Sci. 25: 423–426. [DOI] [PubMed] [Google Scholar]
- Preiss, T. 2002. The end in sight: Poly(A), translation and mRNA stability in eukaryotes. In Translation Mechanisms (eds. L. Brakier-Gingras and J. Lapointe), Landes Bioscience, Georgetown, TX. http://www.eurekah.com/chapter.php?chapid=840&bookid=59&catid=54.
- Schell, T., Kulozik, A.E., and Hentze, M.W. 2002. Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol. 3: reviews 1006.1–1006.6 [DOI] [PMC free article] [PubMed]
- Shatkin, A.J. 1985. mRNA cap binding proteins: Essential factors for initiating translation. Cell 40: 223–224. [DOI] [PubMed] [Google Scholar]
- Shen, E.C., Stage-Zimmermann, T., Chui, P., and Silver, P.A. 2000. The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J. Biol. Chem. 275: 23718–23724. [DOI] [PubMed] [Google Scholar]
- Sherman, F. 2002. Getting started with yeast. Methods Enzymol. 350: 3–41. [DOI] [PubMed] [Google Scholar]
- Tarun, S.Z. and Sachs, A.B. 1996. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 15: 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- ———. 1997. Binding of eukaryotic translation initiation factor 4E (eIF4E) to eIF4G represses translation of uncapped mRNA. Mol. Cell. Biol. 17: 6876–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun, S.Z., Wells, S.E., Deardorff, J.A., and Sachs, A.B. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. 94: 9046–9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visa, N., Izaurralde, E., Ferreira, J., Daneholt, B., and Mattaj, I.W. 1996. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell. Biol. 133: 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, M.F. and Shyu, A.B. 2002. RNA surveillance by nuclear scanning? Nat. Cell. Biol. 4: E144–147. [DOI] [PubMed] [Google Scholar]
- Winstall, E., Sadowski, M., Kuhn, U., Wahle, E., and Sachs, A.B. 2000. The Saccharomyces cerevisiae RNA-binding protein Rbp29 functions in cytoplasmic mRNA metabolism. J. Biol. Chem. 275: 21817–21826. [DOI] [PubMed] [Google Scholar]