Abstract
Polycistronic pre-mRNAs from Caenohabditis elegans operons are processed by internal cleavage and polyadenylation to create 3’ ends of mature mRNAs. This is accompanied by trans-splicing with SL2 ∼100 nucleotides downstream of the 3’ end formation sites to create the 5’ ends of downstream mRNAs. SL2 trans-splicing depends on a U-rich element (Ur), located ∼70 nucleotides upstream of the trans-splice site in the intercistronic region (ICR), as well as a functional 3′ end formation signal. Here we report the existence of a novel gene-length RNA, the Ur-RNA, starting just upstream of the Ur element. The expression of Ur-RNA is dependent on 3′ end formation as well as on the presence of the Ur element, but does not require a trans-splice site. The Ur-RNA is not capped, and alteration of the location of the Ur element in either the 5′ or 3′ direction alters the location of the 5′ end of the Ur-RNA. We propose that a 5’ to 3’ exonuclease degrades the precursor RNA following cleavage at the poly(A) site, stopping when it reaches the Ur element, presumably attributable to a bound protein. Part of the function of this protein can be performed by the MS2 coat protein. Recruitment of coat protein to the ICR in the absence of the Ur element results in accumulation of an RNA equivalent to Ur-RNA, and restores trans-splicing. Only SL1, however, is used. Therefore, coat protein is sufficient for blocking the exonuclease and thereby allowing formation of a substrate for trans-splicing, but it lacks the ability to recruit the SL2 snRNP. Our results also demonstrate that MS2 coat protein can be used as an in vivo block to an exonuclease, which should have utility in mRNA stability studies.
Keywords: mRNA stability, trans-splicing, operon, polycistronic pre-mRNA, 3′ end formation, MS2 coat protein
INTRODUCTION
Caenorhabditis elegans has a highly compact genome featuring small introns, close proximity of genes, and, most surprisingly, the presence of operons (Blumenthal et al. 1996). At least 15% of C. elegans genes are contained in operons (Blumenthal et al. 2002). These polycistronic transcription units comprise multiple genes that are generally separated by ∼100 bp. The polycistronic precursors of operon mRNAs are processed into monocistronic mRNAs by conventional cleavage and polyadenylation at the 3′ ends of upstream genes and trans-splicing at the 5′ ends of downstream genes using a snRNP specialized for this process, the SL2 snRNP. These two processes occur very close together on the pre-mRNA, and previous results have indicated a functional dependency of trans-splicing on the 3′ end formation signals. Mutation of the AAUAAA 3′ end formation signal of gpd-2 resulted in reduction of SL2 trans-splicing from >90% to ∼50% (Kuersten et al. 1997). Conversely, changing the rare variant signal, AGUAAA, the natural 3′ end formation signal of another gene in the same operon, mai-1, to the perfect consensus AAUAAA increased the use of SL2 (Liu et al. 2001). In contrast, 3′ end formation is not dependent on trans-splicing downstream: Mutation of the trans-splice site had no effect on upstream 3′ end formation (Kuersten et al. 1997). How the two events are coupled remains unknown.
To fully understand the mechanism of polycistronic pre-mRNA processing, we have used gpd-2/gpd-3 driven by a heat shock promoter as a test operon (Spieth et al. 1993). We discovered a U-rich sequence (Ur) in the intercistronic region that is necessary and sufficient for SL2-specific trans-splicing of downstream gpd-3 (Huang et al. 2001). When the Ur element was mutated, no downstream mRNA accumulated, presumably because it did not receive a cap by trans-splicing, and therefore it was degraded or transcription was terminated. To prevent this termination/degradation, we mutated the 3′ end formation signal along with the Ur element. In this case, downstream RNA did accumulate, but it was trans-spliced entirely to SL1, rather than SL2. Therefore, the Ur element somehow is responsible for directing the SL2 snRNP to the trans-splice site just downstream. The Ur element has the sequence and location expected for a binding site for cleavage stimulation factor (CstF), a multi-subunit protein that has an important role in 3′ end cleavage (Takagaki et al. 1990). The Ur mutations did not appreciably affect 3′ end formation (Huang et al. 2001), however, and we have been unable to detect an interaction between the Ur element and CstF (unpubl.). On the other hand, the SL2 snRNP has been found in a complex with CstF, and the presence of this complex strongly correlates with the ability of the snRNP to function at operon trans-splice sites (Evans et al. 2001). Clearly, CstF might be a protein that acts at the Ur element but it is too soon to conclude that it is.
Here we have identified a possible processing intermediate, a gene-length RNA that we call the Ur-RNA, which accumulates when trans-splicing is inhibited. The 5′ end of Ur-RNA is uncapped and occurs just upstream of the Ur element, even when we move its location a few base pairs upstream or downstream. Formation of Ur-RNA requires a functional 3′ end formation signal and a Ur element. Based on these results, we propose a model for operon pre-mRNA processing in which, following cleavage at the poly(A) site of the upstream gene, a 5′–3′ exonuclease degrades the precursor RNA, stopping when it reaches a protein bound to the Ur element. The Ur-bound protein may recruit SL2 snRNP, causing the downstream gene to be SL2 trans-spliced. We show that MS2 coat protein can replace the Ur element for formation of Ur-RNA, thereby allowing trans-splicing. Coat protein, however, lacks the ability to recruit SL2 snRNP.
RESULTS
Detection of a novel RNA molecule that accumulates under high heat shock temperature
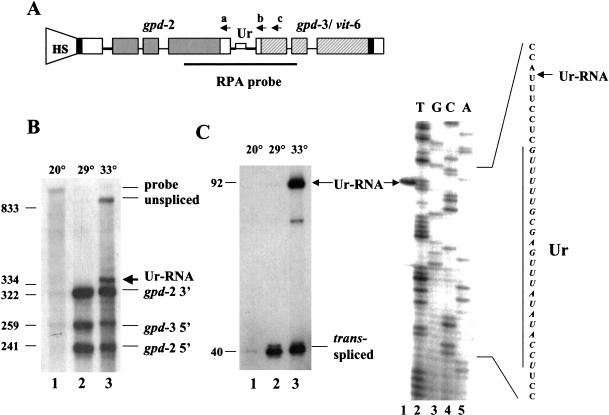
We have described previously an in vivo system to study operon processing using a construct, HS1496, derived from the three-gene operon mai-1/gpd-2/gpd-3 (Spieth et al. 1993). This construct contains gpd-2 and gpd-3 under control of the hsp-16-41 promoter inserted within the last exon of mai-1. RNase protection assays (RPA) were used to analyze the upstream and downstream gene products. Figure 1A ▶ shows the RPA probe, which protects the 3′ end of gpd-2 formed by cleavage and polyadenylation, and the 5′ end of gpd-3 formed by trans-splicing. As shown in Figure 1B ▶, lane 2, on heat shock at 29°C, gpd-2 3′ end and gpd-3 5′ end fragments are expressed and protected as expected. Because the sequences of gpd-2 and gpd-3 are very similar, the 5′ end of gpd-2 also yields a protected fragment even though it is not covered by the probe.
FIGURE 1.
High-temperature heat shock results in accumulation of a novel RNA containing part of the intercistronic region. (A) The HS1496 construct. The position of the heat shock promoter is shown by the shape marked HS. (Shaded bars) Coding regions of gpd-2; (hatched bars) coding regions of gpd-3; (solid bar) coding region of vit-6; (open bars) noncoding regions of exons; (thin lines) introns; (thicker lines) intercistronic sequences. The Ur element is indicated by a narrow bar. The dark line below the operon diagram indicates the region covered by the RNase protection probe. Antisense oligonucleotides a, b, and c are indicated above the positions to which they can anneal. (B) RNase protection assay of total RNA from the transgenic worm strain BL4057, carrying the HS1496 construct. Worms were grown at 20°C and heat shocked at 29°C or 33°C for 2 h. RNA was isolated from 20°C controls as well as the two heat-shocked populations for RPA. The sizes and identifications of the protected fragments were calculated and indicated. Low levels of products seen in lane 1 represent endogenous gpd-2 and gpd-3 transcripts. (C) Primer extension analysis of RNA preparations analyzed in B. Ur-RNA and trans-spliced products are indicated. In the gel at right, location of the 5′ end of Ur-RNA is determined. The gpd-2/gpd-3 intercistronic region was sequenced using the same oligonucleotide used for primer extension and compared with the primer extension product as in the gel shown on the left (lane 3). Part of the intercistronic region sequence is depicted on the right with the Ur element indicated by a vertical line.
To find the optimal heat shock condition, we tried several temperatures. Whereas heat shock is known to inhibit splicing in other organisms, the fact that heat shock genes in C. elegans contain introns suggests that splicing in this organism is relatively resistant to heat shock. Nonetheless, the data in Figure 1B ▶ (lane 3) make it clear that a higher temperature heat shock can inhibit trans-splicing. At 33°C, precursor RNA accumulates, suggesting processing of the polycistronic precursor is incomplete. In addition, a novel fragment slightly bigger than the gpd-2 3′ end fragment is revealed. We hypothesized that this new fragment could be a previously undetected intermediate that is normally processed further to produce gpd-3 mRNA. We suspect it is always formed, but its existence is revealed only by inhibiting trans-splicing by high-temperature heat shock.
To analyze this product further, we performed primer extension using an oligonucleotide capable of detecting both trans-spliced as well as unspliced gpd-3 RNA. Figure 1C ▶ (lane 3) demonstrates that at 33°C, a novel product accumulates significantly above background. It spans the 5′ end of gpd-3 and stops ∼70 nucleotides upstream in the intercistronic region. The lower, fainter band presumably represents a premature reverse transcriptase stop, as it is not routinely observed.
To locate the 5′ end of the novel product precisely, we electrophoresed the primer extension products with the products of sequencing the intercistronic region using the same oligonucleotide (Fig. 1C ▶). The primer extension stop occurs 7 nucleotides upstream of the Ur element, suggesting it has a 5′ end beginning just upstream of the Ur element, so we refer to it as Ur-RNA. Both RNase protection (panel B) and primer extension (panel C) indicate that the 5′ end of this RNA is 75 bp upstream of the gpd-3 trans-splice site.
Ur-RNA is monocistronic and uncapped
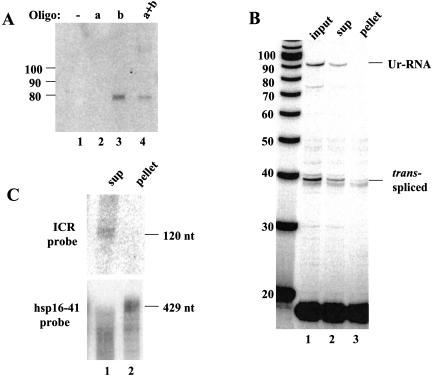
The primer extension stop 7 nucleotides upstream of the Ur element could represent a true RNA 5′ end or it could result from RNA secondary structure or modification. For example, it could be caused by branchpoint formation at this location or a modification of this base. If the stop was attributable to RNA modification or secondary structure, the RNA from which it is reverse transcribed might be polycistronic. The consistency of the RNase protection data of Figure 1 ▶, however, with the primer extension stop, indicates that the 5′ end of the primer extension product represents a true 5′ end. To obtain additional direct evidence, we performed a Northern blot preceded by RNase H treatment. Two oligonucleotides upstream and downstream of the intercistronic region (designated as “a” and “b” in Fig. 1A ▶) were individually or simultaneously annealed to RNA from worms expressing Ur-RNA. RNase H digestion of the RNA/DNA hybrids was followed by Northern blot analysis with a hybridization probe specific for the intercistronic region (ICR probe). If Ur-RNA were monocistronic, oligo b alone would result in a 74-nt product. In contrast, if Ur-RNA were polycistronic, both oligos would be expected to be necessary for formation of a short RNA, and it should be 105 nucleotides long. As shown in Figure 2A ▶, a 74-nt band was seen whenever oligo b was present, indicating that the primer extension stop at 7 nucleotides upstream of the Ur element is a true 5′ end and the Ur-RNA is therefore monocistronic.
FIGURE 2.
Ur-RNA is monocistronic and uncapped. (A) Northern analysis of RNase H digested Ur-RNA. BL4057 RNA (33°C) was annealed with oligonucleotides a, b, or both (Fig. 1A ▶), treated with RNase H, separated on an 8% denaturing polyacrylamide gel and blotted. The blot was probed with a random-primed probe against the intercistronic region (ICR probe). The band just below the 80-nucleotide marker is the size predicted for an RNA beginning at the 5′ end of Ur-RNA and ending at the b oligonucleotide. (B) Fractionation of capped and uncapped RNA on eIF4E resin. BL4057 RNA (33°C) was annealed with oligonucloetide c, treated with RNase H, and affinity purified with GST–eIF4E fusion protein. RNAs from both supernatant and pellet were analyzed by primer extension. (C) Northern blot analysis of the RNase H-digested, eIF4E-fractionated RNA, probed with the ICR probe and with hsp-16-41 probe as control.
Monocistronic Ur-RNA could originate in at least two ways. First, a free 5′ end would be exposed after cleavage and polyadenylation of gpd-2. Such a 5′ end would be expected to be recognized by a nuclear 5′–3′ exonuclease, such as the worm homolog of the yeast RAT1 nuclear exonuclease (Johnson 2001). It would degrade the RNA until it stopped near the Ur element, presumably blocked in its action by a bound protein. If this hypothesis were correct, the Ur-RNA should not have a 5′ cap. A second possibility would be de novo initiation by RNA polymerase. Perhaps after 3′ end formation of gpd-2, RNA polymerase II could remain bound to the DNA and reinitiate transcription just upstream of the Ur element. If this were the case, Ur-RNA would be expected to have an m7G cap. (The possibility that re-initiation in a polycistronic transcription unit might result in an uncapped RNA cannot be eliminated, but production of uncapped RNA by RNA polymerase II has never been observed.) To distinguish between these two possibilities, we took an affinity purification approach using eIF4E, the translational initiation factor that binds the mRNA m7G cap (Rhoads et al. 1993). First the RNA from worms expressing Ur-RNA was shortened by incubation with RNase H in the presence of the “c” oligonucleotide to facilitate binding of capped species to the resin. eIF4E fused with GST (McCracken et al. 1997) was incubated with the cleaved RNA, and the bound (capped) and unbound (uncapped) products were analyzed by primer extension and Northern analysis. As shown in Figure 2B ▶, trans-spliced products are present in both the supernatant and the pellet. The poor precipitation of the trans-spliced product is likely attributable to the fact that it contains a trimethylguanosine cap (Liou et al. 1990) that binds less efficiently to mammalian eIF4E (Darzynkiewicz et al. 1988). In contrast, Ur-RNA is not detected at all in the pellet, suggesting it does not contain a 5′ cap.
This conclusion is further supported by the fact that Ur-RNA was detected exclusively in the supernatant by Northern analysis of this RNA (Fig. 2C ▶). As a positive control, we probed the blot for hsp-16-41 mRNA, which should bind efficiently to the resin. As expected, full-length hsp mRNA was found in the pellet, whereas smaller species are detected as smears in both the supernatant and pellet, as would be expected for partially degraded mRNAs that lack 5′ caps (Fig. 2C ▶). We conclude that Ur-RNA has an uncapped 5′ end 75 nucleotides upstream of the gpd-3 trans-splice site.
Ur-RNA formation is dependent on an intact 3′ end formation signal and the Ur element
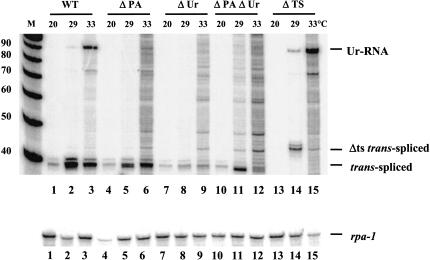
We have reported previously the effects of mutating the poly(A) signal, the Ur element, and the trans-splice site on pre-mRNA processing. To test what effects these mutations had on Ur-RNA accumulation, we subjected transgenic strains carrying the wild-type HS1496 construct along with those containing the mutant constructs to a 2-h 33°C heat shock. RNA was isolated from worms from each strain and used for primer extension. As expected, heat shock was required for induction of the construct, and the Ur-RNA accumulated only when heat shock was performed at elevated temperature, 33°C (Fig. 3 ▶, lanes 1–3).
FIGURE 3.
Both the Ur element and the poly (A) signal are required for Ur-RNA formation. Total RNA from transgenic strains carrying HS1496 (WT) and various mutations of the HS1496 construct were analyzed by primer extension. Ur-RNA and trans-spliced products are indicated. The band above the trans-spliced product that appears in lane 14 is attributable to usage of a cryptic trans-splice site (see text). Similar amounts of RNA were subjected to primer extension in each lane as indicated by the rpa-1 primer extension product (lower panel).
Interestingly, the absence of a functional polyadenylation site prevented formation of Ur-RNA (Fig. 3 ▶, lane 6), suggesting that either 3′ end formation or binding of CPSF to the pre-mRNA is required for Ur-RNA formation. As this mutation still allows SL2 trans-splicing, this result demonstrates that SL2 trans-splicing can occur without formation of Ur-RNA. The Ur element is also required for Ur-RNA formation, with or without the AAUAAA (Fig. 3 ▶, lanes 9,12).
A striking difference was observed when we mutated the trans-splice site. As shown in Figure 3 ▶, lanes 14 and 15, not only was Ur-RNA formed, but its formation did not require elevated temperature. This result strongly supports the idea that formation of Ur-RNA normally requires high temperature because high temperature inhibits trans-splicing, so when trans-splicing is inhibited by mutation, high temperature is no longer required for Ur-RNA formation. We conclude that efficient trans-splicing is not required for Ur-RNA formation. On the contrary, trans-splicing must be inhibited to allow Ur-RNA formation.
We also observe a novel band at a position expected for trans-splicing 4 bp upstream of the normal site of trans-splicing (Fig. 3 ▶, lane 14). We have found that splicing occurs at the sequence TTTTAAT/T, where the slash indicates the exon border (data not shown). Splicing at this site was wholly unexpected. Its occurrence indicates that even in the absence of a good match to the splice site consensus, information exists to cause trans-splicing in the normal region, albeit inefficiently.
The site of the 5′ end of Ur-RNA is dependent on the position of the Ur element
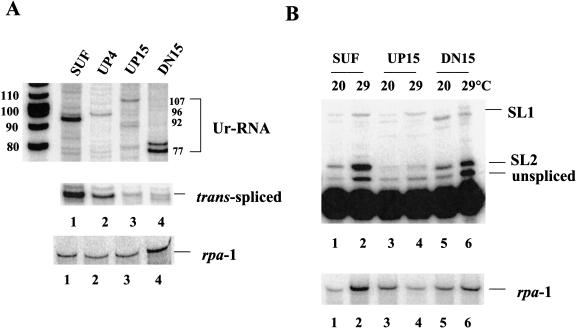
As the 5′ end of Ur-RNA occurs just upstream of the Ur element and the presence of an intact Ur element is required for Ur-RNA formation, we hypothesized that the Ur-element controls the location of the 5′ end of Ur-RNA. To test this idea, we moved the Ur element, testing positions 4 and 15 bp upstream and 15 bp downstream of its normal position, while keeping the length of the intercistronic region unchanged in the context of a synthetic intercistronic region (Huang et al. 2001). In the SUF construct, all of the intercistronic sequences except the Ur element at its normal position were replaced with unrelated polylinker sequence. In this context, accumulation of gpd-3 mRNA was nearly normal, as was the trans-splicing specificity, demonstrating that the Ur element is sufficient to allow SL2 trans-splicing in the absence of other sequences from the gpd-2/gpd-3 intercistronic region (Huang et al. 2001). SUF serves as a positive control in this experiment to show the normal position of trans-spliced product and Ur-RNA, 7 nucleotides upstream of the 5′ end of the Ur element. These RNAs are 40 and 92 nucleotides in length, respectively. It also demonstrates that the Ur element is the only sequence in the intercistronic region required for Ur-RNA formation.
RNA was analyzed by primer extension to locate the 5′ end of the Ur-RNA in each mutant. When Ur was moved 4 or 15 bp upstream, prominent bands are observed 4 or 15 nucleotides longer than the predominant band seen with the SUF construct (Fig. 4 ▶, cf. lanes 2,3 and lane 1). Although other bands are also evident, we believe that the most prominent band in each case represents Ur-RNA at the new, predicted, position. Similarly, when Ur was moved 15 bp downstream, the most prominent band is now 15 nucleotides shorter than the band seen in lane 1 (lane 4 versus lane 1). Although there is another dark band just above it, it seems likely that the 77-nucleotide band represents the Ur-RNA in the DN15 mutant strain. Clearly the location of the Ur element determines the location of the 5′ end of Ur-RNA, as the most prominent band in each case was exactly 7 nucleotides upstream of the new location of the Ur element. We note also that the level of Ur-RNA is influenced by the position of the Ur element. When the Ur element was moved upstream, less Ur-RNA accumulated, perhaps because of competition between exonucleolytic degradation and Ur-binding protein recruitment.
FIGURE 4.
The location of the Ur element determines the location of the 5′ end of Ur-RNA and influences the efficiency of trans-splicing. (A) Transgenic worm strains were grown and heat shocked at 33°C for 2 h. RNA was isolated and analyzed by primer extension. Primer extension was performed on RNA isolated from several strains carrying each construct, but data from only a single representative strain is shown. The predicted sizes of Ur-RNAs from the four constructs are indicated on the right. The trans-spliced products are shown in the lower panel. (B) RNA from strains grown at 20°C and heat shocked at 29°C was subjected to primer extension in the presence of ddGTP. Positions expected for unspliced precursor, SL2, and SL1 trans-spliced products (2-, 3-, and 9-nucleotide extensions, respectively) are shown on the right. Rpa-1 primer extension in the lower panel of both A and B serves as a loading control.
The efficiency and specificity of trans-splicing is dependent on the location of the Ur element
Because the Ur element is required for SL2-specific trans-splicing, we expected that changing its location might alter the efficiency of trans-splicing. The data of Figure 4A ▶ indicate that positioning the Ur element at new locations, either closer to the site of 3′ end formation or of trans-splicing, reduced accumulation of trans-spliced product, with new positions 15 bp in either direction having a more severe effect than the new position only 4 bp upstream. Furthermore, moving the Ur element closer to the site of 3′ end formation reduced the specificity of trans-splicing (Fig. 4B ▶). With the SUF construct almost all of the trans-spliced product is trans-spliced to SL2. With the DN15 mutant, still most trans-splicing is to SL2, whereas with the UP15 construct there is very little trans-splicing to SL2. We conclude that changing the location of the Ur element simultaneously changes the site of the 5′ end of Ur-RNA and the efficiency of trans-splicing downstream. Furthermore, although moving the Ur element downstream still allows SL2-specific trans-splicing, moving the Ur element closer to the 3′ end of the upstream gene appears to severely inhibit SL2 trans-splicing.
MS2 coat protein bound to the intercistronic region can allow trans-splicing
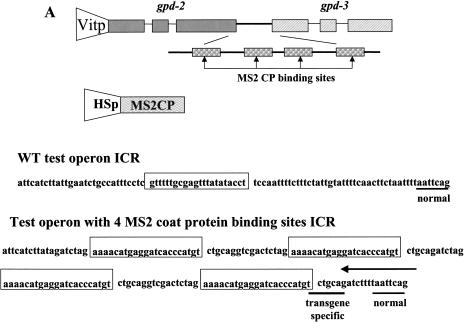
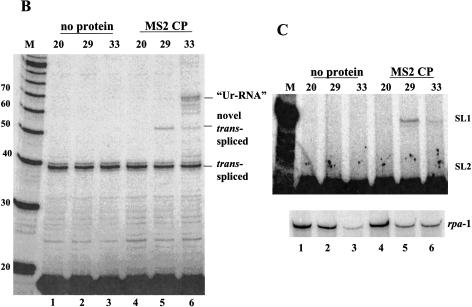
The model predicts that the proteins bound to the Ur element will have two functions, protection against exonucleolytic degradation, thereby allowing time for trans-splicing to occur, and recruitment of the SL2 snRNP. These proposed functions might be separable; that is, it might be possible for a heterologous protein to protect against exonucleolytic digestion, but without attracting the SL2 snRNP. If that were accomplished, we might expect trans-splicing, but using the default snRNP, presumably SL1. We replaced the Ur element between gpd-2 and gpd-3 by four MS2 coat protein-binding sites (Witherell et al. 1991), with the construct under the control of the vit-2 promoter, which expresses exclusively in adult hermaphrodite intestines (Fig. 5A ▶). Transgenic strains were obtained containing this construct along with a second construct expressing the coat protein under control of the hsp-16-41 promoter, which expresses in the intestine as well as elsewhere, following heat shock. The transgenic worms were heat shocked at 29°C or 33°C for 2 h, and RNA was extracted and analyzed by primer extension. Transgenic worms carrying the test operon but not the coat protein construct showed no bands other than the one expected from the endogenous gpd-3 gene (Fig. 5B ▶, lanes 1–3). In contrast, when the coat protein construct was included (lanes 4–6), heat shock at 33°C resulted in expression of a novel band of the size expected for an RNA beginning just upstream of the 3′-most coat protein-binding site. Bands of the sizes expected for primer extension stops just upstream of the other three coat protein-binding sites are also visible in the original autoradiogram (data not shown), but they are not as prominent as the one indicated in the figure labeled “Ur-RNA.” The fact that none of these bands form with a 29°C heat shock supports the idea that these bands represent the Ur-RNA equivalents associated with coat protein binding. Presumably, they result from blockage of exonucleolytic digestion by the bound coat protein. We don’t know why the band resulting from occupation of the binding site closest to the trans-splice site is most prominent.
FIGURE 5.
MS2 coat protein bound to the ICR promotes SL1 trans-splicing. (A) The MS2 constructs. The test operon construct is composed of the gpd-2/gpd-3 operon under control of the vit-2 promoter, with the gpd-2/gpd-3 intercistronic sequence replaced by an ICR containing four MS2 coat protein-binding sites. Exons, introns, and the ICR are shown to scale. The ICR containing the coat protein-binding sites (to scale) is enlarged for clarity. The open reading frame of MS2 coat protein is under control of the hsp-16-41 promoter. The ICR sequence is shown for both the gpd-2/gpd-3 operon and the MS2 construct, with the Ur element and the MS2 coat protein binding sites boxed. The normal and transgene-specific trans-splice sites are underscored. The arrow above the sequence indicates the 3′ end of the primer that targets only the transgene. (B) Primer extension with the primer that spans the 5′ end of gpd-3. “Ur-RNA” and trans-spliced products are indicated. (Lanes 1–3) No coat protein transgene present; (lanes 4–6) with coat protein transgene. (Lanes 1,4) not heat shocked; (lanes 2,3,5,6) heat shocked at the temperatures indicated above the lanes. (M) Markers. (C) Primer extension with the transgene-specific primer in the presence of ddGTP. Positions expected for SL1 and SL2 trans-spliced products are shown on the right. Only SL1 trans-spliced products are observed. Lane designations are as in B. The lower panel shows rpa-1 primer extension as a loading control.
Interestingly, we observed another novel band dependent on heat shock, but this one was more abundant at 29°C than at 33°C, as would be expected for a trans-spliced product. This band might result from trans-splicing at a novel site (TCTGCAG/) 14 nucleotides upstream of the normal one introduced as part of the coat protein-binding site construct. Although this site is present downstream of each of the four binding sites, we observe trans-splicing only to the most 3′ site. To determine whether this truly represents trans-splicing at this site, we designed a transgene-specific primer. Primer extension with the new primer also revealed the novel trans-spliced product, but only when coat protein was induced (data not shown). To determine whether trans-splicing was SL2-specific, as with the wild-type construct, we performed primer extension with the transgene-specific primer in the presence of ddGTP, which terminates primer extension on the predicted SL2 product at the +3 position and the SL1 product at the +9 position. This experiment clearly shows the SL1 product, but provides no evidence for an SL2 trans-spliced product (Fig. 5C ▶). We also performed this experiment with an operon construct containing only two coat protein-binding sites, and essentially identical results were obtained (data not shown).
Taken together, these data indicate that on heat shock, MS2 coat protein can bind to the intercistronic region containing coat protein-binding sites and thereby protect it against degradation following 3′ end formation upstream. This allows trans-splicing to occur. The fact that gpd-3, a normally SL2 trans-spliced RNA, receives only SL1 in such circumstances, demonstrates that SL1 trans-splicing is the default mode, used when no specific signal is present. These results provide strong support for the idea that in polycistronic transcription units, 3′ end cleavage produces a free 5′ end on the downstream RNA that must be protected from exonucleolytic digestion to allow trans-splicing.
DISCUSSION
It is quite unusual for an animal species to have operons. It is now evident, however, that having spliced leader-type trans-splicing has enabled some eukaryotic species to arrange their genes in polycistronic assemblages resembling bacterial operons (Blumenthal et al. 1996). In the case of C. elegans, at least 15% of all genes are transcribed polycistronically (Blumenthal et al. 2002) and the resulting multigene pre-mRNAs are processed to monocistronic mRNAs by 3′ end cleavage accompanied by trans-splicing (Kuersten et al. 1997). In this nematode species and its close relatives, there is a specialized snRNP, the SL2 snRNP for trans-splicing at positions between genes in operons (Nilsen 1995). The precise sequence of events that occur within the short intercistronic regions in operon pre-mRNAs, however, has yet to be elucidated. The results reported in this paper suggest a model by which the monocistronic mRNAs are created.
Function of the Ur element in preventing pre-mRNA degradation
Our results demonstrate the presence of a novel RNA that we call Ur-RNA that accumulates only when trans-splicing is inhibited, either by mutation or high temperature heat shock. Ur-RNA has an uncapped 5′ end that maps to a site 7 nucleotides 5′ of the Ur element, which has been shown previously to be necessary and sufficient for SL2 trans-splicing of operon pre-mRNAs (Huang et al. 2001). By relocating the Ur element within the intercistronic region, we demonstrated that the Ur element controls the location of the 5′ end of Ur-RNA. This result, along with the fact that deletion of the Ur element eliminates Ur-RNA accumulation, implicates the Ur-RNA as a participant in SL2-specific trans-splicing. The fact that the 3′ end formation signal is required for Ur-RNA formation suggests how it may be formed, namely by degradation of a free end created by the 3′ cleavage event. The existence of the un-trans-spliced Ur-RNA indicates that 3′ end formation can precede trans-splicing, and may routinely do so. The simplest model suggests that as the RNA polymerase passes the 3′ end formation site, cleavage occurs, releasing the upstream mRNA and leaving a free 5′ end on the RNA still being transcribed. That RNA is degraded exonucleolytically as the RNA polymerase proceeds down the DNA until the exonuclease encounters a block, perhaps a protein bound to the Ur element.
In this model, the Ur element is required for SL2 trans-splicing downstream because it is necessary to block the progress of the exonuclease long enough to allow trans-splicing. We show here that even a heterologous protein, the MS2 coat protein, can perform this function of the Ur element-binding protein. The Ur-RNA may be an accidental consequence of this sequence of events, rather than a necessary intermediate in the SL2 trans-splicing process. Ur-RNA should only be formed when 3′ end cleavage exposes the free end to exonucleolytic degradation and when a protein is bound to the Ur element to block the exonuclease’s progress. When 3′ end formation is blocked by mutation, SL2 trans-splicing can occur even though no Ur-RNA is formed. Therefore, Ur-RNA may normally be an intermediate in SL2 trans-splicing, but it is clearly not an obligatory one.
What protein acts at the Ur element?
We hypothesize that CstF could be the key protein that acts at the Ur element. The Ur element has the location and sequence expected for a CstF-binding site (Takagaki et al. 1990). Presumably CstF would be present, as it would have just functioned in 3′ end formation. The role of the AAUAAA in facilitating SL2 trans-splicing may be simply to facilitate CstF binding because mutation of the AAUAAA only partially interferes with SL2 trans-splicing. CPSF, the protein that recognizes AAUAAA, and CstF are known to interact cooperatively at 3′ end signals (Gilmartin and Nevins 1991; Murthy and Manley 1992; Wilusz et al. 1990). Furthermore, the SL2 snRNP is known to be present in a complex with CstF-64, and this complex has been shown to be important for SL2 trans-splicing (Evans et al. 2001). Nonetheless, we have not been able to demonstrate binding of CstF to the Ur element (Y. Liu and T. Blumenthal, unpubl.).
Function of the Ur element in recruiting the SL2 snRNP
The Ur element is obligatory for SL2 trans-splicing, as in a double knockout of the 3′ end formation site and the Ur element, only SL1 trans-splicing occurs (Huang et al. 2001). Similarly, when we allow trans-splicing to occur by blocking exonucleolytic digestion with MS2 coat protein in place of the normal Ur element-binding protein, trans-splicing is with SL1, not SL2. Therefore, we conclude that SL1 trans-splicing is the default mode. It occurs at outrons and in cases where the Ur element-binding protein is not present to recruit the SL2 snRNP. The fact that the location of the Ur element seems to be critical for SL2 trans-splicing suggests that attracting the SL2 snRNP may only be part of what is required. The Ur element may serve to precisely coordinate the processes of 3′ end formation and trans-splicing to maximize expression of the downstream gene. The mechanism of SL2 trans-splicing must involve preventing transcription termination, which would normally accompany 3′ end formation (Proudfoot 1989). Perhaps the Ur element serves to prevent transcription termination and must be precisely positioned to do so effectively, as moving the Ur element a short distance up or downstream interfered with SL2 trans-splicing. Whether it serves primarily to prevent transcription termination or exonucleolytic degradation of the pre-mRNA, it seems clear that the Ur element acts to allow trans-splicing to occur before degradation can destroy the pre-mRNA or termination can prevent its synthesis. Recruitment of the SL2 snRNP is a separate function, presumably one that makes splicing at trans-splice sites between genes in polycistronic pre-mRNAs more efficient.
MATERIALS AND METHODS
Worm strains and RNA preparation
Maintenance and growth of worms was as described by Brenner (1974) and by Sulston and Hodgkin (1988). Transgenic worm strains carrying extrachromosomal arrays were generated as described previously (Mello et al. 1991). In all cases, pRF5 was co-injected as marker plasmid. Worms were heat shocked at 29°C or 33°C for 2 h on floating petri plates spread with bacteria. Total RNA was prepared from heat-shocked or nonheat-shocked mixed-stage populations of transgenic worms (Conrad et al. 1991). The strain numbers are as follows: WT (BL4057), ΔPA (BL5422); ΔUr (BL5384); ΔPA/ ΔUr (BL5400); ΔTS (BL4095); SUF (BL8041); UP4 (BL8051); UP15 (BL8067); DN15 (BL8070); 4 MS2CPBS test operon (BL8086); and 4 MS2CPBS test operon + MS2CP (BL8089). BL4057, BL5384, and BL4095 are integrated strains.
For MS2 experiments, the test operon alone or in combination with MS2 coat protein-expressing construct was injected into wild-type worms (N2) to make transgenic strains.
Plasmid construction
The plasmids containing the wild-type operon, HS1496 (diagrammed in Fig. 1A ▶) and mutants of the operon (ΔPA, ΔPA/ΔUr, ΔUr, ΔTS, and SUF) have been described previously (Spieth et al. 1993; Kuersten et al. 1997; Huang et al. 2001). The plasmids UP4, UP15, and DN15 were created by modifying the SUF construct. UP4 was created by cutting the SUF plasmid with BglII, filling in the resulting overhang to form blunt ends, and then religating. UP15 was constructed as follows: the portion of the SUF Ur region was replaced by a PCR product generated using two oligonucleotides, Ur-UP15-US (5′-CGTTAATTAAGATGTTTTTGCGAGTT TATATAC-3′) and Ur-UP15-DS (5′-TATTAATTAAGCTAGCTA TTTAAATGTTTAAACAAGAGAAAGATC-3′) that together reformed the intercistronic region of SUF with the Ur 15 bp upstream, flanked by PacI sites. The PCR fragment was digested with PacI and cloned into the SUF construct from which the PacI fragment had been deleted. The plasmid DN15 was derived from SUFR (Huang et al. 2001) as follows. The ICR between gpd-2 and gpd-3 of SUFR was amplified by PCR using two oligonucleotides that flank the ICR, PAKO-G (5′-TGGTCTCTTCTCAAGAAAG GTTGTATATT-3′) and gpd-3 5′ (5′-GCGTTGAAGCAGTTTCC C-3′). The PCR product of this SUFR ICR was digested with PacI and cloned into the SUF construct from which the PacI fragment had been deleted, and a reverse orientation insert was selected. To create the operon containing the MS2 coat protein-binding sites, a StuI–AgeI fragment was amplified from HS1496 and cloned into pGEM-T-easy vector using two oligonucleotides ICR-UP (5′-CAC CAAGGCCTAAGCTGTCTCAT-3′) and ICR-DN (5′-TTCTACC GGTACCTTACGCTTCT-3′). Then reverse PCR (Coolidge and Patton 1995) was performed to delete the Ur element and introduce a BglII site with MS2-U (5′-TCTATAAGA TGAATAAATA TAC-3′) and MS2-D (5′-TCTTTTAATTCAGGGAAACTGCT-3′) under the following conditions: 96° 2 min, 30 cycles of 96° 1 min, 38° 1 min, and 72° 4.5 min; and an extra extension time of 20 min at 72°. The plasmids containing MS2 coat protein-binding sites pMM2-1 and pET-MS2 were kindly provided by Dr. Jeff Coller (University of Wisconsin, Madison). The MS2 coat protein binding site fragment was amplified by PCR using MS2BglII-up: (5′-GCGCGAAGATCTAGAAAACATGAGG-3′) and MS2PstI–BglII dw: (5′-CGGAAGATCTGCAGACATGGGTGATCC) so that BglII sites were introduced at each end. The MS2 coat protein-binding sites fragment was released by BglII digestion and cloned into pGEM-T-easy-ICR-ΔUr also cut with BglII. Finally, the ICR fragment containing the MS2 coat protein-binding sites was cut with StuI and AgeI, and cloned into pAL1.8, a plasmid with the gpd-2/gpd-3 operon under control of the vit promoter. To create the plasmid expressing MS2 coat protein under HS control, the MS2 coat protein open reading frame was amplified from pET-MS2 using two oligonucleotides: KpnI-MS2-5′ (5′-GCGGGGTACCC CATGGG CAGCAGCCATCATCAT-3′) and MS2-EcoRV-3′ (5′-CCGGATATCGCCCCAAGGGGTTATGCT-3′) so that a KpnI and an EcoRV site was introduced at each end. The PCR product was cloned into pGEM-T-easy vector, then released by KpnI and EcoRV double digestion and cloned into pPD49.83 (kindly provided by A. Fire) in which the KpnI–EcoRV fragment had been deleted.
RNA analysis
RNase protection and primer extension assays were carried out as described previously (Kuersten et al. 1997). All primer extension experiments were performed with the gpd-3 5′ oligonucleotide unless indicated otherwise. In the MS2 experiment, a transgene-specific primer “trans-site-new” was used: 5′-GTTTCCCTGAATT AAAAGATC-3′. For all the loading controls, rpa-1 5′ was used: 5′-TGGCGGCCTTGAGAAGGG-3′. Northern blots were performed according to standard procedures (Conrad et al. 1993). Before electrophoresis, the RNA was cleaved by RNase H at sites indicated in Figure 1A ▶. Thirty μg BL4057 RNA heat shocked at 33°C was mixed with oligonucleotide gpd-YL (designated as “a” in Figure 1A ▶) (5′-CCTTTATTGAGAAGAGACCAT-3′) or gpd-3 5′ (designated as “b”) or both, heated at 72°C for 5 min, slow cooled to room temperature for 30 min. Two units RNase H was added and incubated at 37°C for 30 min. TE was added to 100 μL, the mixture was phenol:chloroform extracted and RNA precipitated. Ten μg of untreated or RNase H-treated RNA samples were electrophoresed on an 8% denaturing polyacrylamide gel, and electroblotted to Hybond-N membrane (Amersham). The intercistronic region probe (ICR probe) was made by random priming from a purified PCR product from plasmid pΔHSGPD2/3VIT6 as the template, P24 US (5′-TTCCTCGTTTTTGCGAGT-3′) and P24 DS (5′-CTGAATTAAAATTAGAAGTTG-3′) as primers.
eIF4E pull down assay
GST-eIF4E was kindly provided by Dr. D. Zorio and Dr. D. Bentley. The protein was prepared according to the method of Edery et al. (1995). Binding of capped mRNA was performed according to the method of McCracken et al. (1997) with the following modifications. BL4057 worms were heat shocked at 33°C and RNA isolated as above. Twenty μg of total RNA was mixed with oligonucleotide gpd-16 (5′-GAATCCGTTGATTCCGAC-3′) (designated as “c” in Fig. 1A ▶), heated at 72°C for 5 min, and slow cooled to room temperature for 30 min. Two units RNase H was added and incubated at 37°C for 30 min. TE buffer was then added to 100 μL, the mixture was phenol:chloroform extracted and the RNA precipitated. Ten μg RNase H treated RNA was mixed with 100 μL binding buffer [3 mL buffer D (0.1 M NaCl, 0.1% NP-40, 20 mM HEPES pH 7.9, 0.1 mM EDTA, 1 mM DTT, 20% glycerol), 390 μL 10% PVA, 150 μL 5 mg/mL Poly U, 60 μL 1 mg/mL (dG:dC): (dG:dC)], heated at 95°C for 2 min, and chilled on ice for 2 min. Thirty μg GST–eIF4E, together with 0.6 μL 1M DTT and 1 μL RNasin were added and incubated at 4°C for 1 h, the mixture was added to 50 μL packed glutathione sepharose beads that had been washed in binding buffer. After 1 h of mixing at 4°C, the beads were collected by centrifugation. The supernatant was pooled with the first wash in 200 μL of buffer D. The beads were then washed one more time with 1 mL buffer D, and the capped RNA was eluted with 500 μL 0.3 M NaAc and 1% SDS. Glycogen was added to all samples and carrier Torula RNA was added to the pellet fraction to maintain the same nucleic acid concentration as the supernatant. Samples were extracted with phenol/chloroform and precipitated. One-third of the total RNA was used for primer extension and the rest for Northern analysis. The Northern blot was first probed with random-primed ICR probe, stripped, and then probed with hsp-16-41 random-primed probe. The hsp-16-41 probe template was synthesized by PCR using BL4057 strain cDNAs as template, hsp-16-41 US (5′-CGGTTCAACTCGAT GTCTCTC-3′) and hsp-16-41 DS (5′-GTTTGTCTTCTTTG GAGCCTC-3′) as primers, and gel purified.
Acknowledgments
We thank Diego Zorio and David Bentley for providing the GST–eIF4E fusion protein and for their helpful suggestions and discussions. This research was supported by Research Grant GM42432 from the National Institute of General Medical Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2128903.
REFERENCES
- Blumenthal, T., Evans, D., Link, C.D., Guffanti, A., Lawson, D., Thierry-Mieg, J., Thierry-Mieg, D., Chiu, W.L., Duke, K., Kiraly, M., et al. 2002. A global analysis of Caenorhabditis elegans operons. Nature 417: 851–854. [DOI] [PubMed] [Google Scholar]
- Blumenthal, T. and Spieth, J. 1996. Gene structure and organization in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 6: 692–698. [DOI] [PubMed] [Google Scholar]
- Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, R., Thomas, J., Spieth, J., and Blumenthal, T. 1991. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol. Cell. Biol. 11: 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, R., Liou, R.F., and Blumenthal, T. 1993. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. EMBO J. 12: 1249–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolidge, C.J. and Patton, J.G. 1995. Run-around PCR: A novel way to create duplications using polymerase chain reaction. Biotechniques 18: 762–764. [PubMed] [Google Scholar]
- Darzynkiewicz, E., Stepinski, J., Ekiel, I., Jin, Y., Haber, D., Sijuwade, T., and Tahara, S.M. 1988. β-globin mRNAs capped with m7G, m2.7(2)G or m2.2.7(3)G differ in intrinsic translation efficiency. Nucleic Acids Res. 16: 8953–8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery, I., Chu, L.L., Sonenberg, N., and Pelletier, J. 1995. An efficient strategy to isolate full-length cDNAs based on an mRNA cap retention procedure (CAPture). Mol. Cell. Biol. 15: 3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, D., Perez, I., MacMorris, M., Leake, D., Wilusz, C.J., and Blumenthal, T. 2001. A complex containing CstF-64 and the SL2 snRNP connects mRNA 3′ end formation and trans-splicing in C. elegans operons. Genes & Dev. 15: 2562–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin, G.M. and Nevins, J.R. 1991. Molecular analyses of two poly(A) site-processing factors that determine the recognition and efficiency of cleavage of the pre-mRNA. Mol. Cell. Biol. 11: 2432–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, T., Kuersten, S., Deshpande, A.M., Spieth, J., MacMorris, M., and Blumenthal, T. 2001. Intercistronic region required for polycistronic pre-mRNA processing in Caenorhabditis elegans. Mol. Cell. Biol. 21: 1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A.W. 2001. Rat1p nuclease. Methods Enzymol. 342: 260–268. [DOI] [PubMed] [Google Scholar]
- Kuersten, S., Lea, K., MacMorris, M., Spieth, J., and Blumenthal, T. 1997. Relationship between 3′ end formation and SL2-specific trans-splicing in polycistronic Caenorhabditis elegans pre-mRNA processing. RNA 3: 269–278. [PMC free article] [PubMed] [Google Scholar]
- Liou, R.F. and Blumenthal, T. 1990. Trans-spliced Caenorhabditis elegans mRNAs retain trimethylguanosine caps. Mol. Cell. Biol. 10: 1764–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Huang, T., MacMorris, M., and Blumenthal, T. 2001. Interplay between AAUAAA and the trans-splice site in processing of a Caenorhabditis elegans operon pre-mRNA. RNA 7: 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken, S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., Hessel, A., Foster, S., Shuman, and Bentley, D.L. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 11: 3306–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C.C., Kramer, J.M., Stinchcomb, D., and Ambros, V. 1991. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy, K.G. and Manley, J.L. 1992. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J. Biol. Chem. 267: 14804–14811. [PubMed] [Google Scholar]
- Nilsen, T.W. 1995. Trans-splicing: An update. Mol. Biochem. Parasitol. 73: 1–6. [DOI] [PubMed] [Google Scholar]
- Proudfoot, N.J. 1989. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem. Sci. 14: 105–110. [DOI] [PubMed] [Google Scholar]
- Rhoads, R.E., Joshi-Barve, S., and Rinker-Schaeffer, C. 1993. Mechanism of action and regulation of protein synthesis initiation factor 4E: Effects on mRNA discrimination, cellular growth rate, and oncogenesis. Prog. Nucleic Acid Res. Mol. Biol. 46: 183–219. [DOI] [PubMed] [Google Scholar]
- Spieth, J., Brooke, G., Kuersten, S., Lea, K., and Blumenthal, T. 1993. Operons in C. elegans: Polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions. Cell 73: 521–532. [DOI] [PubMed] [Google Scholar]
- Sulston, J. and Hodgkin, J. 1988. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [DOI] [PubMed]
- Takagaki, Y., Manley, J.L., MacDonald, C.C., Wilusz, J., and Shenk, T. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes & Dev. 4: 2112–2120. [DOI] [PubMed] [Google Scholar]
- Wilusz, J., Shenk, T., Takagaki, Y., and Manley, J.L. 1990. A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol. Cell. Biol. 10: 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherell, G.W., Gott, J.M., and Uhlenbeck, O.C. 1991. Specific interaction between RNA phage coat proteins and RNA. Prog. Nucleic Acid Res. Mol. Biol. 40: 185–220. [DOI] [PubMed] [Google Scholar]