Abstract
Ribosomal RNAs contain a number of modified nucleotides. The most abundant nucleotide modifications found within rRNAs fall into two types: 2′-O-ribose methylations and pseudouridylations. In eukaryotes, small nucleolar guide RNAs, the snoRNAs that are the RNA components of the snoRNPs, specify the position of these modifications. The 2′-O-ribose methylations and pseudouridylations are guided by the box C/D and box H/ACA snoRNAs, respectively. The role of these modifications in rRNA remains poorly understood as no clear phenotype has yet been assigned to the absence of specific 2′-O-ribose methylations or pseudouridylations. Only very recently, a slight translation defect and perturbation of polysome profiles was reported in yeast for the absence of the Ψ at position 2919 within the LSU rRNA. Here we report the identification and characterization in yeast of a novel intronic H/ACA snoRNA that we called snR191 and that guides pseudouridylation at positions 2258 and 2260 in the LSU rRNA. Most interestingly, these two modified bases are the most conserved pseudouridines from bacteria to human in rRNA. The corresponding human snoRNA is hU19. We show here that, in yeast, the presence of this snoRNA, and hence, most likely, of the conserved pseudouridines it specifies, is not essential for viability but provides a growth advantage to the cell.
Keywords: Stable RNA, ribosome, Saccharomyces cerevisiae, H-ACA snoRNA, pseudouridylation
INTRODUCTION
The biogenesis of rRNA includes the modifications of nucleotides in pre-rRNA. Conversion of uridine to pseudouridine (Ψ) and 2′-O-ribose methylations are the two types of modifications most commonly found in rRNAs. The extent and complexity of modifications differ between species, eukaryotic rRNAs being more extensively modified than their bacterial counterparts (for review, see Decatur and Fournier 2002). Yet, all studied rRNAs are modified to some extent. In eukaryotes, small nucleolar RNAs (snoRNAs) have been shown to play a key role in specifying the positions 2′-O-ribose methylations and Ψ modifications. The 2′-O-ribose methylations are guided by box C/D snoRNAs, whereas pseudouridylations are guided by box H/ACA snoRNAs (for review, see Kiss 2002). Although almost all methylation guide box C/D snoRNAs have been identified in Saccharomyces cerevisiae, in particular by computational approaches (Lowe and Eddy 1999), only about half of the pseudouridylation guides are currently known. Thirty pseudouridines located in the large subunit (LSU) rRNA (Ofengand and Bakin 1997) and 13 located in small subunit (SSU) (Bakin and Ofengand 1995) have been mapped, and guides are known for, respectively, 18 pseudouridylations in LSU and 5 pseudouridylations in SSU. It was noticed that the number of modifications increases with evolution (Ofengand et al. 1995; Ofengand and Bakin 1997) and that modifications are heavily concentrated in conserved, functionally important regions of the ribosome (Decatur and Fournier 2003). These data suggest an important role of rRNA modifications for faithful ribosome function. Yet, the function of these modified nucleotides, and in particular of pseudouridines (Ψ), remains poorly understood. Blocking 2′-O-ribose methylation (Tollervey et al. 1993) or Ψ modification (Zebarjadian et al. 1999) at a global level in rRNA strongly inhibits growth in yeast. In mice, DCK1, a putative pseudouridine synthase associated with H/ACA snoRNPs, is required for proper pseudouridine modification in rRNA. Recent data suggest that dyskeratosis congenita caused by mutations within DCK1 results from impairment in ribosome maturation and function (Ruggero et al. 2003). Yet, until very recently, the only phenotypes associated with the absence of factors involved in particular Ψ modifications were shown to result from the lack of additional functions associated with these factors and distinct from the Ψ modifications themselves. For example, in Escherichia coli, RluD is the pseudouridine synthase responsible for the pseudouridylation at positions 1911, 1915, and 1917 of the LSU rRNA (Raychaudhuri et al. 1998), the Ψs at positions 1915 and 1917 being among the most highly conserved Ψs and position 1917 being, thus far, the only totally conserved Ψ in cytoplasmic ribosomes (Ofengand 2002). Deletion of RluD confers a slow growth phenotype to E. coli cells but, unfortunately, it was later shown that RluD has another important, yet uncharacterized, function precluding the importance of modifications at these positions to be tested separately (Gutgsell et al. 2001). In yeast, snR10 is responsible for Ψ modification at position 2919 of the LSU rRNA, but snR10 also plays an additional role during ribosomal maturation (Tollervey 1987). Very recently, during the course of the revision of the present article, a work was published describing the uncoupling of the two functions played by snR10. Introduction of a point mutation within the snR10 guide sequence precludes the Ψ formation but does not interfere with the additional function. Most interestingly, this mutation did not exhibit a strong growth phenotype, in contrast to the deletion of snR10, yet resulted in slightly altered polysome profiles and in a measurable [35S]methionine incorporation defect in vivo (King et al. 2003). This is the first phenotype specifically attributed to the lack of a given Ψ.
In humans, the snoRNA specifying the modification of the two most highly conserved Ψs has been identified as hU19 (Bortolin and Kiss 1998), but the phenotype associated with the lack of this snoRNA has not been tested. In other model organisms, such as yeast, the role of these two highly conserved Ψs has not been studied because the snoRNA specifically required for Ψ modification at these positions was not known. In this study, we identified this snoRNA in S. cerevisiae. We propose to call it snR191. It is encoded within the intron of NOG2/YNR053C, a gene encoding a protein involved in ribosome maturation (Saveanu et al. 2001). SnR191 shares highly conserved structures with small RNAs potentially encoded within the intron of the NOG2 homologs in other yeast species. These highly conserved structures are very reminiscent of the hU19 structures. Finally, we show that, although snR191 is not essential for cell viability, it confers a small growth advantage to the cells.
RESULTS AND DISCUSSION
The NOG2 intron encodes a new H/ACA snoRNA
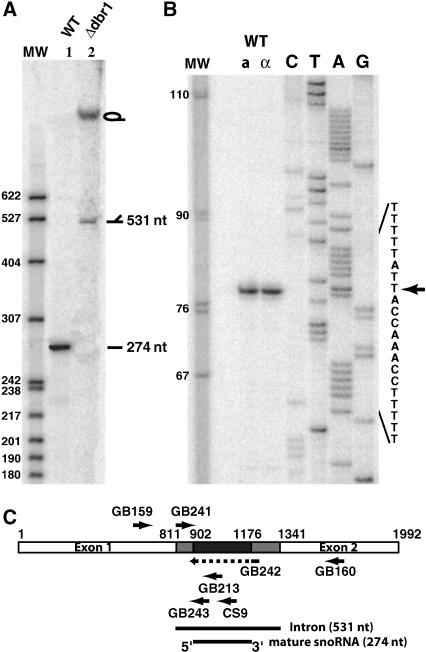
NOG2 encodes an intron that is somewhat atypical for yeast. In contrast to the majority of introns found in S. cerevisiae genes that are short (<250 nt) and located close to the 5′ end, the intron of NOG2 is unusually long (531 nt) and is located quite far (811 nt) from the ATG initiator codon. Northern blot analysis with a probe specific for this intron reveals an abundant small RNA of 274 (±2) nt (Fig. 1A, lane 1). All of the previously known introns encoded small RNAs require the debranching enzyme Dbr1 for their maturation from the lariat excised intron (Ooi et al. 1998). In a strain in which the DBR1 gene has been deleted, the ~274-nt-long RNA is no longer visible and, in contrast, large RNA species, consistent with the lariat and the broken form of the excised intron, are now visible (Fig. 1A, lane 2). We conclude that this small RNA also requires the Dbr1 factor for its maturation from the excised intron and thus behaves, in this matter, as other known intron encoded small RNAs.
FIGURE 1.
Characterization of snR191. (A) Northern blot analysis of snR191. Total RNAs (10 μg) from wild-type strain (lane 1) or Δdbr1 strain (lane 2), separated on a 5% acrylamide-urea gel, were hybridized, after transfer to a nylon membrane, with [32P] 5′ end-labeled oligonucleotide CS9 (see Materials and Methods). The marker is pBR322 DNA digested with MspI and [32P] 5′ end-labeled. (B) The 5′ end of snR191 was mapped by reverse transcription using [32P] 5′ end-labeled oligonucleotide GB213 as primer and total RNAs extracted from Mat a or Mat α (as indicated) wild-type BMA64 strains as template. (Lanes C,T,A,G) Dideoxy sequencing reactions using the same oligonucleotide primer as for reverse transcription and, as template, DNA amplified by PCR using primers MFR207 and MFR208 (see Materials and Methods). The reverse transcription and sequencing reactions were performed as previously described (Saveanu et al. 2001). An arrow indicates the 5′ start position. (C) Schematic representation of the NOG2 gene indicating the relative positions of the different oligonucleotides used for RNA analyses (arrows). The GB242 oligonucleotide overlaps the snoRNA deletion represented by the dotted line within the corresponding arrow. (White boxes) exons; (gray and black boxes) intron sequences. The black box represents the small RNA sequence.
The 5′ end of this small RNA was defined by primer extension analysis (Fig. 1B). The mature small RNA begins at nucleotide 91 of the intron (+902 nt from the ATG). Because we estimate that this RNA to be 274 (±2) nt long, we conclude that it terminates at position 365 (±2) of the intron (+1176 nt from the ATG). A schematic representation of the NOG2 locus, with the position of sequence encoding the small RNA, is shown in Figure 1C.
To test whether this small RNA could belong to one of the C/D box or the H/ACA box types of snoRNAs, we performed a coimmunoprecipitation with protein A-tagged version of proteins known to be specifically associated with members of each snoRNA families: Gar1 and Nop1 for H/ACA and box C/D snoRNAs, respectively (Ganot et al. 1997b). We found the small 274-nt-long RNA to be associated with Gar1 but not Nop1 (data not shown). This suggests that it is a H/ACA snoRNA.
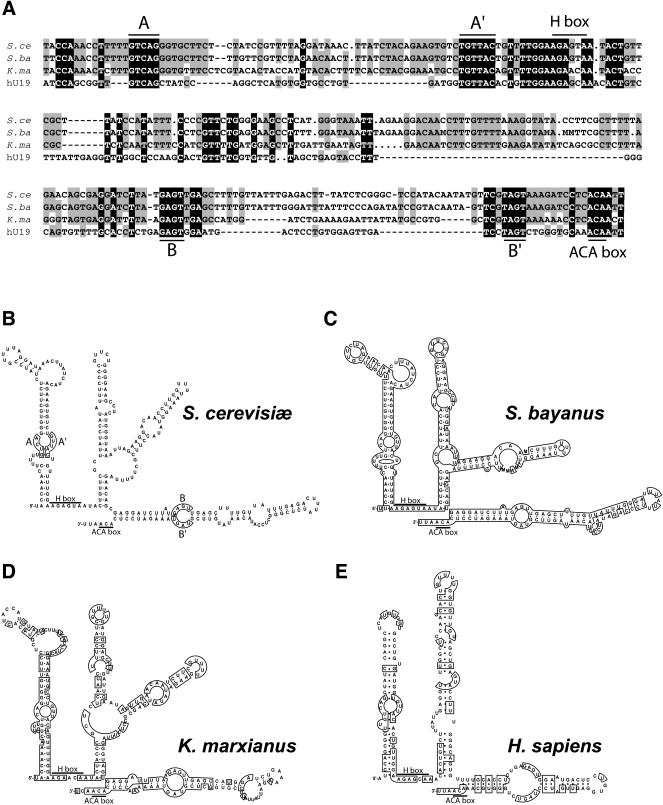
Box H/ACA snoRNAs share common secondary structures together with very small conserved sequences (Balakin et al. 1996). To apply comparative sequence analyses (for review, see Eddy 2002) to predict the secondary structure of the 274-nt-long small RNA, we looked for other NOG2 intron sequences within the partial genomic sequences of other hemiascomycetous yeasts (Souciet et al. 2000). We found complete intron sequences of NOG2 homologs for Saccharomyces bayanus and Kluyveromyces marxianus. Figure 2A shows an alignment of the region corresponding to the 274-nt-long RNA. Using the Mfold program (Walter et al. 1994), we found a potential RNA secondary structure conserved in all three yeast sequences (Fig. 2B–D). As expected, these structures possess the features characteristic of canonical H/ACA snoRNAs, with the H and ACA boxes following long stem–loop structures.
FIGURE 2.
Structure of snR191. (A) Alignment of the Saccharomyces cerevisiae snR191 sequence with that of other yeast species and human hU19. (S. ce) S. cerevisiae; (S. ba) Saccharomyces bayanus (EMBL accession number AL398402); (K. ma), Kluyveromyces marxianus (EMBL accession number AL422984); (hU19) human hU19 (Kiss et al. 1996). Note that the S. bayanus and K. marxianus sequences are issued from the Génolevure project (Souciet et al. 2000) and thus correspond to single reads that are prone to contain some errors or ambiguities (e.g., M stands for A or C). The alignments were performed with the program clustalW (Thompson et al. 1994). Nucleotides conserved in all four sequences are shaded in black, nucleotides conserved in three species are shaded in gray. H and ACA boxes are indicated. Guiding sequences for pseudouridylations are labeled AA′ and BB′. (B, C, and D) Secondary structure of yeast snR191s. Among all snR191 potential secondary structures predicted using the MFOLD program (Walter et al. 1994), the ones that are conserved between S. cerevisiae (B), S. bayanus (C), and K. marxianus (D) are shown. (E) Secondary structure of human hU19 RNA as determined in Kiss et al. (1996). The potential guide sequences are boxed and labeled AA′ and BB′ in the S. cerevisiae structure in B. In C and D, and E, boxes represent conserved nucleotides with regard to S. cerevisiae. The H and ACA motifs are indicated. Note that the 3′ end of snR191 has been experimentally determined with a precision of ±2 nt from the determined 5′ end and the measured length of the RNA (274 ± 2 nt). The actual position of the 3′ end of snR191 is thus known at ±2 nt. Nevertheless, using a length of 274 nt, as presented on the figure, leads to predict a 3′ end 3 nt downstream of the ACA box, precisely as in the consensus H/ACA snoRNA structure (Ganot et al. 1997b).
snR191 specifies the two most conserved pseudouridines of LSU rRNA
According to the canonical H/ACA snoRNA structure, if this new small RNA indeed specifies the position of pseudouridines, the position of each of these modifications should be guided by two short recognition loops complementary to short target RNA sequences overlapping the uridines to be pseudouridylated. These pseudouridines should be located, within the target RNA base-paired sequences, 14–16 nt upstream of the H or ACA boxes (Ganot et al. 1997a,b; Ni et al. 1997). Searching for such complementarities within yeast rRNA sequences reveals only two potential uridine targets within these RNAs, at positions 2258 and 2260 of the LSU rRNA for the box H and ACA, respectively (positions according to the Saccharomyces Genome Database; Cherry et al. 2002). These two positions are close to each other and were previously known to be pseudouridylated (Ofengand and Bakin 1997). However, the snoRNAs specifying these modifications were not yet known in yeast. These results strongly suggest that snR191 is indeed the snoRNA specifying these pseudouridylations. Most interesting, these two pseudouridines are the most highly conserved pseudouridines in cytoplasmic rRNA: Position 2260 (position 1917 in the E. coli 23S rRNA) is so far totally conserved in cytoplasmic ribosomes, whereas position 2258 (position 1917 in E. coli) is conserved in all organisms examined except for Sulfolobus acidocaldarius (Bakin and Ofengand 1995; Ofengand and Bakin 1997; Ofengand 2002).
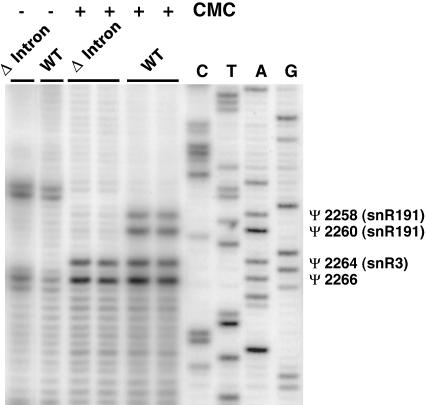
To test directly whether snR191 guides the pseudouridylation at these two positions, we constructed a strain deleted for NOG2 and complemented with a centromeric plasmid carrying the NOG2 gene with (wild-type control) or without its intron (NOG2–Δintron; see Materials and Methods). We verified by reverse transcription that the strain complemented with the wild-type NOG2 gene expressed snR191, whereas the strain complemented with the NOG2–Δintron gene did not (not shown). Total RNAs were prepared from these strains and the presence of pseudouridines at positions 2258 and 2260 of the LSU rRNA was assayed by CMC treatment followed by reverse transcription, as described (Bakin and Ofengand 1993). Figure 3 shows that pseudouridines can be revealed at the expected positions in the strains complemented with the wild-type NOG2 gene but not in the strains complemented with the NOG2–Δintron gene, confirming that snR191 is responsible for pseudouridylations at these two positions. Figure 3 also shows that Ψs at positions 2264 and 2266 of the LSU rRNA, as well as other Ψs (not shown), are not affected by the absence of snR191. This demonstrates that this snoRNA is specific for the modifications at positions 2258 and 2260, as expected.
FIGURE 3.
Mapping pseudouridines targeted by snR191. Primer extension with [32P] 5′ end-labeled oligonucleotide GB150 (complementary to 25S rRNA; see Materials and Methods) and RNAs from wild-type (LMA5–1D) and Δintron (LMA194) strains treated (+) or not treated (−) with CMC (see Materials and Methods). (Lanes C,T,A,G) Dideoxy sequencing reaction using the same oligonucleotide and, as template, 25S rDNA amplified by PCR using primers GB130 and GB131. The positions of pseudouridines are marked by Ψs and the corresponding snoRNA guides, when known, are indicated between brackets.
snR191 is the yeast homolog of the human U19 snoRNA
One known human snoRNA, hU19 (Kiss et al. 1996), has been previously proposed, on the basis of its structure and sequence, to specify these two highly conserved pseudouridines in the human LSU rRNA (Bortolin and Kiss 1998). The alignment of the hU19 snoRNA sequence with yeast snR191 is shown in Figure 2A and the comparison of the potential RNA secondary structures for these RNAs is shown in Figure 2B, E. It shows that, although the secondary structures of these small RNAs are highly conserved, the sequence similarities are mostly restricted to the H/ACA boxes and the guiding loops and extend little beyond these positions. Noticeably, although a human homolog for NOG2 is known, NGP-1 (Racevskis et al. 1996), hU19 is encoded within the intron of another gene, unrelated to NOG2, and whose unique function appears to be encoding hU19 (Bortolin and Kiss 1998).
snR191 confers a growth advantage to the cells
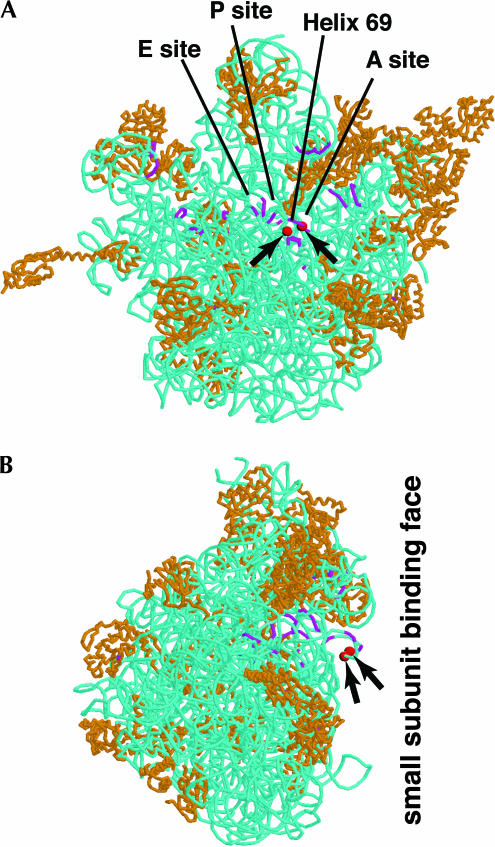
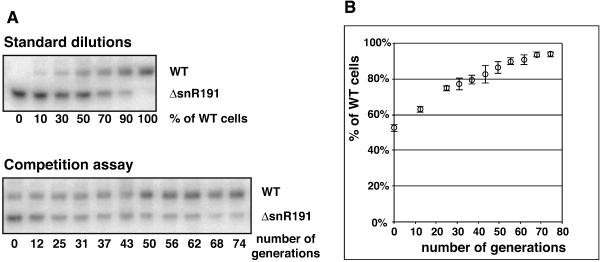
As mentioned, pseudouridines at positions 2258 and 2260 of the yeast LSU rRNA are the two pseudouridines most conserved throughout evolution (Ofengand 2002). They are located within the IVth domain, in the loop of helix H69 (according to the nomenclature used in Yusupov et al. 2001). This loop is remarkable as it is protruding from the large subunit face contacting the small subunit. This loop is also one of the few structural features of the large subunit that change conformation upon binding to the small subunit (Yusupov et al. 2001). Moreover, this loop not only directly contacts the small subunit but also directly contacts the acceptor-site tRNA (Yusupov et al. 2001; Fig. 4). These two highly conserved pseudouridines are at the heart of the LSU/SSU interface, directly next to the tRNA acceptor site (for review, see Decatur and Fournier 2002; see Fig. 4). This strategic position, together with the fact that they are highly conserved, make these two pseudouridines probably among the best candidates to induce a visible phenotype when absent. We had the unique opportunity, with strains specifically lacking snR191, to test the importance of these two pseudouridines. Deletion of the NOG2 intron (strain NOG2–Δintron: LMA194) or of the sequence specifically encoding snR191 within the NOG2 intron (strain NOG2–ΔsnR191: LMA247; see Materials and Methods) does not induce any visible growth defect when serial dilutions were plated on rich medium at 16°C, 25°C, 30°C, or 37°C or on paromomycin or cycloheximide containing mediums at 30°C (not shown). Nevertheless, we tested whether the presence of snR191 could confer a growth advantage to cells placed in competition with cells lacking it. We set up a competition assay in which wild-type cells were mixed, in a proportion of about one to one, with cells in which snR191 was deleted, but otherwise completely isogenics (strain LMA247: NOG2–ΔsnR191). The mixed strains were submitted to repeated cycles of growth to saturation followed by a hundredfold dilution. Altogether, the cultures were submitted to 12 cycles of growth and dilution, corresponding to ~70 generations. Aliquots were taken at different stages of these cultures and the ratio of wild-type versus ΔsnR191 strains within each aliquot was measured using a PCR assay (see Materials and Methods; Fig. 5). Figure 5 shows that, in these conditions, the presence of snR191 gives a small yet reproducible growth advantage to the cells: in six independent cultures, the wild type represented ~50% of the population at time zero and, in average, 94% of the population after 70 generations. To verify that the growth advantage of the wild-type cells did not result from an impairment of splicing of the NOG2 gene, we first verified that the NOG2 mRNA was correctly processed in the ΔsnR191 strains (data not shown). We then repeated the competition experiment but now between wild-type cells and cells in which the entire NOG2 intron had been cleanly deleted (strain LMA194: NOG2–Δintron) and observed the same growth advantage for the wild-type over the mutant cells. We thus conclude that the observed phenotype results from the absence of snR191 and not from impairment in NOG2 expression. Although it cannot be excluded that snR191 plays another unidentified function aside from guiding the modification of the two conserved Ψs, we believe that these observations most likely result from the presence of Ψs at positions 2258 and 2260 providing a selective advantage to yeast cells.
FIGURE 4.
Positions of the two highly conserved Ψs in the large subunit rRNA structure. The position of the two highly conserved Ψs is represented by red spheres (and pointed to by arrows) on the crystal structure (5.5 Å resolution) of the large ribosomal subunit of Thermus thermophilus as determined in Yusupov et al. (2001) (PDB identification number: 1GIY). A backbone representation is used, with rRNAs in blue and proteins in orange. The rRNA regions that are in contact with tRNAs, according to Yusupov et al. (2001), are shown in violet. The structure was displayed using the program RasMol v2.6 (Sayle and Milner-White 1995). (A) View from the small subunit binding face. The approximate regions of the A, P, and E tRNA sites, as well as of helix 69 that carries the two highly conserved Ψs, are pointed to by straight lines. (B) View from the left side of the large subunit.
FIGURE 5.
Competition assay between wild-type cells and cells lacking snR191. The amount of wild-type cells (LMA5–1D strain) relative to cells lacking snR191 (LMA247 strain) during the course of cultures is measured by the relative quantification of PCR products arising from LMA5–1D (wild type) cells (PCR product generated by the GB241 and GB243 oligonucleotides) or LMA247 (ΔsnR191 mutant) cells (PCR product generated by the GB241 and GB242 oligonucleotides) (see Materials and Methods). (A) PCR samples from standard (top) or competition experiments (bottom) were loaded on an 8% acrylamide-urea gel. The figure shows PhosporImager images of the gels. For calibration, standard reference values were generated by quantifying, with a PhosphorImager, the relative amount of the PCR products generated from known mixed fractions of wild-type and ΔsnR191 yeast cells. The evolution of the fraction of wild-type cells during the competition experiment was measured by quantification of the PCR products and normalization using the values determined in the standard experiment. (B) The averaged quantifications of wild-type over mutant cells (% of wild-type cells), averaged from six independent cultures, are plotted to the number of generations. Standard deviations are shown.
In conclusion, despite their presence in rRNAs of all analyzed species, it has been difficult to assign a role to pseudouridines. Here, we show that a phenotype is very likely associated with the absence of two almost universally conserved Ψs, providing new perspectives to determine their function. Additional studies will be required to understand the biochemical basis of the observed effect. Simple assays, such as differential sensitivities to a series of antibiotics, did not reveal more specific phenotypes associated with their absence. Also, polysome profiles are identical in the wild type and the Δ-intron or Δ-SNR191 strains (data not shown), suggesting that the absence of the two Ψs does not result in obvious ribosomal assembly defects. Additional experiments are ongoing to try to narrow the precise biochemical step at which these two Ψs are involved. Note that, because they are located only two nucleotides away from each other, it is reasonable to think that these two Ψs act at the same biochemical step. Nevertheless, it will be interesting to construct more specific mutants impaired in the modification of only one of these two Ψs to determine whether they have a synergetic function or whether the phenotype observed in the present study results from the absence of only one of them.
MATERIALS AND METHODS
Media, strains, oligonucleotides, and plasmids
Yeast media used are standard YPD and YPD supplemented with 4 g/L adenine. E. coli DH5α was used for cloning and propagation of plasmids.
Yeast strains used and constructed for this study listed below:
BMA64: MATa, ura3–1, Δtrp1, ade2–1, leu2–3,112, his3–11,15 (from F. Lacroute, CNRS)
LMA5–1D: BMA64 containing nog2Δ::TRP1 and [pFL38/NOG2] (see below)
LMA163: BMA64 containing dbr1Δ::HIS3
LMA194: BMA64 containing nog2Δ::TRP1 and [pFL38/NOG2–Δintron]
LMA247: BMA64 containing nog2Δ::TRP1 and [pFL38/NOG2–ΔsnR191]
Yeast cell transformations were performed as described (Gietz et al. 1995). All the strains were constructed by homologous recombination using PCR product to transform BMA64. Oligonucleotides used for this study are listed below:
CS9: TAAAGTCTCAAATAACAAAAGCTCAACT
GB130: ACCTTGAATGCTAGAACGTGGA
GB131: AGAACTGGTACGGACAAGGGGA
GB150: GGGAATCTCGTTAATCCATTCA
GB159: TATGAAGAAGGAAACACCACAC
GB160: TCTGACAACACCTCTGAATAAA
GB213: CCAAAACAGTAACAGACACTTC
GB232: TTCGTCTGACGTGGTAATACATGTTC
GB233: TCTCGTCTCTCGAAACTCTATCGAAAAAAAAAATCTC
GB234: TCTCGTCTCTTTCGACTCTTCGAAACAATCAATC
GB235: GAAACAGCTATGACCATGATTACGCC
GB241: CAACCAAGAGGATAGCACGAG
GB242: GATTGATTGTTTCGAAGAGTCG
GB243: CCCTGACAAAAAGGTTTGGTA
MFR207: GATTCTTTCCAACCAAGAGGA
MFR208: CTACTTGTAAAGTTAGAATTG
GB252: GGAATTCCTCTGGGTCCAAAGACTTG
GB253: GACGCGTCGACGGAATGACTGGTATAATGAG.
Plasmid construction
Plasmid pFL38/NOG2: A genomic PCR fragment was synthesized using GB252 hybridizing 290 nt upstream the ATG and GB253 hybridizing 240 nt downstream of the stop codon of the NOG2 gene. This PCR fragment was digested with EcoRI and SalI and cloned into plasmid pFL38 at the corresponding restriction sites, generating plasmid pFL38/NOG2. Plasmid pFL38/NOG2– Δintron: A NcoI–SalI fragment encompassing the NOG2 gene without intron from pASΔΔ/NOG2 (Fromont-Racine et al. 1997) was cloned into a modified pGEX4-T vector and then transferred into plasmid pFL38 as a BamHI–SalI fragment. Plasmid pFL38/NOG2–ΔsnR191: A PCR fragment corresponding to sequence +660 to +888 from the ATG was synthesized using oligonucleotides GB232 and GB233 and pFL38/NOG2 as a matrix and digested with BamHI and BsmBI. A second PCR fragment corresponding to sequence +1207 from the start codon to downstream the stop codon was synthesized using oligonucleotides GB234 and GB235 and pFL38/NOG2 as a matrix, and digested with BsmBI and SphI. The two PCR products were ligated between the BamHI and SphI site of pFL38/NOG2, generating the pFL38/NOG2–ΔsnR191 plasmid. Constuctions were verified by sequencing the region between the BamHI and SphI sites.
RNA extraction and Northern blot
Total RNAs extractions, Northern blots analyses, and primer extensions were performed as described (Saveanu et al. 2001).
Mapping pseudouridines position on 25S rRNA
Total dried RNAs (10 μg) were treated with (or without for control) N-cyclohexyl-N′-β-(4-methylmorpholinium)ethylcarbodiimide p-tosylate (CMC) for 20 min at 37°C and subjected to alkali hydrolysis in the presence of 50 mM Na2CO3 (pH 10.4) at 37°C for 4 h according to Bakin and Ofengand (1993). Five micrograms of these RNAs were then reverse transcribed, as previously described (Saveanu et al. 2001), with 0.2 picomoles of [32P] 5′ end-labeled oligonucleotide GB150.
Competition assay
Six separate cultures, each containing ~50% LMA5–1D and 50% LMA247 cells, were subjected to repeated cycles of growth and dilution: growth was carried out at 30°C in 2 ml of YPD supplemented with 4 g/L adenine until stationary phase (~6.2 generations per cycle) and the culture was then diluted one hundredfold in fresh medium. Adenine is added to avoid [ADE2+] revertants to emerge. One hundred microliters of each culture were pelleted, treated with 50 μL of zymolyase at 300 U/mL for 30 min at 37°C and denatured 10 min at 95°C. PCR was then performed in the presence of 2.5 μL of yeast lysate, 20 pmoles each of GB241, GB242, and GB243 oligonucleotides and 0.02 pmoles of [32P] 5′ end-labeled oligonucleotide GB241 in 25 μL for 30 cycles (30 sec at 94°C, 30 sec at 52°C, and 30 sec at 72°C).
Acknowledgments
G. Badis was supported by a CIFRE (Convention Industrielle de Formation par la Recherche en Enterprise) contract with Hybrigenics SA and the Association Nationale de la Recherche Technique (ANRT).
We thank L. Decourty for her contribution to the competition assay experiment and all members of the laboratory for helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5240503.
REFERENCES
- Bakin, A. and Ofengand, J. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: Analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Mapping of the 13 pseudouridine residues in Saccharomyces cerevisiae small subunit ribosomal RNA to nucleotide resolution. Nucleic Acids Res. 23: 3290–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin, A.G., Smith, L., and Fournier, M.J. 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834. [DOI] [PubMed] [Google Scholar]
- Bortolin, M.L. and Kiss, T. 1998. Human U19 intron-encoded snoRNA is processed from a long primary transcript that possesses little potential for protein coding. RNA 4: 445–454. [PMC free article] [PubMed] [Google Scholar]
- Cherry, J.M., Ball, C., Dolinski, K., Dwight, S., Harris, M., Matese, J.C., Sherlock, G., Binkley, G., Jin, H., Weng, S., et al. 2002. Saccharomyces Genome Database. http://genome-www.stanford.edu/Saccharomyces/. [DOI] [PMC free article] [PubMed]
- Decatur, W.A. and Fournier, M.J. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27: 344–351. [DOI] [PubMed] [Google Scholar]
- ———. 2003. RNA-guided nucleotide modification of ribosomal and other RNAs. J. Biol. Chem. 278: 695–698. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. 2002. Computational genomics of noncoding RNA genes. Cell 109: 137–140. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Rain, J.C., and Legrain, P. 1997. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet. 16: 277–282. [DOI] [PubMed] [Google Scholar]
- Ganot, P., Bortolin, M.L., and Kiss, T. 1997a. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809. [DOI] [PubMed] [Google Scholar]
- Ganot, P., Caizergues-Ferrer, M., and Kiss, T. 1997b. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes & Dev. 11: 941–956. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., Schiestl, R.H., Willems, A.R., and Woods, R.A. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360. [DOI] [PubMed] [Google Scholar]
- Gutgsell, N.S., Del Campo, M.D., Raychaudhuri, S., and Ofengand, J. 2001. A second function for pseudouridine synthases: A point mutant of RluD unable to form pseudouridines 1911, 1915, and 1917 in Escherichia coli 23S ribosomal RNA restores normal growth to an RluD-minus strain. RNA 7: 990–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, T.H., Liu, B., McCully, R.R., and Fournier, M.J. 2003. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 11: 425–435. [DOI] [PubMed] [Google Scholar]
- Kiss, T. 2002. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148. [DOI] [PubMed] [Google Scholar]
- Kiss, T., Bortolin, M.L., and Filipowicz, W. 1996. Characterization of the intron-encoded U19 RNA, a new mammalian small nucleolar RNA that is not associated with fibrillarin. Mol. Cell Biol. 16: 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171. [DOI] [PubMed] [Google Scholar]
- Ni, J., Tien, A.L., and Fournier, M.J. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573. [DOI] [PubMed] [Google Scholar]
- Ofengand, J. 2002. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett. 514: 17–25. [DOI] [PubMed] [Google Scholar]
- Ofengand, J. and Bakin, A. 1997. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266: 246–268. [DOI] [PubMed] [Google Scholar]
- Ofengand, J., Bakin, A., Wrzesinski, J., Nurse, K., and Lane, B.G. 1995. The pseudouridine residues of ribosomal RNA. Biochem. Cell Biol. 73: 915–924. [DOI] [PubMed] [Google Scholar]
- Ooi, S.L., Samarsky, D.A., Fournier, M.J., and Boeke, J.D. 1998. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: Intron length effects and activity of a precursor snoRNA. RNA 4: 1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racevskis, J., Dill, A., Stockert, R., and Fineberg, S.A. 1996. Cloning of a novel nucleolar guanosine 5′-triphosphate binding protein autoantigen from a breast tumor. Cell Growth Differ. 7: 271–280. [PubMed] [Google Scholar]
- Raychaudhuri, S., Conrad, J., Hall, B.G., and Ofengand, J. 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero, D., Grisendi, S., Piazza, F., Rego, E., Mari, F., Rao, P.H., Cordon-Cardo, C., and Pandolfi, P.P. 2003. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science 299: 259–262. [DOI] [PubMed] [Google Scholar]
- Saveanu, C., Bienvenu, D., Namane, A., Gleizes, P.E., Gas, N., Jacquier, A., and Fromont-Racine, M. 2001. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J. 20: 6475–6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayle, R.A. and Milner-White, E.J. 1995. RASMOL: Biomolecular graphics for all. Trends Biochem. Sci. 20: 374. [DOI] [PubMed] [Google Scholar]
- Souciet, J., Aigle, M., Artiguenave, F., Blandin, G., Bolotin-Fukuhara, M., Bon, E., Brottier, P., Casaregola, S., de Montigny, J., Dujon, B., et al. 2000. Genomic exploration of the hemiascomycetous yeasts: 1. A set of yeast species for molecular evolution studies. FEBS Lett. 487: 3–12. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D. 1987. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 6: 4169–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey, D., Lehtonen, H., Jansen, R., Kern, H., and Hurt, E.C. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72: 443–457. [DOI] [PubMed] [Google Scholar]
- Walter, A.E., Turner, D.H., Kim, J., Lyttle, M.H., Muller, P., Mathews, D.H., and Zuker, M. 1994. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. 91: 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]
- Zebarjadian, Y., King, T., Fournier, M.J., Clarke, L., and Carbon, J. 1999. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol. Cell Biol. 19: 7461–7472. [DOI] [PMC free article] [PubMed] [Google Scholar]