Abstract
The universal genetic code links the 20 naturally occurring amino acids to the 61 sense codons. Previously, the UAG amber stop codon (a nonsense codon) has been used as a blank in the code to insert natural and unnatural amino acids via nonsense suppression. We have developed a selection methodology to investigate whether the unnatural amino acid biocytin could be incorporated into an mRNA display library at sense codons. In these experiments we probed a single randomized NNN codon with a library of 16 orthogonal, biocytin-acylated tRNAs. In vitro selection for efficient incorporation of the unnatural amino acid resulted in templates containing the GUA codon at the randomized position. This sense suppression occurs via Watson-Crick pairing with similar efficiency to UAG-mediated nonsense suppression. These experiments suggest that sense codon suppression is a viable means to expand the chemical and functional diversity of the genetic code.
Keywords: Translation, tRNA, unnatural amino acid, sense codon suppression, mRNA display, selection
INTRODUCTION
The universal genetic code comprises of a degenerate set of 61 sense codons that can be translated into polypeptides containing the natural 20 amino acids. In translation, fidelity is ensured primarily through tRNA recognition events, during both aminoacylation and codon:anticodon pairing (Giege et al. 1998; Ibba and Soll 1999; Ogle et al. 2002). The aminoacyl-tRNA is therefore a key player in the execution of the genetic code, tethering nucleic acid and amino acid information together in a single bipartite molecule. Correspondingly, efforts to expand the genetic code and incorporate unnatural amino acids into proteins have converged on re-engineering the biochemistry surrounding tRNAs. To date, these efforts have been focused on altering or using the specificity of aminoacyl-tRNA synthetases to charge unnatural amino acids onto tRNAs and generating novel codon:anticodon interactions that result in frameshift or nonsense suppression (Dougherty 2000; Hohsaka and Sisido 2002; Wang and Schultz 2002). The majority of work has explored insertion of unnatural residues in response to an amber nonsense codon where incorporation of the unnatural residue competes with translation release factors (Nakamura et al. 2000). Surprisingly, little work has been done to explore genetic code expansion by sense codon suppression (Hohsaka et al. 1994; Forster et al. 2001), where incorporation of the unnatural residue would compete with the endogenous pool of aminoacyl-tRNAs.
In principle, some sense codons should be more easily suppressed than others because of a variety of factors that are difficult to predict a priori, including (1) the concentration of competing aminoacyl-tRNAs; (2) binding to elongation factors and the ribosome (LaRiviere et al. 2001); and (3) base-pairing strength. The ability to suppress sense codons efficiently would enable the facile expansion and potential rewriting of the genetic code with unnatural amino acids and holds great potential promise in creating original polymers and chemically diverse display libraries.
Recent work in our laboratory has demonstrated that the unnatural residue biocytin (biotinyl-lysine, a biotin coupled via an amide linkage to the ɛ-amino group of lysine) can be incorporated into mRNA display libraries using the THG73 amber suppressor tRNA (Saks et al. 1996) chemically-acylated with the unnatural amino acid (Li et al. 2002). In mRNA display, mRNA molecules are covalently attached to the peptide or protein they encode using a natural translation system (Roberts and Szostak 1997; Roberts 1999). Our selection experiments demonstrated that mRNA display could be used to select mRNA templates capable of efficiently incorporating an unnatural amino acid, namely those mRNAs that contain a UAG codon complementary to the anticodon in THG73 (Li et al. 2002). There, biocytin was chosen as the unnatural amino acid because of its facile selection on a streptavidin-linked matrix.
We reasoned that a similar mRNA display-based strategy could be used to probe which of 16 GNN sense codons could be efficiently suppressed from a pool of competing aminoacyl-tRNAs. In vitro selection for streptavidin binding resulted in templates containing the GUA codon at the randomized position out of 64 possible sequences. This sense suppression is comparable with UAG-mediated nonsense suppression, suggesting that sense codons can be used to expand the chemical and functional diversity of the genetic code for a multitude of in vitro applications.
RESULTS AND DISCUSSION
Sense suppression selection
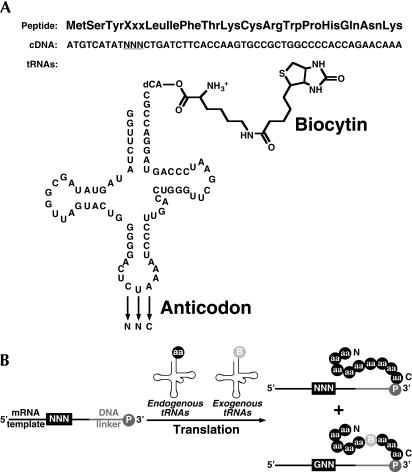
We have developed a selection methodology to examine whether the unnatural amino acid biocytin could be incorporated into an mRNA display library at sense codons. In these experiments, we probed a single randomized NNN codon with a library of 16 orthogonal, biocytin-acylated tRNAs (Heckler et al. 1984; Robertson et al. 1991; Li et al. 2002), each containing an anticodon that can recognize one of 16 corresponding GNN codons via Watson-Crick base-pairing (Fig. 1A). To perform the in vitro selection, the template library and the tRNA pool were added to a commercially-available rabbit reticulocyte lysate (Novagen, Inc.) for translation (Jackson and Hunt 1983) to generate mRNA–peptide fusions (Fig. 1B) (Roberts and Szostak 1997). The mRNA–peptide fusion population contains a mixture of templates, some of which bear natural peptides and others that bear biocytin. The population of molecules that bears biocytin can be purified specifically on streptavidin-agarose. Our selection protocol, therefore, effectively contains two selective steps: (1) a competition during translation for incorporation of the unnatural residue over the endogenous pool of aminoacyl-tRNAs (Fig. 1B) and (2) a selection caused by purification using streptavidin-agarose. The affinity purification provides a quantitative means to measure the percentage of all mRNA–peptide fusions that have incorporated biocytin. We predicted that the randomized triplet sequence would be enriched for a subset of the GNN codons after a number of selection rounds.
FIGURE 1.
Selection scheme for sense suppression. (A) Translation reactions in rabbit reticulocyte lysate contain a template library and exogenous, orthogonal tRNAs. The amber suppressor THG73 was mutated at the anticodon position to Watson-Crick base pair to all 16 GNN codons. The pool of 16 tRNAs were then acylated in one batch with the unnatural amino acid biocytin. (B) Incorporation of the unnatural amino acid into an mRNA–peptide fusion is a competition between the endogenous tRNA population charged with natural amino acids (aa) and the exogenous tRNA pool charged with biocytin (B). The sequence present in the peptide is then encoded in the covalently-attached mRNA through its 3′ puromycin (P), allowing the sequence information in the protein to be read and recovered via the attached RNA. Selecting for streptavidin binding through biocytin incorporation after several rounds enriches the template library at the randomized position for GNN codons.
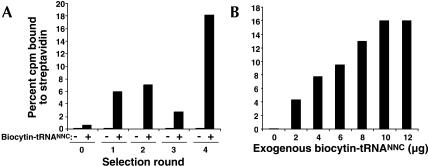
A total of four rounds of selection were performed on the template library. In each successive round the increase in streptavidin binding was dependent on the presence of the biocytin–tRNANNC pool (Fig. 2A). Even in round 0, a small but measurable amount of fusion binds streptavidin-agarose over the no-tRNA control. We controlled the stringency of the selection with the amount of exogenous tRNAs added to the translation reaction. In rounds 0, 1, 2, and 4, 4.0 μg of the biocytin–tRNANNC pool was used, and selection on streptavidin produced a steady increase in the incorporation of biocytin into the library. In round 3, the stringency was increased by using only 1.0 μg of acylated tRNA. This resulted in an overall decrease in the amount of biocytin-bearing product made. The benefit of this high stringency step can be seen in comparing rounds 2 and 4, as a roughly threefold increase in streptavidin binding is observed.
FIGURE 2.
Iterative selection rounds result in the enrichment of sense suppression by orthogonal tRNAs. (A) The binding of [35S]Met-labeled mRNA–peptide fusions to streptavidin-agarose for rounds 0–4 were measured by scintillation counting. Negative controls indicate the percent of mRNA–peptide fusions bound to immobilized streptavidin without any exogenous tRNAs added for each round during translation. The addition of 4.0 μg of the biocytin–tRNANNC pool to the translation reaction for each round is also shown. For a higher stringency selection in round 3, only 1.0 μg of the biocytin–tRNANNC pool was added. (B) Dose-response curve of the biocytin–tRNANNC pool on the translation of round 3 clones. Values for A and B are expressed as the percent of [35S]Met-labeled mRNA–peptide fusions bound to immobilized streptavidin.
Incorporation of biocytin should compete with the endogenous pool of aminoacyl–tRNAs. Likewise, the percent of the fusion that contains biocytin should depend on the concentration of biocytin–tRNA used in the experiment. To test the biocytin–tRNANNC pool dose-response on translation, the template library from round 3 was used to generate mRNA–peptide fusions (Fig. 2B). These results reveal that modest amounts of aminoacyl–tRNAs (e.g., 4.0 μg) are sufficient to drive incorporation and that lower tRNA concentrations (e.g., 1.0 μg) likely provide increased selection stringency, consistent with our observations during the selection experiment.
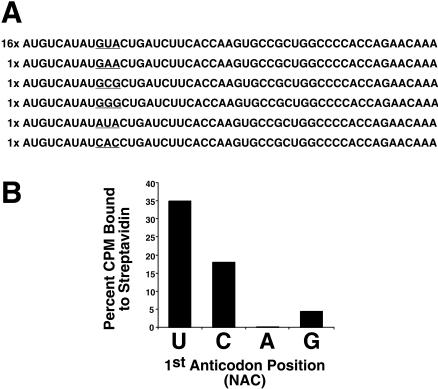
Sequence analysis of the round 4 clones reveals a clear enrichment for GUA, a valine codon, at the randomized position (Fig. 3A) and no other changes to the template sequence (data not shown). Codons that occurred only once in the round-4 pool include GCG alanine, GGG glycine, GAA glutamic acid, CAC histidine, and AUA isoleucine. This observation implies that GUA codons are preferentially recognized by one of the 16 tRNAs in our mixture during translation. We wished to address which of the 16 tRNAs is responsible for inserting biocytin in response to GUA codons. In principle, the GUA codon could be recognized by standard Watson-Crick pairing, via wobble interaction (Crick 1966) or via noncanonical pairing. To test this, we probed a discrete GUA template based on the selected mRNA–peptide fusion with four different biocytin-acylated tRNAs, each of which contained a different base in the wobble position (anticodon = UAC, CAC, AAC, or GAC in Fig. 4). These sense suppression experiments allow us to compare codon:anticodon recognition from orthogonal sets of tRNAs. Incorporation results demonstrate that Watson-Crick recognition is by far the most efficient of the possible pairing schemes (Fig. 3B). The codon:anticodon wobble pairing of A:A proves unfavorable for GUA suppression, which is in accordance with Crick’s wobble rules (Crick 1966). The level of GUA suppression by biocytin–tRNAAAC is roughly equivalent to the background level of suppression (<0.2%) observed for biocytin-acylated THG73 on the same template (data not shown). Both the A:C and A:G pairings support GUA suppression to a lesser extent than the canonical A:U interaction consistent with a loss of one hydrogen bond in the misread wobble position (Parker 1989; Lim and Curran 2001).
FIGURE 3.
(A) Sequences of the template library (21 clones) after four rounds of selection on streptavidin-agarose. (B) Incorporation of biocytin via canonical and noncanonical base-pair interactions at GUA codons. Approximately 30 pmole of template with a fixed GUA codon was translated in the presence of 2.0 μg biocytin-acylated tRNAs containing the anticodon NAC, where the first position N is either U, C, A, or G. Values are expressed as the percent of [35S]Met-labeled mRNA–peptide fusions bound to immobilized streptavidin.
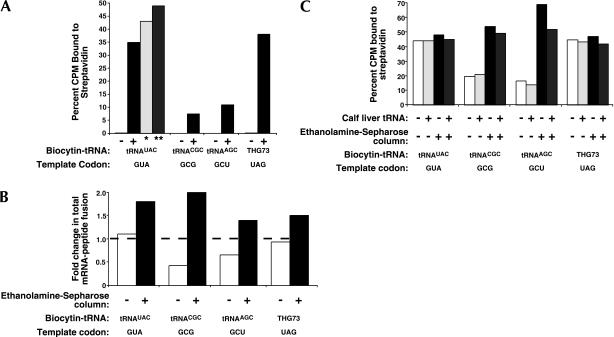
FIGURE 4.
(A) Sense and nonsense suppression comparisons were performed in translation reactions of commercial rabbit reticulocyte lysate containing ~30 pmole of each template with fixed codons as indicated. The percent of [35S]Met-labeled mRNA–peptide fusions bound to immobilized streptavidin with or without translation reactions containing 2.0 μg of the indicated biocytin-acylated tRNA are shown. The binding results in which the translation lysate was bound to ethanolamine-Sepharose in batch mode to reduce endogenous tRNA concentrations before the suppression reaction is denoted by a single asterisk (*). The double asterisk (**) shows binding results from the same translation lysate supplemented with 0.5 μg calf liver tRNA. (B) The tRNA dependence of translation reactions as measured by the fold change in total mRNA–peptide fusion on the addition of 1.5 μg calf liver tRNA is used to assess the level of tRNA depletion of our in-house rabbit reticulocyte lysate preparation by column chromatography using ethanolamine-Sepharose. These values are calculated by dividing total [35S]-labeled mRNA–peptide fusions translated with supplemented calf liver tRNA by those fusions translated without supplementation. The broken line at 1.0 indicates no change in total mRNA–peptide fusion formation on the addition of calf liver tRNA. (C) The mRNA–peptide fusions described in B were then bound to streptavidin-agarose to determine the level of biocytin incorporation at the indicated template codons and expressed as the percent of [35S]Met-labeled mRNA–peptide fusions bound to immobilized streptavidin.
Sense suppression is tRNA-dependent
We sought to compare the efficiency of biocytin incorporation at our selected GUA codon with UAG-based nonsense suppression, as well as to two other arbitrarily chosen GNN type codons. To do this, we synthesized four discrete templates based on our library sequence containing (1) the GUA valine codon (above); (2) the GCG alanine codon; (3) the GCU alanine codon; and (4) the UAG nonsense codon. We then probed each of these templates with a complementary biocytin-acylated THG73 variant (Fig. 4A). This analysis indicates that the efficiency of GUA-mediated sense suppression is similar to UAG-mediated nonsense suppression. This result is striking given that the sense suppression is competing with a natural tRNA pool present in the lysate. (Here, calf liver tRNA is supplemented in the lysate to give efficient translation on many different templates [Jackson and Hunt 1983].) It should be noted that a fraction of rabbit reticulocyte lysate tRNA does survive micrococcal nuclease treatment in our own lysate preparations; data not shown.) The GCG and GCU alanine codons show reduced incorporation relative to the GUA valine codon that we have selected. Nonetheless, the substantial incorporation present in even these codons contends generally that supplanting natural codon pairing may not be as difficult as assumed previously.
Our experiments raise the issue of why GUA codons are efficiently competed by exogenous tRNA. Lack of competition at GUA codons may result from low gene-dosage of this particular valine tRNA. The number of tRNA genes encoding UAC anticodons in rabbit and cow is unknown, but in the closely-related human, only one of the 157 tRNA genes recognizes GUA via Watson-Crick pairing, whereas 11 genes code for tRNAs that recognize the GCU alanine codon (Marck and Grosjean 2002). Correspondingly, sense suppression of GCU codons by biocytin–tRNAAGC (Fig. 4A) resulted in a threefold lower level of biocytin incorporation as compared to GUA suppression. Sense suppression of GCG, an alanine codon with one complementary human tRNA gene and one occurrence in the round 4 pool, gave a fivefold lower signal than GUA-based suppression. Whereas GUA codons can be recognized only via canonical Watson-Crick pairing, GCG and GCU codons can both be recognized canonically and via wobble interactions (Lim and Curran 2001), making them less likely to serve as blanks in the genetic code in commercial rabbit reticulocyte lysate.
The GUA codon has one of the lowest uses of the form GNN in higher eukaryotes and correspondingly a low abundance of its isoacceptor tRNAs (Ikemura and Ozeki 1983; Kanaya et al. 2001), which is our primary assertion for why it is selected for sense suppression. If this is true, then reducing the overall tRNA concentration should increase suppression at all sense codons. Previously, Jackson and coworkers developed a protocol using ethanolamine-Sepharose to specifically deplete tRNAs from a translation extract (Jackson et al. 2001). We subjected commercial rabbit reticulocyte lysate to ethanolamine-Sepharose to create a lysate whose endogenous tRNA population has been partially depleted, and then assayed GUA suppression by biocytin–tRNAUAC (Fig. 4A). In the commercially available lysate, lowering the endogenous tRNA concentration produces a modest increase in GUA suppression (Fig. 4A). This result indicates that an optimal concentration or balance of endogenous tRNAs to the sense suppressor likely exists. Lysates totally lacking tRNA will not support translation (Jackson et al. 2001), whereas lysates that contain large quantities of endogenous tRNA will require high concentrations of suppressor to incorporate an unnatural amino acid. In line with this hypothesis, adding back calf liver tRNA to 20 μg/mL (a lower concentration than the untreated, commercial lysate concentration of 60 μg/mL; Jackson and Hunt 1983) results in improved sense suppression approaching 50% (Fig. 4A). Other calf liver tRNA concentrations were tested (data not shown), resulting in a titration curve optimized at the value shown. Therefore, it appears that partially removing the complex endogenous tRNA population can tip the balance in favor of sense suppression by orthogonal tRNAs. It is then likely that a more stringent elimination of competing tRNAs could allow for replacement of all the natural amino acids, enabling synthesis of trillion-member unnatural mRNA display libraries.
In an effort to determine the impact of endogenous tRNA depletion in translation reactions on sense codon suppression, we passed our in-house rabbit reticulocyte lysate (prepared as described in Materials and Methods) over an ethanolamine-Sepharose column (Jackson et al. 2001). The level of endogenous tRNA depletion is assessed by the fold change in total mRNA–peptide fusion (i.e., [35S]-labeled material eluted from dT25-cellulose) on the addition of 1.5 μg calf liver tRNA (Fig. 4B). Before passing lysate through the ethanolamine-Sepharose column, translation appears to be independent of calf liver tRNA for GUA and UAG templates, and in the cases for GCG and GCU templates, the amount of mRNA–peptide fusion formed is actually inhibited by added tRNA. This inhibitory effect is reduced on pre-incubation (5–20 min) of the calf liver tRNA in the translation reaction before adding templates (data not shown), suggesting that the calf liver tRNA is in large part deacylated and that GCG and GCU isoacceptor tRNAs from this pool exert a larger effect than those for GUA because of differences in concentrations. Once passed through the ethanolamine-Sepharose column, the translation lysate exhibits a tRNA-dependence to form mRNA–peptide fusions for all templates tested (Fig. 4B), thereby demonstrating the efficiency with which the chromatographic step removes endogenous tRNAs. It should be noted that we did not observe a complete elimination of endogenous tRNAs, which suggests that the background counts measured by Jackson and coworkers for tRNA-dependent lysate without tRNA supplementation (Jackson et al. 2001) can still support efficient translation of mRNA–peptide fusions.
We next measured biocytin incorporation at sense and nonsense codons to compare rabbit reticulocyte lysate before and after ethanolamine-Sepharose treatment to remove tRNA. Before removal of tRNAs from our in-house lysate, the four templates give biocytin incorporation levels (Fig. 4C) similar to those seen in the commercial lysate (Fig. 4A). Suppression of the GCG and GCU codons, however, jumps dramatically when tRNA is removed from the in-house lysate preparation lacking tRNA, and is comparable with suppression at GUA and UAG codons (Fig. 4C). Interestingly, the levels of suppression do not change significantly for GUA and UAG templates on endogenous tRNA depletion. Suppression at the amber and GUA codons appears to be independent of tRNA depletion. This observation is in line with our conclusion that the GUA codon was selected for biocytin incorporation because its isoacceptor tRNA concentrations in the lysate (both commercial and in-house preparations) are very low. The marked improvement that we observe at GCG and GCU codons represents a clear demonstration that removing endogenous and competing tRNAs produces blanks in the genetic code for sense suppression at a variety of sense codons.
Closing remarks
In focusing on the GNN quadrant of the universal genetic code, we were able to design a selection experiment using mRNA display to identify sense codons that could be efficiently suppressed. Although the suppressor tRNA THG73 has been used to incorporate both natural and unnatural amino acids (Dougherty 2000; Saks et al. 1996), the ability of biocytin to be selected on streptavidin-linked matrices underscores its utility in the iterative selections and direct measurements for sense suppression. This general strategy should allow us to identify codons in the other three quadrants (UNN, CNN, and ANN codons) that can be easily suppressed in the rabbit reticulocyte translation system. Our results show that sense suppression efficiency depends on the concentration of both the endogenous tRNA pool and the suppressor tRNA used. The elimination of competing tRNAs from the translation lysate, as demonstrated by ethanolamine-Sepharose chromatography, creates readily suppressible sense codons—synthetic blanks in the genetic code that permit high-efficiency insertion of unnatural amino acids into mRNA display libraries. We would predict that easily suppressed sense codons should depend on the translation system or organism used, and that suppression efficiency would be inversely proportional to codon usage and tRNA abundance (Ikemura and Ozeki 1983; Hohsaka et al. 1994; Tsai and Curran 1998; Navarro and Thuriaux 2000). Translation systems that are partially or totally reconstituted (Forster et al. 2001; Shimizu et al. 2001), where tRNA concentrations (Jackson et al. 2001) or synthetase activity can be controlled (Tao and Schimmel 2000), should therefore provide useful platforms for rewriting large blocks of the universal code, and enable the creation of trillion-member unnatural display libraries.
MATERIALS AND METHODS
Preparation of biocytin-acylated tRNAs
The pUC19-based plasmid (a gift from Dennis Dougherty, California Institute of Technology) harboring the gene for THG73 was mutated at the tRNA anticodon position by QuikChange (Stratagene) with 16 appropriate sets of primers (self-complementary pairs 42 bp in length). Resulting clones were verified by DNA sequencing before synthesizing individual tRNAs by in vitro transcription with T7 polymerase from Fok I-linearized plasmids. Transcribed tRNAs were then gel-purified and desalted by ethanol precipitation.
Purified tRNAs were ligated to a molar excess of NVOC-protected biocytin-dCA with T4 RNA ligase (New England Biolabs) as described previously (Li et al. 2002). Reaction mixtures were extracted in an equal volume of phenol:CHCl3:isoamyl alcohol (25:21:1, pH 5.0), and precipitated with 2.5 volumes ethanol (−20°C). After drying, the pellets were resuspended in 1.0 mM sodium acetate, pH 5.2, and adjusted to 1.0 mg/mL for each acylated tRNA. Before adding to translation reactions, biocytin–tRNAs were deprotected by a xenon lamp outfitted with a 315-nm cut-off filter for 5 min to remove the NVOC group.
Generation of mRNA–peptide fusions
The synthetic DNA template 5′-GGACAATTAC TATTTACAAC CACCATGTCA TATNNNCTGA TCTTCACCAA GTGCCGC TGG CCC-3′ (where N is T, C, A, or G) was PCR-amplified with synthetic primers T7-E (5′-AGTGAATTCTAATACGACTCACTA TAGGGACAATTACTATTTACAACCACCATG-3′) and 3P (5′-[2′ O-methyl U] [2′ O-methyl U] TGTTCTGGTGGGGCCAGCG GCACTTGGT-3′). Discrete templates (i.e., DNA without the randomized codon, NNN) were also amplified in a similar fashion. The PCR product was transcribed by T7 polymerase to generate mRNA that was subsequently ligated with T4 DNA ligase (New England Biolabs) to a flexible DNA linker containing puromycin, pF30P (5′-dA21[C9]3dAdCdCP that was 5′-phosphorylated by phophorylation reagent II, Glen Research; C9 = triethylene glycol phosphate, and P = CPG-puromycin, Glen Research) with a DNA splint (5′-T15GTTCTGGTGGGGCCA-3′) as described previously (Li et al. 2002). After ligation, the fusion template was gel-purified and desalted by ethanol precipitation. The purified fusion template was translated in rabbit reticulocyte lysate (Novagen, Inc.) using standard conditions (30°C for 60 min). mRNA–peptide fusion formation was stimulated by the addition of MgCl2 and KCl to 50 mM and 0.6 M, respectively, before overnight incubation at −20°C.
Selection via sense suppression
The mRNA–peptide fusions were initially isolated from translation reactions by dT25-cellulose binding in 5 mL isolation buffer (1M NaCl, 100 mM Tris-HCl pH 8.0, 0.2% Triton X-100) at 4°C for 45 min, washed in 700 μL isolation buffer seven times at 4°C, and eluted in water (ambient temperature). Purified mRNA–peptide fusions were concentrated via ethanol precipitation in the presence of 30 μg linear acrylamide (Ambion) and reverse transcribed by Superscript II RNase H− reverse transcriptase (Invitrogen) per manufacturer’s instructions before binding to streptavidin-agarose (Pierce) in 1.0 mL 50 mM sodium phosphate pH 7.0 at 4°C for 1 h. Bound material was washed in 700 μL 50 mM sodium phosphate pH 8.0, 100 mM NaCl, 0.1% SDS five times. PCR amplification of the bound fusions with primers T7-E and 3P created products used for cloning with the TOPO Clone kit (Invitrogen) to sequence the library pool, and for transcribing mRNA for the next selection round.
Chromatographic depletion of endogenous tRNAs
Ethanolamine-Sepharose resin was prepared from epoxy-activated Sepharose 6B (Sigma) as described previously (Jackson et al. 2001). This resin was used to deplete endogenous tRNA in commercial and in-house preparations of rabbit reticulocyte lysate. All procedures were performed at 4°C unless otherwise specified. For tRNA depletion in commercial translation lysate 100 μL of a 50% ethanolamine-Sepharose slurry in buffer A (25 mM KCl, 10 mM NaCl, 1.1 mM MgCl2, 0.1 mM EDTA, 10 mM HEPES-KOH, pH 7.2) with additional 50 mM KCl and 0.25 mM MgCl2 was incubated for 45 min with rabbit reticulocyte lysate (Novagen, Inc.) before brief centrifugation at 1500g to clarify the desired supernatant.
In-house preparations of rabbit reticulocyte lysate were prepared as described previously (Jackson and Hunt 1983). Briefly, 10 mL of completely clarified rabbit reticulocyte lysate was incubated for 30 min at ambient temperature with 0.2 mL 1.0 mM hemin (Sigma) in 85% ethylene glycol, 0.1 mL 5 mg/mL creatine kinase solution (50% vol/vol aqueous glycerol; Boehringer Mannheim), 0.1 mL 0.1 M CaCl2, and 0.1 mL micrococcal nuclease (15,000 U/mL; Boehringer Mannheim). The reaction was then quenched by the addition of 0.1 mL 0.2 M EGTA-KOH, pH 7.5, and placing the solution on ice. Salts were added to a concentration of 50 mM KCl and 0.25 mM MgCl2, and then 2 mL of this material was passed over an ethanolamine-Sepharose column (0.5-mL bed volume; 0.4-cm inner diameter × 10-cm height) pre-equilibrated in 5 mL buffer A with additional 50 mM KCl and 0.25 mM MgCl2 (Jackson et al. 2001). The first 0.5-mL fraction of lysate that passed through the column was discarded and the subsequent 4 × 0.5-mL fractions were pooled and saved in 250 μL aliquots. Rabbit reticulocyte lysates depleted of endogenous tRNA were either used immediately for translation reactions or snap-frozen in an ethanol-dry ice bath and stored at −80°C.
Acknowledgments
We thank Prof. Dennis Dougherty (Caltech) for the use of his xenon lamp and the pUC19 plasmid encoding THG73. We are grateful to the Roberts group for helpful discussions; Terry Takahashi for insights into mRNA display library design; and Shuwei Li and Steven Millward for biocytin-acylated dCA. This work was funded by National Institutes of Health grant GM60416 (R.W.R) and an American Cancer Society Postdoctoral Fellowship (A.F.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5350303.
REFERENCES
- Crick, F.H. 1966. Codon–anticodon pairing: The wobble hypothesis. J. Mol. Biol. 19: 548–555. [DOI] [PubMed] [Google Scholar]
- Dougherty, D.A. 2000. Unnatural amino acids as probes of protein structure and function. Curr. Opin. Chem. Biol. 4: 645–652. [DOI] [PubMed] [Google Scholar]
- Forster, A.C., Weissbach, H., and Blacklow, S.C. 2001. A simplified reconstitution of mRNA-directed peptide synthesis: Activity of the epsilon enhancer and an unnatural amino acid. Anal. Biochem. 297: 60–70. [DOI] [PubMed] [Google Scholar]
- Giege, R., Sissler, M., and Florentz, C. 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26: 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckler, T.G., Chang, L.H., Zama, Y., Naka, T., Chorghade, M.S., and Hecht, S.M. 1984. T4 RNA ligase mediated preparation of novel “chemically misacylated” tRNAPheS. Biochemistry 23: 1468–1473. [DOI] [PubMed] [Google Scholar]
- Hohsaka, T. and Sisido, M. 2002. Incorporation of non-natural amino acids into proteins. Curr. Opin. Chem. Biol. 6: 809–815. [DOI] [PubMed] [Google Scholar]
- Hohsaka, T., Sato, K., Sisido, M., Takai, K., and Yokoyama, S. 1994. Site-specific incorporation of photofunctional nonnatural amino acids into a polypeptide through in vitro protein biosynthesis. FEBS Lett. 344: 171–174. [DOI] [PubMed] [Google Scholar]
- Ibba, M. and Soll, D. 1999. Quality control mechanisms during translation. Science 286: 1893–1897. [DOI] [PubMed] [Google Scholar]
- Ikemura, T. and Ozeki, H. 1983. Codon usage and transfer RNA content: Organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harbor Symp. Quant. Biol. 47: 1087–1097. [DOI] [PubMed] [Google Scholar]
- Jackson, R.J. and Hunt, T. 1983. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 96: 50–74. [DOI] [PubMed] [Google Scholar]
- Jackson, R.J., Napthine, S., and Brierley, I. 2001. Development of a tRNA-dependent in vitro translation system. RNA 7: 765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya, S., Yamada, Y., Kinouchi, M., Kudo, Y., and Ikemura, T. 2001. Codon usage and tRNA genes in eukaryotes: Correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J. Mol. Evol. 53: 290–298. [DOI] [PubMed] [Google Scholar]
- LaRiviere, F.J., Wolfson, A.D., and Uhlenbeck, O.C. 2001. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science 294: 165–168. [DOI] [PubMed] [Google Scholar]
- Li, S., Millward, S., and Roberts, R. 2002. In vitro selection of mRNA display libraries containing an unnatural amino acid. J. Am. Chem. Soc. 124: 9972–9973. [DOI] [PubMed] [Google Scholar]
- Lim, V.I. and Curran, J.F. 2001. Analysis of codon:anticodon interactions within the ribosome provides new insights into codon reading and the genetic code structure. RNA 7: 942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck, C. and Grosjean, H. 2002. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8: 1189–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, Y., Ito, K., and Ehrenberg, M. 2000. Mimicry grasps reality in translation termination. Cell 101: 349–352. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., and Ramakrishnan, V. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732. [DOI] [PubMed] [Google Scholar]
- Parker, J. 1989. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 53: 273–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R.W. 1999. Totally in vitro protein selection using mRNA-protein fusions and ribosome display. Curr. Opin. Chem. Biol. 3: 268–273. [DOI] [PubMed] [Google Scholar]
- Roberts, R.W. and Szostak, J.W. 1997. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. 94: 12297–12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, S.A., Ellman, J.A., and Schultz, P.G. 1991. A general and efficient route for chemical aminoacylation of transfer RNAs. J. Am. Chem. Soc. 113: 2722–2729. [Google Scholar]
- Saks, M.E., Sampson, J.R., Nowak, M.W., Kearney, P.C., Du, F., Abelson, J.N., Lester, H.A., and Dougherty, D.A. 1996. An engineered Tetrahymena tRNAGln for in vivo incorporation of unnatural amino acids into proteins by nonsense suppression. J. Biol. Chem. 271: 23169–23175. [DOI] [PubMed] [Google Scholar]
- Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K., and Ueda, T. 2001. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19: 751–755. [DOI] [PubMed] [Google Scholar]
- Tao, J. and Schimmel, P. 2000. Inhibitors of aminoacyl-tRNA synthetases as novel anti-infectives. Expert Opin. Investig. Drugs 9: 1767–1775. [DOI] [PubMed] [Google Scholar]
- Tsai, F. and Curran, J.F. 1998. tRNA2Gln mutants that translate the CGA arginine codon as glutamine in Escherichia coli. RNA 4: 1514–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. and Schultz, P.G. 2002. Expanding the genetic code. Chem. Commun. (Camb.): 1–11. [DOI] [PubMed]