Abstract
Assembly helpers exist for the formation of ribosomal subunits. Such a function has been suggested for the DnaK system of chaperones (DnaK, DnaJ, GrpE). Here we show that 50S and 30S ribosomal subunits from an Escherichia coli dnaK-null mutant (containing a disrupted dnaK gene) grown at 30°C are physically and functionally identical to wild-type ribosomes. Furthermore, ribosomal components derived from mutant 30S and 50S subunits are fully competent for in vitro reconstitution of active ribosomal subunits. On the other hand, the DnaK chaperone system cannot circumvent the necessary heat-dependent activation step for the in vitro reconstitution of fully active 30S ribosomal subunits. It is therefore questionable whether the requirement for DnaK observed during in vivo ribosome assembly above 37°C implicates a direct or indirect role for DnaK in this process.
Keywords: DnaK chaperone, in vitro reconstitution, ribosome assembly, assembly helpers
INTRODUCTION
Extrinsic factors are likely to be involved in assembly of bacterial ribosome subunits (Alix 1993; for a recent extensive review of the 30S assembly, see Culver 2003). Such factors should be necessary to circumvent the heat-dependent conformational change of ribosomal precursor intermediates required during in vitro reconstitution of ribosomes (Nierhaus 1991; Williamson 2003). The chaperones DnaK/DnaJ/GrpE (Alix and Guerin 1993; Sbai and Alix 1998) and GroEL/GroES (El Hage et al. 2001) are prime candidates for such a function, but their role, direct or not, in ribosome assembly is still mysterious. A defective DnaK system does not affect ribosome assembly at 30°C but seriously disturbs assembly at temperatures above 35°C (El Hage et al. 2001). In vitro reconstitution of ribosomes at different temperatures in the presence or absence of chaperones offers a tool to explore the role of chaperones, and has been recently exploited (Maki et al. 2002). In these studies, the authors concluded that the DnaK chaperone system facilitates 30S ribosomal subunit assembly. In contrast to these results, we show here that addition of chaperones to in vitro reconstitution could not substitute the necessity of the heat-activating step, and that ribosomal subunits, rRNAs and ribosomal proteins purified from a mutant that contains a disrupted dnaK gene (ΔdnaK), are as thermostable and active as their dnaK+ counterparts in protein synthesis and in vitro reconstitution.
RESULTS AND DISCUSSION
In vitro reconstitution of 30S ribosomal subunits in the presence and absence of chaperones
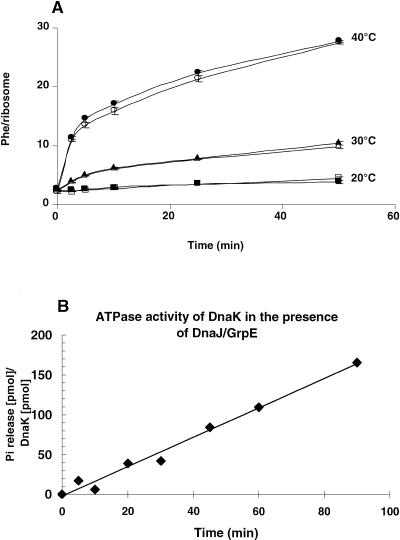
We tested whether or not the presence of chaperones relieves the heat dependence of the 30S reconstitution process and/or accelerates the formation of active particles. To this end, the kinetics of total in vitro reconstitution of 30S ribosomal subunits at various temperatures (20°C, 30°C, and 40°C) were performed in the presence or absence of purified chaperones DnaK, DnaJ, GrpE, and ATP. The activity of the reconstituted 30S ribosomal subunits was assayed in a poly(Phe) synthesis system after complementing with native 50S subunits. The assay was performed at 20°C for 2 h, so that no further heat activation of the reconstituted particles would occur during poly(Phe) synthesis. Figure 1A demonstrates that reconstitution was efficient only at 40°C, in agreement with results reported by Traub and Nomura (1969), and that addition of chaperones and ATP to the reconstitution mixtures at all temperatures did not relieve the requirement for the heat activation of the reconstitution process or accelerate the formation of active particles. In fact, the rates in the presence and absence of chaperones were identical within the errors at each temperature and time point. rRNAs were not degraded during the reconstitution incubation, as indicated by the good activity of the particles formed and judged by SDS-polyacrylamide gel electrophoresis (data not shown), excluding the possibility of contaminating RNase(s) in the chaperone batches.
FIGURE 1.
(A) Kinetics of in vitro reconstitution of 30S ribosomal subunits at the indicated temperatures in the presence (closed symbols) or absence (open symbols) of the chaperones (DnaK, DnaJ, GrpE + ATP). At the indicated times, an aliquot was withdrawn from the reconstitution mixture, complemented with native 50S subunits and assayed for poly(Phe) synthesis at 20°C for 2 h. For details, see Materials and Methods. (B) ATPase activity of the DnaK system. The specific ATPase activity of DnaK (released phosphate/minute/DnaK) at 37°C was determined from the slope of the curve assuming a molecular mass for DnaK of 70 kD.
In an additional control experiment, we tested the activity of the chaperones used in the reconstitution assays concerning ATP hydrolysis. Figure 1B demonstrates that the DnaK system was highly active, with a turnover number of 1.8 min−1, a value comparable to those reported by other groups (Ha et al. 1999).
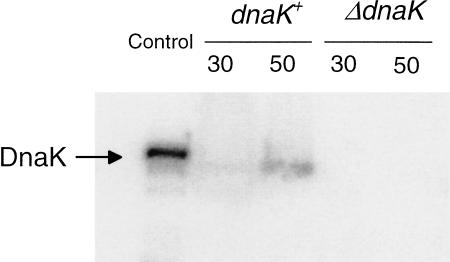
To control for the possibility of endogenous DnaK present in the subunit preparation used for the preparation of TP30 and 16S rRNA, the subunits were analyzed by immunoblotting for the presence of DnaK. This was important because it had been reported that 50S subunits contain endogenous DnaK although 30S subunits did not (Vysokanov 1995). We nevertheless analyzed the DnaK content of the 30S subunits that were prepared from a dnaK+ Escherichia coli strain by immunoblotting. The first lane of Figure 2 shows the signal from 0.5 pmole of DnaK; in the second and third lanes, TP30 derived from 375 pmole of small subunits and TP50 from 160 pmole of large subunits, respectively, were applied. Scanning of the band intensities revealed that the 50S subunits contained only traces of DnaK (molar ratio DnaK:50S = 1:950), whereas DnaK was virtually absent in the 30S preparation (molar ratio 1:20,000; see legend to Fig. 2). The specific association of DnaK with the 50S subunits is reminiscent of the trigger-factor-binding specificity for the 50S subunit (Kramer et al. 2002a; Blaha et al. 2003). This subunit specificity of the binding is probably related to the cotranslational folding exerted by DnaK on nascent polypeptide chains emerging at a specific site (tunnel) on the cytoplasmic surface of the 50S ribosomal subunits, where the trigger factor was located recently (Kramer et al. 2002b; Blaha et al. 2003). The common or similar binding region is reflected by an overlapping function that is exemplified by the interesting interplay that exists between the trigger factor and DnaK. Deletion of either gene is tolerated by the cell, but deletion of both is lethal (Bukau et al. 2000).
FIGURE 2.
Immunoanalysis of DnaK in native 50S and 30S ribosomal subunits. (Lane 1) Control, DnaK (30 ng ≅ 0.5 pmole); (lane 2) 5.2 A260 units ≅ 375 pmole of 30S subunits from strain CAN20-12E (dnaK+); (lane 3) 4.4 A260 units ≅ 160 pmole of 50S subunits from strain CAN20-12E; (lane 4) 5.6 A260 units of 30S subunits from strain BB11 (ΔdnaK); (lane 5) 5.6 A260 units of 50S subunits from strain BB11 (ΔdnaK). The subunits were boiled in SDS sample buffer before subjecting them to an SDS-gel electrophoresis followed by the blotting procedure. The relative density of the bands was determined by scanning (Molecular Dynamics) and processing with the ImageQuant program. The pixel numbers for lanes 1, 2, and 3 were 63,840, 2434, and 21,577, yielding a molar ratio of DnaK per 30S and 50S subunits of 1:20,000 and 1:950, respectively. For details, see Materials and Methods.
With regard to the 30S assembly, the practically complete absence of contaminating endogenous DnaK on this subunit means that no traces of DnaK (if not added) were present during the reconstitution of this subunit. It follows that the DnaK chaperone family is not sufficient to facilitate reconstitution of 30S subunits at temperatures at or below 40°C, in striking contrast to the results of Culver and colleagues (Maki et al. 2002).
Thermostability of ribosomes and reconstitution competence of ribosomal components from a ΔdnaK mutant grown at 30°C
In the next experiments, we made use of an E. coli dnaK-null mutant (BB1553) that contains a disrupted gene (ΔdnaK52::cmR), resulting in a lack of DnaK (Fig. 2) and also drastically reduced levels of DnaJ, probably because the transcription of the distal dnaJ gene is interrupted (Sell et al. 1990).
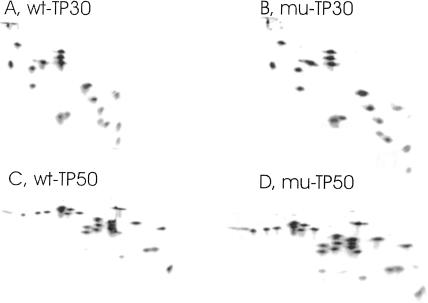
Wild-type E. coli cells containing normal amounts of RNase I are an unsuitable source for ribosomes that should be used for a reconstitution analysis, because this RNase binds tightly to the ribosome, copurifies with the protein fraction, and destroys the rRNA during the reconstitution incubation (Dohme and Nierhaus 1976). Therefore, we set out to transduce the rna-19 allele encoding an inactive RNase I into a dnaK-null mutant (and its wild-type isogene; Materials and Methods) to study the in vitro characteristics of their 30S and 50S ribosomal subunits at normal and high temperatures. The resulting strains, MC41.14 (rna-19, dnaK+) and BB11 (rna-19, ΔdnaK52::cmR), were grown at 30°C, because BB11 is temperature sensitive at 37°C (Table 1A), and their 30S and 50S ribosomal subunits were isolated. Their activities in poly(U)-dependent [14C]-poly(Phe) synthesis were the same, irrespective of the dnaK status of the strain (Table 1B). Similarly, their ribosomal protein composition (TP50 and TP30 patterns) was identical (Fig. 3). It could be argued that the 30S and 50S subunits from the ΔdnaK strain appear perfectly normal, just because they were purified as 70S ribosomes and thus selected as normal ones. However, this is unlikely, because we have previously shown that the corresponding crude extract (S30) shows the same radioactive RNA profile after sucrose-gradient centrifugation as that seen with a wild-type strain, as long as the ΔdnaK strain was grown at temperatures not higher than 30°C (Fig. 1F of El Hage et al. 2001). In other words, no accumulation of assembly intermediates was observed, because of the absence of DnaK. Our findings further agree with the previous observation that no noticeable difference in protein synthesis levels exists between dnaK+ and ΔdnaK52 cells grown at 30°C, except a very low fraction (0.5%–1%) of total proteins aggregated in ΔdnaK52 cells (Tomoyasu et al. 2001).
TABLE 1.
Cell growth and activities of both ribosomal subunits and reconstituted particles of the strains dnaK+ (MC41.14) and ΔdnaK (BB11)
| MC 41.14 dnaK+ | BB11 ΔdnaK52::cmR | ||
| A. Phenotypea | |||
| Growth at 30°C | + | + | |
| Growth at 30°C + 25 μg/mL chloramphenicol | − | + | |
| Growth at 37°C | + | − | |
| B. Ribosome activity (Phe incorporation per 70S)b | |||
| 50S subunits (in the presence of wt30S) | 88 ± 4 | 80 ± 5 | |
| 30S subunits (in the presence of wt50S) | 105 ± 5 | 103 ± 4 | |
| C. Reconstituted 30S subunits (Phe incorporation per 70S) (in the presence of native 50S)c | |||
| TP30 | |||
| dnaK+ | ΔdnaK | ||
| 16S rRNA | dnaK+ | 40 ± 3 | 36 ± 2 |
| ΔdnaK | 47 ± 3 | 48 ± 2 | |
| D. Reconstituted 50S subunits (Phe incorporation per 70S) (in the presence of native 30S)d | |||
| TP50 | |||
| dnaK+ | ΔdnaK | ||
| (23S + 5S) rRNA | dnaK+ | 48 ± 3 | 49 ± 3 |
| ΔdnaK | 46 ± 2 | 42 ± 3 | |
aDistinctive phenotypes of the dnaK+ and ΔdnaK strains used for in vitro reconstitution studies.
bPoly(Phe) synthesis activity of subunits derived from both dnaK+ and ΔdnaK strains in the presence of an excess of the complementary native subunits from the strain CAN20-12E (dnaK+). For details, see Materials and Methods.
c30S reconstitution experiments were performed with TP30 and 16S rRNA from both strains (dnaK+ and ΔdnaK) in all possible combinations. The activity of the reconstituted particles was assessed by poly(Phe) synthesis in the presence of an excess of native 50S subunits from strain CAN20-12E (dnaK+).
dExperiments corresponding to those described in C but involving 50S reconstituted particles. Background values (no reconstituted particles) between 150 and 500 cpm were subtracted. The resulting Phe incorporation values were between 7200 and 21,000 cpm, corresponding to 35 and 105 Phe incorporated per 70S ribosome, respectively.
FIGURE 3.
Two-dimensional polyacrylamide gel electrophoresis of 50S and 30S ribosomal proteins from the dnaK+ (MC41.14) and ΔdnaK (BB11) strains. TP50 and TP30 were prepared and subjected to two-dimensional polyacrylamide gel electrophoresis according to Geyl et al. (1981). (A,B) TP30 from 30S subunits of the MC41.14 dnaK+ strain (wt-TP30) and mutant BB11 (ΔdnaK) strain (mu-TP30), respectively. (C,D) TP50 from 50S subunits of the MC41.14 dnaK+ strain (wt-TP50) and mutant BB11 (ΔdnaK) strain (mu-TP50), respectively.
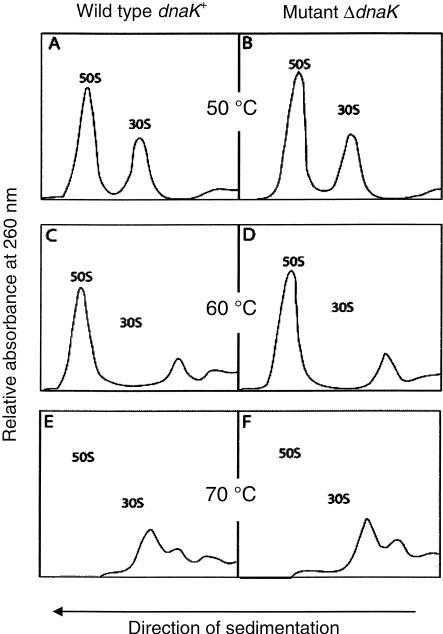
To check whether the ribosomes formed in the absence of DnaK/DnaJ were as thermostable as their counterparts from a dnaK+ strain, 70S ribosomes from both strains ΔdnaK and dnaK+ were subjected to a severe thermal injury (up to 70°C) for 10 min in high salt conditions (1 mM MgCl2, 200 mM NH4Cl), before subjecting them to sucrose-gradient centrifugation under the same ionic conditions. Figure 4 shows that ribosomal subunits from both strains are equally stable until 50°C (Fig. 4A,B), but that thermodestruction occurs at 60°C for 30S subunits (but not for 50S subunits, Fig. 4C,D) and at 70°C for both subunits (Fig. 4E,F), in agreement with studies on wild-type ribosomes by Tal et al. (1977). Ribosomes of the ΔdnaK strain are as thermostable as those from the wild-type strain. Once assembled at 30°C, ribosomes do not need DnaK for thermoprotection, neither in vivo (Alix and Guerin 1993) nor in vitro (Fig. 4).
FIGURE 4.
Sedimentation profiles of heat-treated 70S ribosomes prepared from dnaK+ (MC41.14) and ΔdnaK (BB11) strains. (A,B) Ribosomes from wild-type and mutant strains, respectively, were incubated at various temperatures (0°C, 30°C, 40°C, or 50°C) and then subjected to sucrose-gradient centrifugations. The profiles were practically identical; here those after a 50°C incubation are shown. (C,D) Same as A and B, respectively, but incubation temperature was 60°C; (E,F) same as A and B, respectively, but incubation temperature was 70°C. For details, see Materials and Methods.
In the next experiment, the reconstitution competence of the ribosomal proteins and rRNAs components from the ΔdnaK strain BB11 was tested. 16SrRNAs, (23S + 5S) rRNAs, TP30, and TP50 were extracted from the ribosomal subunits of both dnaK+ and ΔdnaK strains. Homologous reconstitutions (ribosomal proteins and rRNAs from the same strain) and heterologous reconstitutions (ribosomal proteins from one strain and rRNAs from the other, and vice versa) were performed using both types of subunits (50S and 30S). The results shown in Table 1, C and D, reveal that 16S rRNA, (23S + 5S) rRNA, TP50, and TP30 from a ΔdnaK52::cmR strain were as active as their wild-type counterparts, in all different possible combinations. Therefore, we conclude that DnaK is not required for biosynthesis of ribosomal subunits, at least at 30°C, nor for the capabilities of ribosomal components to reconstitute active ribosomal subunits.
Wireman and Sypherd (1975) detected disparity between the physical formation of 30S particles and the acquisition of activity when the reconstitution is performed with mature 16S rRNA: formation of 30S particles occurs at temperatures 15°C lower than the appearance of active subunits. Both features were reported to be improved by the DnaK system (Maki et al. 2002), that is, reconstituted particles became more homogeneous as shown with sucrose-gradient profiles, and tRNA binding occurred in the absence of a heat-activation step. However, tRNA binding was used as the sole functional assay of the reconstituted particles, and the conditions of the applied test include an incubation at 37°C for 15 min (Culver and Noller 1999). Because this incubation could easily provide the heat-activation energy of 38 kcal/mole required for in vitro reconstitution (Traub and Nomura 1969), it raises doubt as to whether the DnaK system does actively stimulate the formation of active particles.
Overexpression of ribosomal protein S4 partially rescued the thermosensitive phenotype of a dnaK756-ts mutant and showed more wild-type profiles of ribosome patterns in sucrose-gradient analyses, and these observations were thought to “suggest that DnaK can functionally interact with ribosomal components in vivo and that this interaction has an effect on ribosome assembly” (Maki et al. 2002). We see an alternative explanation based on the following findings: (1) Binding of the primary (such as S4) and secondary ribosomal proteins to the nascent 16S rRNA is DnaK-independent over a wide range of temperatures tested (El Hage et al. 2001). (2) S4 is a translational autorepressor such that overexpression will decrease expression of other ribosomal proteins in the same operon (Zengel and Lindahl 1994) and also will stimulate rRNA synthesis (Takebe et al. 1985; Torres et al. 2001). It follows that overexpression of S4 does not foster the assembly step from precursors to active particles, but rather has the opposite effect, that is, to increase the amount of precursor particles. The observed rescue of the dnaK756-ts phenotype by overexpression thus asks for another explanation. One possibility is a titration of the misfolded or “sticky” DnaK756 chaperone by S4, which otherwise might sequester one or several cellular components at high temperatures. In line with this interpretation is the observation that the ribosome assembly defect of a dnaK756-ts mutant could not be cured by a roughly equimolar expression of wild-type DnaK (Sbai and Alix 1998).
In any case, the results presented here do not support a role for DnaK during the in vitro formation of functional 30S subunits. Our observations are in agreement with in vivo data demonstrating that biogenesis of the 30S and 50S ribosomal subunits is unaffected in a strain lacking DnaK and grown at 30°C, but is partially and totally defective at 37°C and 42°C, respectively (El Hage et al. 2001). The DnaK chaperone system is therefore necessary for in vivo ribosome assembly at high temperatures, but not at 30°C. Just the opposite would be expected, if DnaK would help to bypass the heat-activation step, namely, a requirement at low temperatures such as 30°C or less. The use of a more sophisticated in vitro reconstitution system, starting from nascent rather than mature ribosomal components, that is, in vitro rRNA transcripts (Semrad and Green 2002), will perhaps be necessary to assign a defined role to extrinsic factors in ribosome assembly.
MATERIALS AND METHODS
Strain BB11 (ΔdnaK52) is chloramphenicol-resistant (cmR, growth in LB medium in the presence of 25 μg/mL chloramphenicol) and temperature-sensitive (ts, does not grow at 37°C), whereas strain MC41.14 (dnaK+) is sensitive to chloramphenicol and thermoresistant (at 30°C–44°C).
Preparations of crude 70S ribosomes and 50S and 30S ribosomal subunits followed Bommer et al. (1996). The ribosomes were derived from the strain CAN20-12E (dnaK+; lacking RNases BN, I, II, and D; Deutscher et al. 1984; Zaniewski et al. 1984) unless otherwise specified. rRNA polyacrylamide-SDS gel electrophoresis, two-dimensional polyacrylamide gel electrophoresis of ribosomal proteins, and poly(U)-directed [14C]-poly(Phe) synthesis were carried out as described (Nierhaus 1990). The presence or absence of RNase I encoded by the rna gene was checked by the methyl-green procedure (Wright 1971). E. coli chaperones DnaK, DnaJ, and GrpE were purchased from StressGen Biotechnologies Corp. The ATPase activity of these chaperones was monitored by the hydrolysis of [γ-32P]ATP according to Seals et al. (1978) with the modifications described by Benaroudj et al. (1994).
The rna-19 allele encoding an inactive form of RNase I and present in strain D10 (Pfennig and Flower 2001) was introduced into strains MC4100 (dnaK+) and BB1553 (MC4100 ΔdnaK52::cmR, sidB1 = mutant rpoH; Bukau and Walker 1990) in two steps. First, the nearby tetR genetic marker zba-601::Tn10 was transduced from the wild-type strain CAG12149 (Singer et al. 1989) into strain D10 with phage P1, selecting for tetracycline resistance, and a tetR, rna-19 P1 transductant was chosen and named D10TET. Second, the rna-19 allele was readily transduced from strain D10TET into MC4100 and BB1553, taking advantage of the tetR marker adjacent to it, yielding strains MC41.14 (dnaK+, tetR, rna-19) and BB11 (ΔdnaK52::cmR, ts, tetR, rna-19), respectively.
Total reconstitution experiments for both 30S and 50S subunits were performed as described (Nierhaus 1990). For reconstitution kinetics (Fig. 1A), 30S reconstitution mixtures were prepared in an ice bath, each containing 4 A260 units of 16S rRNA and 5.6 equivalent units (=0.7 A230 units) of TP30, both extracted from 30S subunits of the strain CAN20-12E (dnaK+) in a final volume of 300 μL, in the presence or absence of 1 mM ATP, 2 μg of DnaK, 1.4 μg of DnaJ, and 1.6 μg of GrpE. Negative controls without 16S rRNA, or TP30, or both, and a positive control with native 30S ribosomal subunits were included. The samples were then incubated at 20°C, 30°C, or 40°C; at the indicated times; aliquots of 30 μL were withdrawn and checked for activity in a poly(U)-directed [14C]-poly(Phe) synthesis assay containing an excess of native 50S ribosomal subunits at 20°C for 2 h.
Quantitative immunodetection of DnaK in native 50S and 30S ribosomal subunits was performed as follows. Protein samples were prepared by boiling aliquots of DnaK, 30S, or 50S ribosomal subunits with SDS gel-loading buffer, and subjected to an electrophoresis on SDS-polyacrylamide (8%) gels. Serial dilutions of the samples allowed quantification of the signals in the linear range as demonstrated before (Lopes-Ferreira and Alix 2002). Immunoblots were then prepared as described in Sambrook et al. (1989). The blots were probed with polyclonal rabbit antibody raised against E. coli HSP70 (DnaK; from Dako, S.A.), and reactive bands were visualized with [125I]-labeled Protein A (Amersham). Protein concentrations in the bands were quantified using the PhosphorImager.
Sedimentation profiles of heat-treated 70S ribosomes
For sedimentation profiles, 5 A260 units of crude 70S ribosomes prepared from strains MC41.14 (dnaK+) and BB11 (ΔdnaK52::cmR) were diluted in the buffer H20M1N200SH4 (20 mM HEPES-KOH at pH 7.6 and 0°C, 1 mM MgCl2, 200 mM NH4Cl, 4 mM β-mercaptoethanol), which promotes dissociation of ribosomal subunits, and incubated at 0°C, 30°C, 40°C, 50°C, 60°C, or 70°C for 10 min. They were then loaded on 12-mL 10% → 30% linear sucrose gradients prepared in the same buffer. After centrifugation at 4°C in a Beckmann SW40 rotor for 24 h at 18,000 rpm, the optical density (A260/mL) profile of each gradient was recorded.
Acknowledgments
We thank Nicolas Lopes-Ferreira for the immunoblot experiment; Moncef Ladjimi for helping to measure the ATPase activity of chaperones; and Daniel Wilson, Madina Iskakova, Yoshika Teraoka, and Viter Marquez for help and discussions. This work was supported by the Centre National de la Recherche Scientifique (U.P.R. 9073), the University Paris 7–Denis Diderot, and by a grant from the Deutsche Forschungsgemeinschaft to K.H.N. (Ni174/8-3).
This is paper No. 4 in the series “Extrinsic Factors in Ribosome Assembly”; paper No. 3 is El Hage et al. (2001).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
rRNA, ribosomal RNA
TP30, total ribosomal proteins of the 30S subunit
TP50, total ribosomal proteins of the 50S subunit
ts, thermosensitive for growth
tetR and cmR, genetic determinants conferring resistance to tetracycline and chloramphenicol, respectively
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5360203.
REFERENCES
- Alix, J.-H. 1993. Extrinsic factors in ribosome assembly. In The translational apparatus: Structure, function, regulation, evolution (eds. K.H. Nierhaus et al.), pp. 173–184. Plenum, New York.
- Alix, J.-H and Guerin, M.F. 1993. Mutant DnaK chaperones cause ribosome assembly defects in Escherichia coli. Proc. Natl. Acad. Sci. 90: 9725–9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benaroudj, N., Fang, B., Triniolles, F., Ghelis, C., and Ladjimi, M.M. 1994. Overexpression in Escherichia coli, purification and characterization of the molecular chaperone HSC70. Eur. J. Biochem. 221: 121–128. [DOI] [PubMed] [Google Scholar]
- Blaha, G., Wilson, D.N., Stoller, G., Fischer, G., Willumeit, R., and Nierhaus, K.H. 2003. Localization of the trigger factor binding site on the ribosomal 50S subunit. J. Mol. Biol. 326: 887–897. [DOI] [PubMed] [Google Scholar]
- Bommer, U., Burkhardt, N., Jünemann, R., Spahn, C.M.T., Triana-Alonso, F.J., and Nierhaus, K.H. 1996. Ribosomes and polysomes. In Subcellular fractionation. A practical approach (eds. J. Graham and D. Rickwoods), pp. 271–301. IRL Press, Oxford University Press, Oxford, UK.
- Bukau, B. and Walker, G.C. 1990. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 9: 4027–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau, B., Deuerling, E., Pfund, C., and Craig, E.A. 2000. Getting newly synthesized proteins into shape. Cell 101: 119–122. [DOI] [PubMed] [Google Scholar]
- Culver, G.M. 2003. Assembly of the 30S ribosomal subunit. Biopolymers 68: 234–249. [DOI] [PubMed] [Google Scholar]
- Culver, G.M. and Noller, H.F. 1999. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA 5: 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher, M.P., Marlor, C.W., and Zaniewski, R. 1984. Ribonuclease T: New exoribonuclease possibly involved in end-turnover of tRNA. Proc. Natl. Acad. Sci. 81: 4290–4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohme, F. and Nierhaus, K.H. 1976. Total reconstitution and assembly of 50S subunits from E. coli ribosomes in vitro. J. Mol. Biol. 107: 585–599. [DOI] [PubMed] [Google Scholar]
- El Hage, A., Sbai, M., and Alix, J.H. 2001. The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli. Mol. Gen. Genet. 264: 796–808. [DOI] [PubMed] [Google Scholar]
- Geyl, D., Böck, A., and Isono, K. 1981. An improved method for 2-D gel electrophoresis: Analysis of mutationally altered ribosomal proteins of E. coli. Mol. Gen. Genet. 181: 309–312. [DOI] [PubMed] [Google Scholar]
- Ha, J.H., Johnson, E.-R., McKay, D.B., Sousa, M.C., Takeda, S., and Wilbanks, S.M. 1999. Structure and mechanism of HSP70 proteins. In Molecular chaperones and folding catalysts: Regulation, cellular function and mechanisms (ed. B. Bukau), pp. 573–607. Harwood Academic Publishers, Amsterdam.
- Kramer, G., Ramachandrian, V., Horowitz, P.M., and Hardesty, B. 2002a. The molecular chaperone DnaK is not recruited to translating ribosomes that lack trigger factor. Arch. Biochem. Biophys. 403: 63–70. [DOI] [PubMed] [Google Scholar]
- Kramer, G., Rauch, T., Rist, W., Vorderwulbecke, S., Patzelt, H., Schulze-Specking, A., Ban, N., Deuerling, E., and Bukau, B. 2002b. L23 protein functions as a chaperone docking site on the ribosome. Nature 419: 171–174. [DOI] [PubMed] [Google Scholar]
- Lopes-Ferreira, N. and Alix, J.-H. 2002. The DnaK chaperone is necessary for α-complementation of β-galactosidase in Escherichia coli. J. Bacteriol. 184: 7047–7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki, J.A., Schnobrich, D.J., and Culver, G.M. 2002. The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol. Cell 10: 129–138. [DOI] [PubMed] [Google Scholar]
- Nierhaus, K.H. 1990. Reconstitution of ribosomes. In Ribosomes and protein synthesis. A practical approach (ed. G. Spedding), pp. 161–189. IRL Press, Oxford University Press, Oxford, UK.
- ———. 1991. The assembly of prokaryotic ribosomes. Biochimie 73: 739–755. [DOI] [PubMed] [Google Scholar]
- Pfennig, P.L. and Flower, A.M. 2001. BipA is required for growth of Escherichia coli K12 at low temperature. Mol. Genet. Genomics 266: 313–317. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sbai, M. and Alix, J.H. 1998. DnaK-dependent ribosome biogenesis in Escherichia coli: Competition for dominance between the alleles dnaK756 and dnaK+. Mol. Gen. Genet. 260: 199–206. [DOI] [PubMed] [Google Scholar]
- Seals, J.R., McDonald, J.M., Bruns, D., and Jarett, L. 1978. A sensitive and precise isotopic assay of ATPase activity. Anal. Biochem. 90: 785–795. [DOI] [PubMed] [Google Scholar]
- Sell, S.M., Eisen, C., Ang, D., Zylicz, M., and Georgopoulos, C. 1990. Isolation and characterization of dnaJ null mutants of Escherichia coli. J. Bacteriol. 172: 4827–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrad, K. and Green, R. 2002. Osmolytes stimulate the reconstitution of functional 50S ribosomes from in vitro transcripts of Escherichia coli 23S rRNA. RNA 8: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, M., Baker, T.A., Schnitzler, G., Deischel, S.M., Goel, M., Dove, W., Jaacks, K.J., Grossman, A.D., Erickson, J.W., and Gross, C.A. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53: 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe, Y., Miura, A., Bedwell, D.M., Tam, M., and Nomura, M. 1985. Increased expression of ribosomal genes during inhibition of ribosome assembly in Escherichia coli. J. Mol. Biol. 184: 23–30. [DOI] [PubMed] [Google Scholar]
- Tal, M., Silberstein, A., and Moyner, K. 1977. In vivo reassembly of 30S ribosomal subunits following their specific destruction by thermal shock. Biochim. Biophys. Acta 479: 479–496. [DOI] [PubMed] [Google Scholar]
- Tomoyasu, T., Mogk, A., Langen, H., Goloubinoff, P., and Bukau, B. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40: 397–413. [DOI] [PubMed] [Google Scholar]
- Torres, M., Condon, C., Balada, J.M., Squires, C., and Squires, C.L. 2001. Ribosomal protein S4 is a transcription factor with properties remarkably similar to NusA, a protein involved in both non-ribosomal and ribosomal RNA antitermination. EMBO J. 20: 3811–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub, P. and Nomura, M. 1969. Structure and function of the Escherichia coli ribosome. Mechanism of assembly of 30S ribosomes studied in vitro. J. Mol. Biol. 40: 391–413. [DOI] [PubMed] [Google Scholar]
- Vysokanov, A.V. 1995. Synthesis of chloramphenicol acetyltransferase in a coupled transcription–translation in vitro system lacking the chaperones DnaK and DnaJ. FEBS Lett. 375: 211–214. [DOI] [PubMed] [Google Scholar]
- Williamson, J.R. 2003. After the ribosome structures: How are the subunits assembled? RNA 9: 165–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wireman, J.W. and Sypherd, P.S. 1975. Temperature dependence for physical and functional reconstitution of 30S ribosomes of E. coli. Biochem. Biophys. Res. Commun. 66: 570–577. [DOI] [PubMed] [Google Scholar]
- Wright, M. 1971. Mutants of Escherichia coli lacking endonuclease I, ribonuclease I, or ribonuclease II. J. Bacteriol. 107: 87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaniewski, R., Petkaites, E., and Deutscher, M.P. 1984. A multiple mutant of Escherichia coli lacking the exoribonucleases RNase II, RNase D, and RNase BN. J. Biol. Chem. 259: 11651–11653. [PubMed] [Google Scholar]
- Zengel, J.M. and Lindahl, L. 1994. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucl. Acid Res. Mol. Biol. 47: 331–370. [DOI] [PubMed] [Google Scholar]