Abstract
The extreme halophile Halobacterium species NRC-1 overcomes external near-saturating salt concentrations by accumulating intracellular salts comparable to those of the medium. This raises the fundamental question of how halophiles can maintain the specificity of protein–nucleic acid interactions that are particularly sensitive to high salts in mesophiles. Here we address the specificity of the essential aminoacylation reaction of the halophile, by focusing on molecular recognition of tRNACys by the cognate cysteinyl-tRNA synthetase. Despite the high salt environments of the aminoacylation reaction, and despite an unusual structure of the tRNA with an exceptionally large dihydrouridine loop, we show that aminoacylation of the tRNA proceeds with a catalytic efficiency similar to that of its mesophilic counterparts. This is manifested by an essentially identical Km for tRNA to those of the mesophiles, and by recognition of the same nucleotide determinants that are conserved in evolution. Interestingly, aminoacylation of the halophile tRNACys is more closely related to that of bacteria than eukarya by placing a strong emphasis on features of the tRNA tertiary core. This suggests an adaptation to the highly negatively charged tRNA sugar-phosphate backbone groups that are the key elements of the tertiary core.
Keywords: Halobacterium species NRC-1, tertiary core, sugar-phosphate backbone, protein–RNA interactions, cysteine
INTRODUCTION
The Halobacterium species NRC-1 is an obligatory extreme halophile that grows and thrives in conditions of near-saturating salinity (4–5 M; Ng et al. 2000). This microorganism of the Archaea domain is widespread in the Dead Sea and Great Salt Lake and constitutes an important member of the microbial population (Mevarech et al. 2000). To cope with the external extreme osmotic pressure, Halobacterium sp. NRC-1 contains correspondingly high concentrations of salts internally, primarily K+ as the cation and Cl− as the anion (Madigan and Oren 1999). The presence of molar concentrations of inorganic ions in the cytoplasm thus requires far-reaching adaptations of the intracellular enzymatic machinery. Of particular interest are enzymes that recognize nucleic acids as substrates. The highly negatively charged phosphate backbone of nucleic acids generates strong electrostatic interactions, which together with the highly acidic nature of the NRC-1 proteins (Kennedy et al. 2001), can make protein–nucleic acid interactions difficult. In mesophiles, these interactions are extremely sensitive to salt (Ha et al. 1992; Lohman et al. 1996). Thus, how the extreme halophile adapts its protein–nucleic acid interactions to high salts is an important question that needs to be addressed to gain insights into the fundamentals of the extreme cellular life.
As an initial step to address the question, we focused on the aminoacylation reaction catalyzed by aminoacyl-tRNA synthetases (aaRS). Aminoacylation is the underlying basis of the genetic code because it provides a direct link between amino acids and trinucleotide sequences encoded as anticodons in tRNAs (Ibba and Söll 2000). There are typically 20 synthetases in an organism, and each of these enzymes activates the cognate amino acid with ATP and transfers the activated aminoacyl-adenylate to the 3′ end of the cognate tRNAs. The conservation of the genetic code in all life forms demands that the specificity of aminoacylation be maintained at the highest possible levels, regardless of cellular environments. This specificity must be executed at both the adenylate synthesis and tRNA aminoacylation steps. Of the two, the tRNA aminoacylation step is particularly challenging because of the macromolecular nature of tRNA and a similar L-shaped tertiary structure that docks tRNAs similarly on surfaces of synthetases (Giegé et al. 1998).
Aminoacylation of tRNACys by CysRS is of special interest in Halobacterium sp. NRC-1. Not only will this reaction provide a model to study synthetase–tRNA interactions in hypersalinity, but it will also shed light on an unusual tRNA and the functional relationship of this tRNA with its counterparts in bacteria and eukarya. Analysis of the sequenced genome of Halobacterium sp. NRC-1 (Ng et al. 2000) has revealed that tRNACys, while having U73 and the GCA anticodon common to all cysteine tRNAs (Sprinzl et al. 1998), features an exceptionally large dihydrouridine (D) loop of 15 nt and only two possible Watson-Crick base pairs in the D-stem (Fig. 1). This unusual secondary structure, also found in the sequenced tRNACys of Haloferax volcanii (Gupta 1984), is distinct from all other halophile tRNAs and does not conform to the standard tRNACys structure of either the bacterial type or eukaryotic type. Specifically, all bacterial tRNACyss can fold into a D3V4 conformation (3 bp in the D-stem and 4 nt in the variable, V, loop), whereas eukaryotic tRNACyss fold into the more common D4V5 conformation (Fig. 1; Hou et al. 1999; Ming et al. 2002). Interestingly, the difference in tRNA secondary structure is correlated with a difference in aminoacylation. Whereas aminoacylation of both types is strictly dependent on the conserved U73 and the GCA anticodon (Lipman and Hou 1998), that of the bacterial type also depends on recognition of the tRNA tertiary core (Hamann and Hou 1997, 2000). The tertiary core, consisting of the D-stem–loop, V-loop, and TΨC-stem–loop, contains extensive stacking and hydrogen-bonding interactions that stabilize joining of the two long arms of the tRNA L shape. Recognition of the tertiary core in bacterial tRNAs is focused, not on specific bases, but on the sugar-phosphate backbone, which is highly negatively charged (Ming et al. 2002). This emphasis on the tertiary core is not used in aminoacylation of eukaryotic tRNACys (Ming et al. 2002).
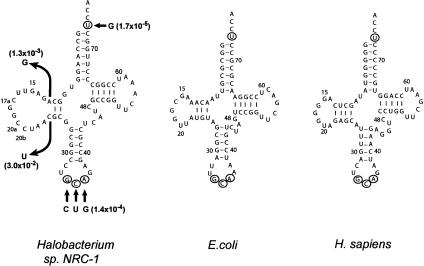
FIGURE 1.
Sequence and cloverleaf structure of Halobacterium species NRC-1, Escherichia coli, and Homo sapiens tRNACys, in which the conserved U73 and the GCA anticodon are circled. Nucleotides of the Halobacterium sp. NRC-1 that have been tested by mutations are indicated by arrows, and the effects of mutation in reducing the catalytic efficiency of aminoacylation are indicated in parentheses.
Although tRNACys of Halobacterium sp. NRC-1 is neither of the bacterial nor the eukaryotic type, the CysRS of the extreme halophile is clearly of the bacterial type. Extensive sequence alignment has shown that the bacterial type CysRS differs from the eukaryotic type by lacking a large insertion peptide of >100 amino acids in the catalytic domain (Fig. 2; Newberry et al. 2002). Although the function of this insertion peptide is not known, it is clearly absent from the halophile CysRS, indicating that the enzyme is of the bacterial type (Fig. 2). This has raised the question of whether the halophile enzyme can recognize the unusual tRNACys using the bacterial strategy with an emphasis on features of the tertiary core. To address this question, and to develop a system for analysis of the salt adaptation of synthetase–tRNA interactions, we cloned and expressed both the halophile CysRS and tRNACys in Escherichia coli for analysis of aminoacylation in vitro. Because of the extreme halophilicity of the CysRS, which renders the enzyme insoluble in E. coli when expressed, we have developed a procedure to reconstitute the enzyme. Our studies indicate that, despite the unusual structural feature of the tRNA and the extreme halophilic aminoacylation condition, the kinetic parameters of aminoacylation of the halophile tRNACys are quantitatively similar to those of bacteria and eukarya, represented by the E. coli and human species, respectively. In addition, aminoacylation of the halophile tRNA is sensitive to mutations in the tertiary core, implying a resemblance to the bacterial type and a molecular adaptation to the negatively charged sugar-phosphate backbone.
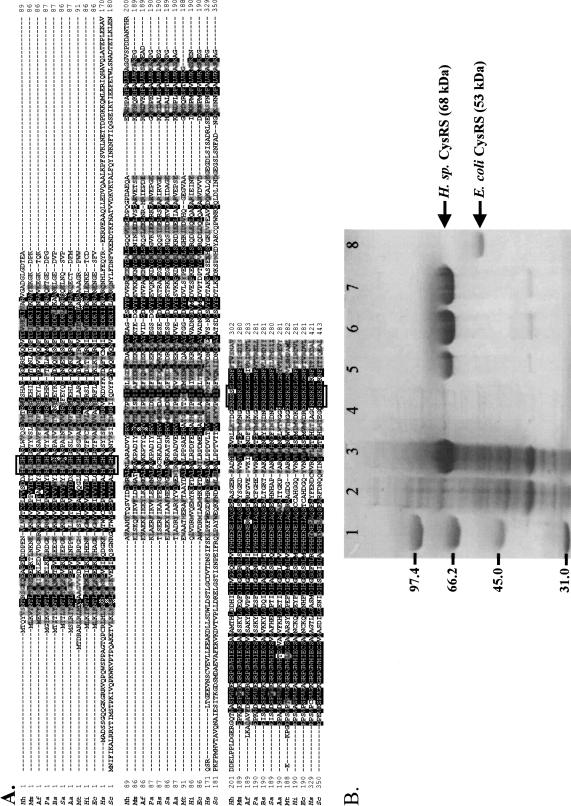
FIGURE 2.
(A) A multiple sequence alignment of CysRS of Hh (Halobacterium species NRC-1, also known as H. halobium), Mm (Methanococcus maripaludis), Af (Archaeoglobus fulgidus), Pa (Pyrococcus abyssi), Bs (Bacillus subtilis), Sa (Staphylococcus aureus), Aa (Aquifex aeolicus), Mt (Mycobacterium tuberculosis), Hi (Haemophilus influenzae), Ec (Escherichia coli), Hs (Homo sapiens), and Sc (Saccharomyces cerevisiae), showing division of the bacterial and eukaryotic type. (B) An SDS-PAGE analysis of expression of Halobacterium sp. NRC-1 CysRS in E. coli and purification by a metal-affinity resin. (Lane 1) Molecular markers; (lane 2) whole cell lysate before induction with IPTG; (lane 3) whole cell lysate after induction; (lane 4) the soluble fraction of the cell lysate; (lane 5) the insoluble fraction of the cell lysate; (lane 6) the bound fraction of the metal affinity resin; (lane 7) the reconstituted CysRS, and (lane 8) the cloned and expressed E. coli CysRS as a marker.
RESULTS AND DISCUSSION
Overexpression of Halobacterium sp. NRC-1 CysRS and reconstitution of activity in vitro
The gene for the halophile CysRS was identified from the genome (Ng et al. 2000), based on conserved motifs in the predicted protein sequence that are common to other class I CysRSs (Fig. 2A). To obtain sufficient quantities of the enzyme for biochemical studies, efforts were made to overexpress the gene as a recombinant enzyme fused to an N-terminal His-tag. The homologous expression in the halophile itself was an attractive idea, because Halobacterium sp. was genetically tractable, plasmids for overexpression have been described (Turner et al. 1999; Peck et al. 2000), and protein products of expression can be naturally folded in the hypersaline conditions. However, initial attempts of expressing CysRS under the control of the bacterio-odopsin (bop) promoter were not successful. There was no easily detectable CysRS in the whole cell lysate, as probed by antisera against the His-tag. In contrast, the heterologous expression in E. coli, using the plasmid pET19b under control of the inducible lac promoter, yielded easily detectable enzyme on a Coomassie blue-stained gel at levels of at least 2 mg/L of cell culture. The apparent molecular weight of the His-tagged enzyme on SDS-PAGE was 68 kD (Fig. 2B), although the calculated molecular weight is 58 kD.
The heterologous expression in E. coli, however, segregated the CysRS into insoluble cellular fractions, indicating inappropriate folding. A screen with different reagents (urea, guanidium-HCl, guanidium-thiocyanate, and Triton X-100) identified that 8 M urea was necessary to completely solubilize the enzyme. The urea-denatured enzyme was then bound to a metal-affinity resin, eluted with imidazole, and immediately dialyzed in solution D, which closely approximates the intracellular salt concentrations of the halophile (Gupta 1984). To evaluate the reconstitution method, the aminoacylation activity of the reconstituted enzyme was compared with that of the native enzyme. The latter was prepared by fractionation of the cell lysate of Halobacterium sp. over the phenyl Sepharose resin using principles of hydrophobic interaction chromatography. The native CysRS was identified by its ability to catalyze aminoacylation from a decreasing concentration gradient of ammonium sulfate.
The aminoacylation activity of the reconstituted CysRS was comparable to that of the native enzyme. For example, with 1 μM tRNACys transcript of the halophile as the substrate, the rate of synthesis of cysteinyl-tRNACys by the reconstituted enzyme (0.36 pmole/min per nM enzyme) was within twofold of that of the native enzyme (0.73 pmole/min per nM enzyme). This validated the reconstitution method and provided an experimental framework to study aminoacylation under hypersaline conditions. The ability to use the unmodified tRNA transcript as the substrate also facilitated rapid analysis of tRNA features that are important for aminoacylation. All known class I CysRSs in eukarya and bacteria recognize tRNACys without the need of specific nucleotide modifications (Hou et al. 1993; Lipman and Hou 1998).
Aminoacylation by the reconstituted CysRS and salt dependence
The kinetic parameters of aminoacylation by the reconstituted CysRS were determined to compare the halophilic enzyme with its bacterial and eukaryotic counterparts, represented by the previously studied E. coli and human enzymes (Christian et al. 2000; Davidson et al. 2001), respectively (Table 1). Here, the halophilic aminoacylation was performed at 2.8 M KCl, using Cl− as the sole source of the anion. Although the intracellular concentrations of the halophile have been estimated as 1.4 M Na+, 4.6 M K+, and 3.6 M Cl−, these were difficult to reproduce without definitive knowledge of organic anions that joined with Cl− to balance cations (Bayley and Morton 1978). Under our assay conditions, the steady-state parameter Km for tRNACys was virtually identical to those of the E. coli and human, indicating an overall similar affinity to the tRNA substrate. The kcat of the halophilic reaction was fourfold to eightfold lower, resulting in a mildly reduced catalytic efficiency of aminoacylation. The comparable parameters indicate that, when assayed under conditions close to the natural, the halophilic reaction can be almost as efficient as those of the mesophilic enzymes.
TABLE 1.
Kinetic parameters of aminoacylation with unmodified transcripts of tRNACys
| tRNA | CysRS | Km (μM) | kcat (sec−1) | kcat/Km (M−1 sec−1) | Fold |
| Ec tRNACys | E. coli | 1.16 ± 0.01 | 2.46 ± 0.06 | 2.12 × 106 | 8.0 |
| Hs tRNACys | Human | 1.4 ± 0.1 | 1.5 ± 0.1 | 1.01 × 106 | 3.8 |
| Hsp tRNACys, wt | Hsp | 1.39 ± 0.20 | 0.36 ± 0.13 | 2.65 × 105 | 1.0 |
| Hsp, U73G | Hsp | — | — | 4.3 | 1.7 × 10−5 |
| Hsp, CUG anticodon | Hsp | 53.1 ± 15.6 | (1.7 ± 0.4) × 10−3 | 35.9 | 1.4 × 10−4 |
| Hsp, A12G | Hsp | 27.2 ± 0.85 | (9.0 ± 0.9) × 10−3 | 3.33 × 102 | 1.3 × 10−3 |
| Hsp, C23U | Hsp | 9.92 ± 1.05 | (7.8 ± 0.2) × 10−2 | 7.96 × 103 | 3.0 × 10−2 |
The steady-state parameters Km and kcat were determined with catalytic enzyme concentrations that were at least 20-fold below the lowest concentrations of a given tRNA substrate. This amounted to 0.5–1.0 nM E. coli (Ec) or human (Hs) CysRS with 0.2–6 μM tRNA and 3–7 nM Halobacterium sp. NRC-1 CysRS with 0.5–8 μM wild-type tRNA and 5–100 μM mutant tRNAs. The parameters for E. coli CysRS (Christian et al. 2000) and human CysRS (Davidson et al. 2001) were published previously.
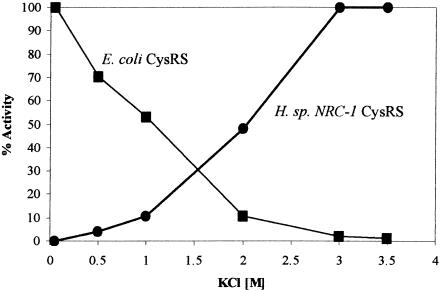
The salt dependence of the halophilic aminoacylation was tested. One purpose was to test if the catalytic efficiency of aminoacylation could improve further as the KCl concentration in the assay buffer approached that of the natural. Also, the halophilic enzyme should respond to salt differently than its mesophilic counterparts, based on studies of other halophilic proteins (Bergqvist et al. 2002; Wright et al. 2002). This difference could provide the basis for further investigation of the halophilicity of the synthetase–tRNA interaction. To test these ideas, the reconstituted CysRS and E. coli CysRS were compared for their sensitivity to KCl concentrations in the aminoacylation reaction (Fig. 3). The difference between the two enzymes was dramatic. Whereas the activity of the E. coli enzyme decreased with increasing salt concentrations and virtually disappeared at near 3 M, that of the halophile enzyme displayed the opposite trend and reached its maximum. Thus, the activity of the halophilic enzyme can improve by raising KCl from 2.8 to 3.0 M. Also, as the activity of the E. coli enzyme at 2.8 M KCl in the regular assay buffer was nearly completely inhibited, this indicates that the regular assay reaction measures only the activity of the halophilic enzyme. Even if there might be a contamination of the endogenous E. coli CysRS during in vitro reconstitution of the halophilic enzyme, the contaminant would not be active.
FIGURE 3.
The aminoacylation activity of the Escherichia coli and Halobacterium species NRC-1 CysRS as a function of the concentration of KCl. The activity of each enzyme (▪ E. coli; • NRC-1) is expressed as a fraction of the maximum.
Nucleotide determinants for aminoacylation of the halophile tRNACys
Nucleotides important for aminoacylation of the halophile tRNACys were identified by mutational analysis. Because this analysis has been performed for the E. coli and human tRNACys, which provided a basis for comparison, mutations of the halophile tRNA were created by selected substitutions rather than the complete and exhaustive spectrum of substitutions. The universally conserved U73 and GCA anticodon for tRNACys were tested first (Fig. 1; Table 1). These elements play an essential role in aminoacylation of both the bacterial and eukaryotic types (Komatsoulis and Abelson 1993; Lipman and Hou 1998). Indeed, alteration of U73 to G in the halophile tRNA had a major deleterious effect, which prevented accurate measurement of individual parameters. Based on an estimation of Km at 1.5–2.0 mM, the catalytic efficiency was determined by measuring the initial rate at 0.1 mM tRNACys, which showed a decrease by nearly 5 orders of magnitude from that of the wild type. Higher tRNACys concentrations were not practical in the assay. The significant effect of the mutation is quantitatively similar to that in E. coli and human tRNACys, emphasizing a conserved role of U73 in the acceptor stem (Komatsoulis and Abelson 1993; Hamann and Hou 1995; Lipman and Hou 1998). The importance of the GCA anticodon was tested in a single mutant that carried mutations of the three anticodon nucleotides at once: G34C, C35U, and A36G. The activity of the mutant was also severely defective, because of an increase of Km by 50-fold, a decrease of kcat by 102–103-fold, and an overall reduction in efficiency by 104-fold. Previous analysis of the anticodon of E. coli tRNACys, although in single substitutions, indicated a similar effect. Specifically, mutations of G34 in E. coli tRNACys alone were sufficient to reduce the activity by 103-fold, whereas mutations of C35 and A36, respectively, reduced the activity by 100–300-fold (Komatsoulis and Abelson 1993). Conceivably, each of these individual reductions in E. coli tRNACys might be compromised upon a complete substitution of all anticodon nucleotides to produce the effect of 104-fold as observed in the halophile tRNACys.
The functional significance of U73 and the anticodon in the halophile tRNACys indicates conservation of a similar mode of tRNA recognition as in all other cysteine tRNAs. To determine if this mode in the halophile is more closely related to the bacterial or eukaryotic type, mutations in the unusual D-stem–loop were tested. The bacterial type is particularly sensitive to mutations in this region. For example, mutations in E. coli tRNACys that introduce a fourth base pair in the D-stem (creating a D4V4 structure), or those that alter the conserved purine 15:pyrimidine 48 tertiary base pair, can reduce aminoacylation by 102-fold (Hamann and Hou 1997, 2000). In contrast, mutations that alter the D-stem structure in the eukaryotic type have little effect on aminoacylation (Lipman and Hou 1998). To test the effect of mutation, the D-stem of the halophile tRNA was first mapped by the chemical probe dimethyl sulfate (DMS), which modifies the N1 and N3 positions of the adenine and cytosine bases, respectively. This mapping showed that A12 and C23 were both accessible to the modification, as can be visualized by an effect of the modification on primer extension (C. Evilia and Y.-M. Hou, unpubl.). The chemical accessibility of A12 and C23 indicates that they are not base-paired, thus confirming two Watson-Crick base pairs (10:25, 11:24) in the D-stem.
Two mutants, the A12G and C23U, were created in the halophile tRNACys (Fig. 1; Table 1). These mutations extended the D-stem by creating a third base pair as G12:C23 and A12:U23, respectively. Both mutants were reduced in aminoacylation by 102–103-fold relative to the wild type, which were more significant than those observed in E. coli tRNACys in the range of 10–100-fold (Hamann and Hou 1997). Of the two mutants, the A12G mutant showed a stronger defect in both Km and kcat, indicating that a G12:C23 base pair is potentially more harmful to aminoacylation than A12:U23. Notably, although both mutations can reduce the flexibility of the tertiary core by introducing an extra base pair in the D-stem, a G12:C23 base pair would create three consecutive G:C base pairs that could reduce the flexibility even further. This analysis indicates that the flexibility of the core is important for aminoacylation. The wild-type tRNACys, having only two base pairs in the D-stem, would have the highest flexibility. Interestingly, extensive studies of the core of E. coli tRNACys have indicated that the flexibility of the tertiary core is an important factor for aminoacylation of the bacterial-type tRNA (Christian et al. 2000; Sherlin et al. 2000).
Thus, despite an unusual tRNA structure and an extremely halophilic condition, aminoacylation of the halophile tRNACys is of the bacterial type. This has confirmed the functional significance of the bacterial-like CysRS of the halophile (Fig. 2). One major feature of the bacterial-type recognition is a strong emphasis on the sugar-phosphate backbone of tRNA that is not seen in the eukaryotic type (Ming et al. 2002). For example, based on a phosphorothioate-interference assay, backbone groups that are important for the bacterial type have been identified. These include phosphate groups not only in the tertiary core, but in the anticodon loop adjacent to the three anticodon bases. In contrast, the same interference assay identified no phosphates of significance in aminoacylation of the eukaryotic type (Ming et al. 2002). The interference assay has not been successfully tested on the halophile tRNACys, because of the technical difficulty of applying the assay in hypersaline conditions, where the phosphorothioate-modified tRNA is extremely unstable. Thus, the possibility exists that aminoacylation of the halophile tRNA, although sensitive to mutations in the tertiary core, may not directly depend on the sugar-phosphate backbone as in the bacterial type. However, enzymes that recognize the tRNA tertiary core typically recognize the sugar-phosphate backbone, rather than specific nucleotide sequences, as shown in the crystal structure of EF-Tu complexed with E. coli tRNACys (Nissen et al. 1999). The lesser role of specific nucleotide sequences in aminoacylation of the halophile tRNA is also consistent with the fact that this tRNA shares no common sequence in the tertiary core with those of its E. coli and human counterparts (Fig. 1). The implication therefore is that aminoacylation of the halophile tRNA would depend on selected phosphate groups of the tRNA backbone. Some of these phosphate groups are likely to be located in the tertiary core, as evidenced in the sensitivity to mutations in the core, but they may not be completely identical to those in E. coli tRNACys, because of the distinct tertiary core in the halophile tRNA.
Importantly, the bacterial-type recognition by the halophile CysRS indicates an adaptation that not only must overcome the extreme halophilicity to bring the synthetase and tRNA together, but also must discriminate among highly negatively charged phosphate groups to achieve the aminoacylation specificity necessary for a cellular life. This most likely requires closer contact between the halophile CysRS and the backbone of tRNACys for better discrimination, thus making the challenge of the molecular adaptation to the extreme halophilicity even stronger. This report, therefore, presents a fascinating biological situation that will be addressed in future studies to better understand a synthetase–tRNA interaction that achieves specificity in extreme salinity.
MATERIALS AND METHODS
Construction of the genes for tRNACys and CysRS of Halobacterium sp. NRC-1
The gene for tRNACys was identified from the sequenced genome of Halobacterium sp. NRC-1 and was constructed by enzymatic extension of two overlapping oligonucleotides. The synthesized gene was cloned into the HindIII and BamHI restriction sites of the plasmid pTFMa under the control of the bacterial phage T7 RNA polymerase promoter. The gene for CysRS was also identified from the genome and was amplified by PCR and cloned into the NdeI and BamHI sites of the plasmid pET19b under the control of the inducible lac promoter. This construction enables expression of an N-terminally His-tagged fusion protein.
Expression of tRNACys of Halobacterium sp. NRC-1
The tRNA gene was transcribed by T7 RNA polymerase from the tRNA plasmid that has been restricted at the BstNI site (Hou et al. 1993). The transcribed tRNA was gel-purified, resuspended in TE, and folded into the native form by heating at 80°C for 3 min and reannealing at 37°C in solution D (3.5 M KCl, 100 mM Mg acetate, 10 mM Tris-HCl at pH 7.5, and 5 mM β-mercaptoethanol). Mutations in the tRNA gene were created by the Kunkel method and confirmed by sequence analysis (Kunkel et al. 1987).
Purification of Halobacterium sp. NRC-1 CysRS
The native CysRS was isolated from sonicated cell lysate (0.1 vol solution D/vol of cell paste). After a low-speed (20,000g, 30 min) and a high-speed (100,000g, 1 h) spin, the cleared cell lysate was dialyzed overnight against solution D, 5 mM DTT, and 40% glycerol. The lysate (~100 mg) was treated twice with an equal volume of 3 M (NH4)2SO4, each time centrifuged to remove precipitation, adjusted to 2.5 M of (NH4)2SO4, and loaded onto a phenyl Sepharose column (1 × 5 cm) in 3 M (NH4)2SO4, 0.5 M KCl, 20 mM Tris-HCl (pH 8.0), 10 mM β-mercaptoethanol, and 10% glycerol. After washing the column with the starting buffer, bound proteins were eluted by step gradients of decreasing concentrations of (NH4)2SO4 (3, 2.5, 2.0, 1.5, 1.0, 0.5, and 0 M), two column volumes of each step. The CysRS activity was identified by the aminoacylation assay, using the total tRNA of Halobacterium sp. NRC-1 as the substrate. It was pooled and dialyzed in solution D, 5 mM DTT, and 40% glycerol before storage at −20°C.
For heterologous expression and purification, the CysRS gene in pET19b was expressed in the E. coli strain BL21(DE3)pSJS grown in LB medium containing ampicillin at 100 μg/mL, spectinomycin at 50 μg/mL, and induced at OD600 0.6–0.8 for 3 h by 1 mM IPTG. Cells were collected, resuspended in a buffer (20 mM Tris-HCl at pH 8.0, 1 M KCl, 0.1% Triton-X, and 5 mM β-mercaptoethanol) and sonicated in the presence of lysozyme (100 μg/mL). The sonicated cell lysate was centrifuged, and the insoluble pellet was resuspended in the denaturing buffer (8 M urea, 20 mM Tris-HCl at pH 7.5, 5 mM β-mercaptoethanol) at 4°C for 8–16 h to solubilize CysRS. After removal of insoluble materials by a spin of 10,000g for 30 min, the soluble fraction was applied to the Co+2-based Talon affinity resin (ClonTech), and the unbound fraction was removed by the wash buffer (8 M urea, 50 mM HEPES at pH 7.5, 500 mM NaCl, 5 mM β-mercaptoethanol). The CysRS was eluted with 150 mM imidazole in the wash buffer, dialyzed in solution D, and concentrated in the dialysis bag by using PEG1500-2000 outside the dialysis membrane to absorb water. Protein concentration was determined by the Bradford method (BioRad), and the active fraction was determined by a modified active-site titration burst assay in solution D (Fersht et al. 1975).
Assay for aminoacylation of tRNACys by Halobacterium sp. NRC-1 CysRS
The assay reaction (24 μL), mixed with 6 μL of a 4× cocktail (3.5 M KCl, 40 mM MgCl2, 8 mM ATP, 80 mM Tris-HCl at pH 7.5, 50 μM cysteine, and 5 mM DTT), 14 μL of a reannealed tRNA, and 4 μL of enzyme in solution D, contained the final KCl at 2.85 M. The reaction was incubated at 40°C, and aliquots were quenched by carboxymethylation and trichloroacetic acid precipitation on filter pads as described (Cole and Schimmel 1970).
Acknowledgments
This work was supported by the NIH (grant GM56662 to Y.M.H.), an NSF postdoctoral fellowship (DBI-0074388 to C.E.), and the NSF (grant MCB-0296017 to S.D.). We thank Rich Lipman and members of the Hou laboratory for discussion and Brian Berquist for experimental assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5320603.
REFERENCES
- Bayley, S.T. and Morton, R.A. 1978. Recent developments in the molecular biology of extremely halophilic bacteria. CRC Crit. Rev. Microbiol. 6: 151–205. [DOI] [PubMed] [Google Scholar]
- Bergqvist, S., Williams, M.A., O’Brien, R., and Ladbury, J.E. 2002. Reversal of halophilicity in a protein–DNA interaction by limited mutation strategy. Structure (Camb.) 10: 629–637. [DOI] [PubMed] [Google Scholar]
- Christian, T., Lipman, R.S., Evilia, C., and Hou, Y.M. 2000. Alternative design of a tRNA core for aminoacylation. J. Mol. Biol. 303: 503–514. [DOI] [PubMed] [Google Scholar]
- Cole, F.X. and Schimmel, P.R. 1970. On the rate law and mechanism of the adenosine triphosphate–pyrophosphate isotope exchange reaction of amino acyl transfer ribonucleic acid synthetases. Biochemistry 9: 480–489. [DOI] [PubMed] [Google Scholar]
- Davidson, E., Caffarella, J., Vitseva, O., Hou, Y.M., and King, M.P. 2001. Isolation of two cDNAs encoding functional human cytoplasmic cysteinyl-tRNA synthetase. Biol. Chem. 382: 399–406. [DOI] [PubMed] [Google Scholar]
- Fersht, A.R., Ashford, J.S., Bruton, C.J., Jakes, R., Koch, G.L., and Hartley, B.S. 1975. Active site titration and aminoacyl adenylate binding stoichiometry of aminoacyl-tRNA synthetases. Biochemistry 14: 1–4. [DOI] [PubMed] [Google Scholar]
- Giegé, R., Sissler, M., and Florentz, C. 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 26: 5017–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R. 1984. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 259: 9461–9471. [PubMed] [Google Scholar]
- Ha, J.H., Capp, M.W., Hohenwalter, M.D., Baskerville, M., and Record Jr., M.T. 1992. Thermodynamic stoichiometries of participation of water, cations and anions in specific and non-specific binding of lac repressor to DNA. Possible thermodynamic origins of the “glutamate effect” on protein–DNA interactions. J. Mol. Biol. 228: 252–264. [DOI] [PubMed] [Google Scholar]
- Hamann, C.S. and Hou, Y.M. 1995. Enzymatic aminoacylation of tRNA acceptor stem helices with cysteine is dependent on a single nucleotide. Biochemistry 34: 6527–6532. [DOI] [PubMed] [Google Scholar]
- ———. 1997. An RNA structural determinant for tRNA recognition. Biochemistry 36: 7967–7972. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Probing a tRNA core that contributes to aminoacylation. J. Mol. Biol. 295: 777–789. [DOI] [PubMed] [Google Scholar]
- Hou, Y.M., Westhof, E., and Giegé, R. 1993. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc. Natl. Acad. Sci. 90: 6776–6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y.M., Motegi, H., Lipman, R.S., Hamann, C.S., and Shiba, K. 1999. Conservation of a tRNA core for aminoacylation. Nucleic Acids Res. 27: 4743–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba, M. and Söll, D. 2000. Aminoacyl-tRNA Synthesis. Annu. Rev. Biochem. 69: 617–650. [DOI] [PubMed] [Google Scholar]
- Kennedy, S.P., Ng, W.V., Salzberg, S.L., Hood, L., and DasSarma, S. 2001. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsoulis, G.A. and Abelson, J. 1993. Recognition of tRNACys by Escherichia coli cysteinyl-tRNA synthetase. Biochemistry 32: 7435–7444; Erratum 32: 13374. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A., Roberts, J.D., and Zakour, R.A. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154: 367–382. [DOI] [PubMed] [Google Scholar]
- Lipman, R.S. and Hou, Y.M. 1998. Aminoacylation of tRNA in the evolution of an aminoacyl-tRNA synthetase. Proc. Natl. Acad. Sci. 95: 13495–13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman, T.M., Overman, L.B., Ferrari, M.E., and Kozlov, A.G. 1996. A highly salt-dependent enthalpy change for Escherichia coli SSB protein–nucleic acid binding due to ion–protein interactions. Biochemistry 35: 5272–5279. [DOI] [PubMed] [Google Scholar]
- Madigan, M.T. and Oren, A. 1999. Thermophilic and halophilic extremophiles. Curr. Opin. Microbiol. 2: 265–269. [DOI] [PubMed] [Google Scholar]
- Mevarech, M., Frolow, F., and Gloss, L.M. 2000. Halophilic enzymes: Proteins with a grain of salt. Biophys. Chem. 86: 155–164. [DOI] [PubMed] [Google Scholar]
- Ming, X., Smith, K., Suga, H., and Hou, Y.M. 2002. Recognition of tRNA backbone for aminoacylation with cysteine: Evolution from Escherichia coli to human. J. Mol. Biol. 318: 1207–1220. [DOI] [PubMed] [Google Scholar]
- Newberry, K.J., Hou, Y.M., and Perona, J.J. 2002. Structural origins of amino acid selection without editing by cysteinyl-tRNA synthetase. EMBO J. 21: 2778–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W.V., Kennedy, S.P., Mahairas, G.G., Berquist, B., Pan, M., Shukla, H.D., Lasky, S.R., Baliga, N.S., Thorsson, V., Sbrogna, J., et al. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. 97: 12176–12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Thirup, S., Kjeldgaard, M., and Nyborg, J. 1999. The crystal structure of Cys-tRNACys–EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 7: 143–156. [DOI] [PubMed] [Google Scholar]
- Peck, R.F., Dassarma, S., and Krebs, M.P. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35: 667–676. [DOI] [PubMed] [Google Scholar]
- Sherlin, L.D., Bullock, T.L., Newberry, K.J., Lipman, R.S., Hou, Y.M., Beijer, B., Sproat, B.S., and Perona, J.J. 2000. Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases. J. Mol. Biol. 299: 431–446. [DOI] [PubMed] [Google Scholar]
- Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, G.J., Reusch, R., Winter-Vann, A.M., Martinez, L., and Betlach, M.C. 1999. Heterologous gene expression in a membrane-protein-specific system. Protein Expr. Purif. 17: 312–323. [DOI] [PubMed] [Google Scholar]
- Wright, D.B., Banks, D.D., Lohman, J.R., Hilsenbeck, J.L., and Gloss, L.M. 2002. The effect of salts on the activity and stability of Escherichia coli and Haloferax volcanii dihydrofolate reductases. J. Mol. Biol. 323: 327–344. [DOI] [PubMed] [Google Scholar]