Abstract
The mRNA export pathway is highly conserved throughout evolution. We have used RNA interference (RNAi) to functionally characterize bona fide RNA export factors and components of the exon–exon junction complex (EJC) in Caenorhabditis elegans. RNAi of CeNXT1/p15, the binding partner of CeNXF1/TAP, caused early embryonic lethality, demonstrating an essential function of this gene during C. elegans development. Moreover, depletion of this protein resulted in nuclear accumulation of poly(A)+ RNAs, supporting a direct role of NXT1/p15 in mRNA export in C. elegans. Previously, we have shown that RNAi of CeSRm160, a protein of the EJC complex, resulted in wild-type phenotype; in the present study, we demonstrate that RNAi of CeY14, another component of this complex, results in embryonic lethality. In contrast, depletion of the EJC component CeRNPS1 results in no discernible phenotype. Proteins of the REF/Aly family act as adaptor proteins mediating the recruitment of the mRNA export factor, NXF1/TAP, to mRNAs. The C. elegans genome encodes three members of the REF/Aly family. RNAi of individual Ref genes, or codepletion of two Ref genes in different combinations, resulted in wild-type phenotype. Simultaneous suppression of all three Ref genes did not compromise viability or progression through developmental stages in the affected progeny, and only caused a minor defect in larval mobility. Furthermore, no defects in mRNA export were observed upon simultaneous depletion of all three REF proteins. These results suggest the existence of multiple adaptor proteins that mediate mRNA export in C. elegans.
Keywords: mRNA export, RNA interference, exon–exon junction complex, REF/Aly, C. elegans
INTRODUCTION
The different classes of RNAs are exported as ribonucleoprotein complexes (RNPs) via distinct pathways that depend on different export receptors (for review, see Stutz and Rosbash 1998). Whereas transport of noncoding RNAs, such as tRNAs, rRNAs, and U snRNAs, is dependent on Ran-GTP and is mediated by members of the karyopherin family of nucleocytoplasmic transport receptors, bulk messenger RNA (mRNA) export is not dependent on Ran (for review, see Cullen 2003).
Progress in the characterization of yeast and mammalian mRNA export factors has been reviewed recently (Zenklusen and Stutz 2001; Lei and Silver 2002). Different experimental approaches such as genetic screens in yeast and biochemistry in higher eukaryotes have been used to identify mRNA export factors. For instance, Mex67p was identified through its genetic interaction with the nucleoporin Nup85p, and was shown to be essential for poly(A)+ RNA export (Segref et al. 1997) and able to mediate nuclear export of a variety of RNA polymerase II transcripts (Hurt et al. 2000). Other protein factors that are essential for mRNA export include the nuclear pore-associated proteins Gle1p and Gle2p (Murphy et al. 1996; Watkins et al. 1998), and Dbp5p, an RNA helicase of the DEAD-box family (Snay-Hodge et al. 1998; Tseng et al. 1998). A variety of mRNA-binding proteins, such as nucleocytoplasmic shuttling hnRNP proteins may also have a role in mRNA export. For instance, in yeast, export of poly(A)+ RNA is blocked in a temperature-sensitive mutant of the hnRNP protein Npl3p (Lee et al. 1996). More recently, two shuttling SR proteins, SRp20 and 9G8, have been found to promote the export of intronless mRNAs and to function as mRNA export adaptors (Huang and Steitz 2001; Huang et al. 2003). Nuclear retention sequences have been identified in the nonshuttling proteins, hnRNP C and SC35 (Nakielny and Dreyfuss 1996; Cazalla et al. 2002), and removal of these nonshuttling RNA-binding proteins may constitute a requisite for mRNA export.
In simian type-D retroviruses, nuclear export of incompletely spliced mRNAs is mediated by a cis-acting RNA sequence, the constitutive transport element (CTE), that is sufficient to induce nuclear export in the absence of any viral gene product (Bray et al. 1994). An excess of CTE RNA blocks cellular mRNA export in Xenopus oocytes, indicating that a cellular factor that binds the CTE sequence and promotes nuclear export of viral transcripts also mediates mRNA export (Pasquinelli et al. 1997; Saavedra et al. 1997). This factor was identified and proved to be the human homolog of Mex67, termed TAP or NXF1 (for nuclear export factor one; Gruter et al. 1998). NXF1/TAP is a nucleocytoplasmic shuttling protein that can be cross-linked to poly(A)+ RNA and interacts with FG-containing nucleoporins (Bear et al. 1999; Kang and Cullen 1999; Bachi et al. 2000). NXF1 is member of a family of related proteins in higher eukaryotes consisting of two members in C. elegans, four members in Drosophila, and at least five members in humans (Herold et al. 2000). NXF1/TAP heterodimerizes with NXT1/p15, a small protein related to the Ran-GDP-binding nuclear transport factor NTF2, and this heterodimer subsequently binds to the nucleoporins and facilitates export of poly(A)+ RNA (Katahira et al. 1999; Fribourg et al. 2001; Wiegand et al. 2002). There are two NXT/p15 proteins (NXT1/p15-1 and NXT2/p15-2) in humans, both of which bind multiple NXF proteins, and a single protein, Mtr2p, in Saccharomyces cerevisae (Herold et al. 2000). Although Mtr2p is not related to p15 protein in sequence, it heterodimerizes with Mex67p, and is therefore a functional homolog of p15 (Santos-Rosa et al. 1998). The mRNA export pathway is evolutionarily conserved, and human TAP can rescue mRNA export in Mex67/Mtr2p double-mutant deficient cells when coexpressed with its cofactor p15 (Katahira et al. 1999). Furthermore, overexpression of TAP/p15 heterodimers stimulates export of RNAs that are otherwise inefficiently exported in cultured cells and in Xenopus oocytes (Braun et al. 2001; Guzik et al. 2001).
Although NXF1/TAP interacts directly with the CTE element to promote RNA export of retroviral RNAs, adaptor proteins are required to facilitate its interaction with cellular mRNAs (for review, see Izaurralde 2002; Reed and Hurt 2002). It was found that Mex67p interacts with Yra1p, an essential yeast hnRNP-like protein, which belongs to an evolutionary conserved family of hnRNP-like proteins termed REFs (for RNA and export factor-binding proteins; Strasser and Hurt 2000; Stutz et al. 2000). The REF proteins (also known as Aly) are characterized by the presence of one RNP-motif RNA-binding domain. Yra1p was originally isolated in a screen for yeast genes that cause overexpression-mediated growth arrest (Espinet et al. 1995), and was also shown to possess RNA–RNA annealing activity (Portman et al. 1997). The level of Yra1p is tightly regulated by a negative feedback mechanism that involves splicing of its unusual intron (Preker et al. 2002). Depletion of Yra1p results in nuclear accumulation of poly(A)+ RNA, supporting its direct role in mRNA export in yeast (Strasser and Hurt 2000; Stutz et al. 2000). Furthermore, anti-REF antibodies inhibit mRNA export in Xenopus oocytes, whether or not the mRNAs are generated by splicing, and also recombinant REF proteins stimulate the export of mRNAs (Koffa et al. 2001; Rodrigues et al. 2001).
It has been suggested that pre-mRNA splicing stimulates mRNA export as a result of a coupling between the splicing and the mRNA export machineries, perhaps by removing nuclear retention factors and/or actively recruiting mRNA export factors (Luo and Reed 1999; Cullen 2000a). However, pre-mRNA splicing is not essential for mRNA export, as naturally occurring vertebrate mRNAs that lack introns are efficiently exported. In addition, in yeast in which only a small percentage of genes contain introns, mRNA export proceeds efficiently (for review, see Cullen 2000b; Izaurralde 2002). Two recent studies showed that mRNA splicing enhances gene expression, but its stimulatory effect is not related mainly to facilitating mRNA export; rather, pre-mRNA splicing stimulates 3′end processing, and can also markedly enhance the cytoplasmic translation of an mRNA (Lu and Cullen 2003; Nott et al. 2003). The coupling between splicing and mRNA export is proposed to be mediated by a multicomponent complex, the exon-junction complex (EJC), which is deposited on the mRNA as a consequence of the splicing reaction in a sequence-independent, position-dependent manner. The presence of REF/Aly, a bona fide component of the splicing-dependent EJC complex, promotes recruitment of the heterodimer TAP/p15 to cellular mRNPs (Le Hir et al. 2000, 2001b; Zhou et al. 2000). It was proposed that the conserved DEAD-box helicase UAP56, which functions during spliceosome assembly, facilitates REF/Aly recruitment to the spliced mRNP complex (Luo et al. 2001; Strasser and Hurt 2001). Together, these observations have led to a model in which REF/Aly provides the molecular link between pre-mRNA splicing and mRNA export (Zhou et al. 2000). However, it has been shown recently that, whereas HEL/UAP56 is essential for mRNA export in Drosophila cells, REF proteins are dispensable, suggesting that additional adaptor proteins can recruit NXF1/p15 to cellular mRNPs in Drosophila (Gatfield et al. 2001; Gatfield and Izaurralde 2002).
In this study, we have investigated the mRNA export pathway in C. elegans. The expression of mRNA export factors and components of the EJC complex has been inhibited by dsRNA interference. We show that CeNXT1/p15, the binding partner of CeNXF1/TAP, has an essential function during C. elegans development, and is required for the export of mRNA. Depletion of the EJC component CeRNPS1 results in no discernible phenotype; in contrast, RNAi of the EJC Y14 protein results in embryonic lethality, although this protein is not directly involved in mRNA export. We also show that the three members of the REF/Aly family of adaptor proteins in C. elegans are neither essential for C. elegans development, nor do they affect mRNA export. Our findings strongly suggest the existence of multiple adaptor proteins that mediate mRNA export in C. elegans.
RESULTS
The mRNA export pathway is highly conserved throughout evolution. We have used RNA interference (RNAi) in the nematode C. elegans to ask whether selectively interfering with the expression of genes encoding the homologs of different bona fide export factors and EJC components affects the development of this organism. This technique has now been established as a rapid and convenient method for selectively interfering with gene expression, not only in C. elegans, but also in other organisms and in cultured mammalian cells (for review, see McManus and Sharp 2002). For instance, RNAi has been used in a systematic functional analysis of the C. elegans genome, whereby the function of ~86% of the predicted genes was inhibited (Kamath et al. 2003). Introduction of double-stranded RNA results in a drastic reduction in the level of mRNA of the corresponding endogenous gene in a highly sequence-specific manner, and has been shown to phenocopy strong loss of function or null alleles of the targeted gene (Fire et al. 1998). To elicit RNA interference in C. elegans, we resorted to microinjection of dsRNA into the gonads or the gut of young adult hermaphrodites, because this method has been proven to be more effective than feeding worms with dsRNA or simply soaking the worms in dsRNA (Tabara et al. 1998; Timmons and Fire 1998). Injected animals (Bristol strain N2) were left to recover and to lay any eggs present in utero prior to injection, for 16 h, and were then transferred onto individual plates and allowed to egg lay. The effect of RNAi was observed in the F1 progeny on days 3–5 after injection.
The TAP/p15 heterodimer
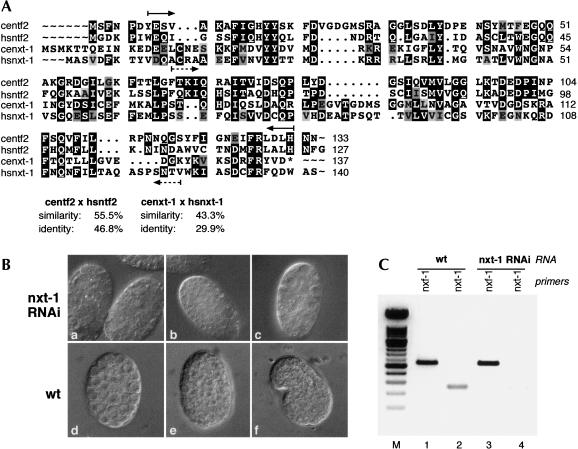
Two homologs of human NXF1/TAP were identified previously in the C. elegans genome (Herold et al. 2000; Tan et al. 2000). Analysis of the C. elegans genome revealed one predicted homolog of NXT1/p15, termed Cenxt-1. Interestingly, proteins of the NXF/TAP family, as well as NXT/p15, share a domain of high homology to the human nuclear transport factor 2 (NTF2), a cytosolic factor for nuclear import that interacts with the nuclear pore complex (Paschal and Gerace 1995; Fribourg et al. 2001). The C. elegans genome also encodes a candidate homolog of NTF2, termed ceran-4. The CeNXT1/p15 ORF (Cenxt-1) displays 43.3% similarity and 29.9% identity to the human p15 protein sequence (Fig. 1A). This homology is almost entirely attributed to the NTF2-like domain present in the p15 sequence in both species. RNA interference of CeNXF1/TAP resulted in lethality for both embryos and adult nematodes, and in nuclear accumulation of poly(A)+ RNA as it was described previously by the Felber laboratory (Tan et al. 2000; data not shown). In addition, we show that RNAi depletion of CeNXT1/p15 mRNA results in early embryonic lethality, with development of affected embryos arrested probably around gastrulation (Fig. 1B). Efficiency of the RNAi treatment was confirmed by RT–PCR analysis, showing that levels of CeNXT1/p15 mRNA were greatly depleted in injected animals (Fig. 1C, cf. lanes 2 and 4). In contrast, a control mRNA was present at levels comparable with wild-type animals (Fig. 1C, cf. lanes 1 and 3). In addition, we found that RNAi of CeNTF2-like gene (Ceran-4) also results in embryonic lethality, most likely due to defects in RanGDP import into the nucleus (data not shown).
FIGURE 1.
RNA interference with the CeNXT1/p15 gene results in embryonic lethality. (A) Sequence comparison between human and C. elegans NXT1 proteins, and between human and C. elegans NTF-2 proteins. Sequences were compared using the GAP program (GCG10 software), and output was produced using PRETTYBOX (GCG10 software). Identical residues are highlighted in black. The dsRNA fragments used for RNAi correspond to the regions marked by the arrows below and above the sequences, respectively. (B) RNA interference with the CeNXT1/p15 gene leads to early embryonic lethality (a–c). Wild-type embryo developmental stages (d) gastrulated, (e) beginning of morphogenesis, (f) comma are shown for comparison. Each embryo is ~50 μm in length. (C) The effectiveness of RNAi was determined by examining the level of the residual transcripts following dsRNA injections by RT–PCR with specific primers, as described previously (Longman et al. 2000). CeNXT1/p15 mRNA is specifically depleted in RNAi-treated animals (lane 4) compared with wild-type animals (lane 2); whereas the level of a control mRNA, corresponding to CeNXF1/Tap1, is unaffected (lanes 1,3). The figure shows a negative of an ethidium bromide-stained agarose gel. (M) 100-bp ladder DNA size marker.
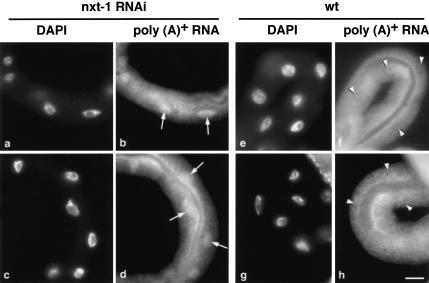
Because CeNXT1/p15 has been identified as a homolog of the human mRNA export factor, we examined the effect of its depletion on poly(A)+ RNA localization in C. elegans. Following RNAi with dsRNA homologous to CeNXT1/p15, adult nematodes were fixed and subjected to RNA in situ hybridization with a Cy3-labeled oligo(dT) probe. Depletion of CeNXT1/p15 resulted in nuclear accumulation of poly(A)-containing RNAs as shown on an example of an adult intestinal tissue (Fig. 2b,d). In contrast, poly(A)-containing RNAs were distributed uniformly in the nuclei and cytoplasm of intestinal tissue of control N2 nematodes (Fig. 2f,h). These results clearly demonstrate a direct role of CeNXT1/p15 in mRNA export in C. elegans.
FIGURE 2.
Depletion of CeNXT1/p15 causes accumulation of poly(A)+ mRNA in the nuclei. Injected animals and control N2 worms were fixed and subjected to RNA in situ hybridization as described. In each panel the intestine is the predominant tissue characterized by the presence of large nuclei. Accumulation of poly(A)-containing RNAs was observed in intestinal nuclei of RNAi-treated animals (indicated by arrows in b and d) and corresponding DAPI-stained nuclei as shown in a and c. Poly(A)+ RNA distribution in intestines of wild-type, untreated worms are shown in f and h (nuclei are indicated by arrowheads), and corresponding DAPI- stained nuclei are shown in e and g. Bar, 25 μm.
UAP56 is an essential protein in C. elegans and is required for mRNA export
The DEAD-box helicase UAP56 is involved in pre-mRNA splicing (Fleckner et al. 1997; Libri et al. 2001), and has also been shown to facilitate REF/Aly recruitment to the spliced mRNP complex (Jensen et al. 2001; Luo et al. 2001; Strasser and Hurt 2001). Interestingly, UAP56 (known as HEL in insects) is recruited cotranscriptionally to the Balbiani ring mRNP particles in Chironomus tentans in an intron-independent manner, suggesting that its recruitment to mRNAs does not necessarily require pre-mRNA splicing (Kiesler et al. 2002). RNAi of UAP56 in Drosophila cells results in growth inhibition and accumulation of polyadenylated RNAs within the nucleus, demonstrating that it is essential for mRNA export in Drosophila (Gatfield et al. 2001). Here, we show that RNAi of CeUAP56 (Cehel-1) resulted in embryonic lethality and in nuclear accumulation of poly(A)-containing RNAs, demonstrating that this protein has an essential role in nematodes and is directly involved in mRNA export (Table 1; MacMorris et al. 2003).
TABLE 1.
Characterization of C. elegans homologs of mRNA export factors, EJC, and EJC-related proteins
| Protein | Gene | RNAI phenotype | poly(A) mRNA accumulation | Reference |
| TAP1/NXF-1 | C15H11.3; Cenxf-1 | Embryonic and adult lethality | + | Tan et al. 2000; this paper |
| TAP2/NXF-2 | C15H11.6; Cenxf-2 | Wild-type phenotype | nd | Tan et al. 2000 |
| NXT-1/p15 | Y71F9AM.5; Cenxt-1 | Early embryonic lethality | + | This paper |
| Y14 | R07E5.14; Cernp-4 | Late embryonic lethality | − | This paper |
| MAGO | R09B3; Cemag-1 | Embryonic lethality, sterility | nd | Li et al. 2000 |
| SRm160 | F28D9.1; Cersr-1 | Wild-type phenotype | nd | Longman et al. 2000 |
| RNPS1 | K02F3.11; Cernp-5 | Wild-type phenotype | nd | This paper |
| UAP56 | C26D10.2; Cehel-1 | Late embryonic lethality | + | This paper; MacMorris et al. (this issue) |
| UPF3 | F46B6.3; Cesmg-4 | Wild-type phenotype | nd | Kamath et al. 2003 |
Note: The REF adaptor proteins which are part of the EJC are discussed in Table 2.
The exon–exon junction complex
The known components of the exon–exon junction complex (EJC) include the splicing-associated factors SRm160, DEK, and RNPS1, the mRNA export adaptor REF/Aly, Y14 and its binding partner mago, and Upf3 (Blencowe et al. 1998; Mayeda et al. 1999; Kataoka et al. 2000; Le Hir et al. 2000; McGarvey et al. 2000; for review, see Dreyfuss et al. 2002). The EJC complex serves as a binding platform for factors involved in mRNA export, and also in nonsense-mediated decay (NMD), thus providing a link between several steps of messenger RNA processing (Le Hir et al. 2001b; Lykke-Andersen et al. 2001). We have shown previously that RNAi of SRm160 results in no obvious phenotype (Longman et al. 2000). In contrast, RNAi of CeSRm160 in combination with individual CeSR family proteins resulted in a distinct phenotype, demonstrating that interactions between SRm160 and multiple SR family proteins are important for proper development in C. elegans (Longman et al. 2001).
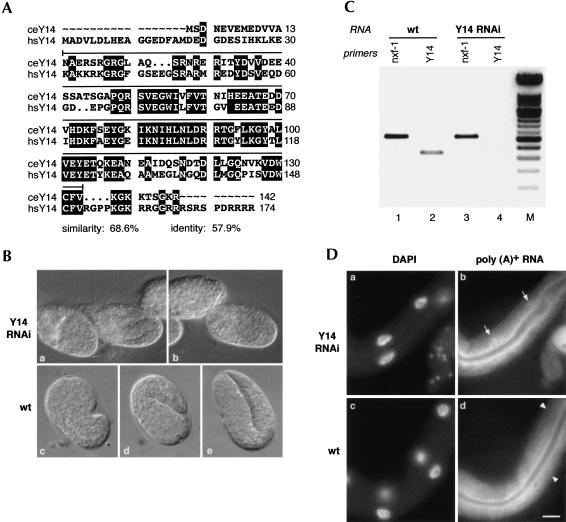
In this study, we analyzed the effect of depleting individual components of this complex in a whole organism (summarized in Table 1). The C. elegans genome contains one candidate homolog of Y14, Cernp-4, an RNA-binding protein that is preferentially recruited to mRNAs generated by splicing (Kataoka et al. 2000; Zhao et al. 2000). The Y14 protein associates with another protein, termed Mago (Zhao et al. 2000) and Y14/Mago heterodimers are essential in cultured Drosophila cells (Le Hir et al. 2001a), and have also been implicated in mRNA localization in Drosophila (Newmark et al. 1997; Hachet and Ephrussi 2001). The C. elegans homolog of Mago, Cemag-1, functions in the regulation of hermaphrodite germ-line sex determination, and its depletion causes lethality in the F1 progeny (Li et al. 2000).
The predicted CeY14 protein is highly homologous throughout its length to the human protein (68.6% similarity, 57.9% identity; Fig. 3A). When CeY14 (Cernp-4) was depleted by dsRNA interference, a late embryonic lethal phenotype was observed in the F1 progeny. Although organogenesis had occurred as indicated by the presence of major tissue types, the embryos were morphogenically defective (Fig. 3B). This clearly demonstrates that CeY14 is an essential protein in C. elegans, being required for at least one nonredundant function. The RNAi effect was specific and extensive; whereas CeY14 mRNA was greatly depleted in the treated animals as shown by RT–PCR analysis, a control mRNA was present at levels comparable with wild-type animals (Fig. 3C, cf. lanes 2 and 4 and lanes 1 and 3). Depletion of CeY14 by RNA interference did not cause nuclear accumulation of poly(A)+ RNAs, suggesting that although CeY14 has an essential role in C. elegans development, it is not directly associated with mRNA export (Fig. 3D). The C. elegans genome also has a single homolog of human RNPS1, Cernp-5. The worm protein displays high homology (55.8% similarity and 47.4% identity) with its human counterpart (Mayeda et al. 1999). Interestingly, depletion of CeRNPS1 resulted in no discernible phenotype (Table 1).
FIGURE 3.
RNA interference with the CeY14 gene results in embryonic lethality but does not cause accumulation of poly(A)+ mRNA in the nuclei. (A) CeY14 predicted protein and human Y14 protein sequences were compared using the GAP program (GCG10 software), and the alignment was generated using PRETTYBOX (GCG10 software). Identical residues are highlighted in black, and the dsRNA fragment used for RNAi corresponds to the solid line above the sequence. (B) RNA interference with the CeY14 gene results in late embryonic lethality (a,b). Wild-type embryonic developmental stages are shown for comparison (c) comma, (d) 1.5-fold, (e) twofold. Each embryo is ~50 μm in length. (C) The effectiveness of RNAi was determined by examining the level of the residual transcripts following dsRNA injections by RT–PCR with specific primers, as described previously (Longman et al. 2000). CeY14 mRNA is specifically depleted in RNAi-treated animals (lane 4) compared with wild-type animals (lane 2); whereas the level of a control mRNA, corresponding to CeNXF1/Tap1, is unaffected (lanes 1,3). The figure shows a negative of an ethidium bromide-stained agarose gel. (M) 100-bp ladder DNA size marker. (D) Depletion of CeY14 does not result in nuclear accumulation of poly(A)+ mRNA. Injected animals and control N2 animals were fixed and subjected to RNA in situ hybridization as described. In each panel, the intestine is the predominant tissue characterized by the presence of large nuclei. No accumulation of poly(A)-containing RNAs was observed in nuclei of intestines of RNAi-treated animals (indicated by arrows in b). (a) The corresponding DAPI-stained nuclei. Poly(A)+ RNA distribution in intestines of wild-type, untreated worms are shown in d (nuclei are indicated by arrowheads), and corresponding DAPI- stained nuclei are shown in c. Bar, 25 μm.
The REF family of mRNA export adaptor proteins
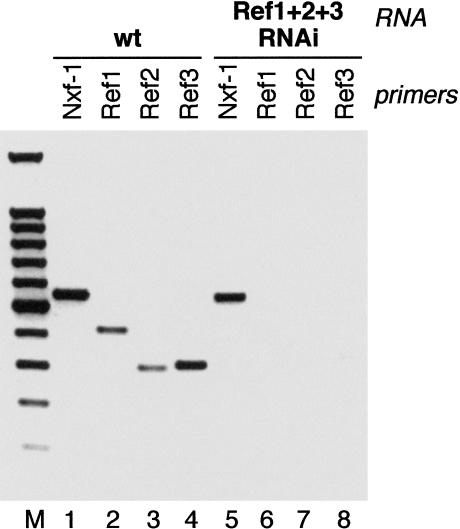
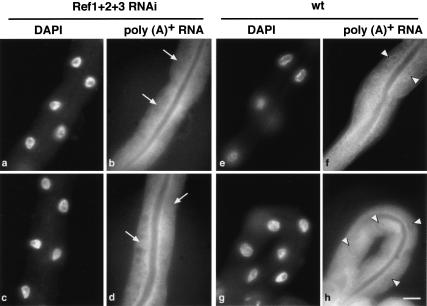
The C. elegans genome contains three homologs of human REF/Aly (Stutz et al. 2000). The CeREF proteins display high homology to the human protein sequences. This high degree of conservation is evident throughout the different motifs of the protein (Stutz et al. 2000). RNAi of individual Ref genes (Cealy-1, Cealy-2, and Cealy-3), or combinations of two Ref genes, showed no obvious phenotype, which suggested the existence of functional redundancy for this family of proteins in C. elegans. Surprisingly, simultaneous suppression of three Ref genes did not compromise viability and only caused a slight decrease in larval mobility, implying that the REF proteins do not fulfill an essential function in C. elegans (Table 2). The effectiveness of the RNAi treatment was assessed by analyzing the level of residual mRNAs corresponding to the targeted genes following dsRNA injections by RT–PCR, as described previously (Longman et al. 2000). Total RNA was prepared from animals injected with dsRNA corresponding to all three C. elegans Ref genes. RT–PCR analysis showed that all three Ref mRNAs were greatly depleted in the treated animals (Fig. 4, cf. lanes 6–8 with lanes 2–4), whereas mRNA for a control gene was present at levels comparable with wild-type animals (Fig. 4, cf. lanes 1 and 5). Interestingly, simultaneous suppression of all three C. elegans Ref genes did not result in accumulation of poly(A)+ mRNA in the nucleus (Fig. 5b,d). Therefore, CeREF proteins are not essential factors for export of bulk mRNA from the nucleus, suggesting the existence of additional mRNA export adaptors.
TABLE 2.
RNAi phenotypes for C. elegans Ref/Aly genes and their codepletion with SR and SR-related genes
| Protein | Gene | RNAi phenotype |
| REF-1 | C01F6.5; Cealy-1 | Wild-type phenotype |
| REF-2 | F23B2.6; Cealy-2 | Wild-type phenotype |
| REF-3 | M18.7; Cealy-3 | Wild-type phenotype |
| REF-1 + REF-2 | Wild-type phenotype | |
| REF-1 + REF-3 | Wild-type phenotype | |
| REF-2 + REF-3 | Wild-type phenotype | |
| REF-1 + REF-2 + REF-3 | Reduced mobility | |
| REF-1 + REF-2 + REF-3 + SRp20 | Reduced mobility | |
| REF-1 + REF-2 + REF-3 + SC35 | Reduced mobility | |
| REF-1 + REF-2 + REF-3 + SRm160 | Reduced mobility |
FIGURE 4.
RNAi depletion of C. elegans Ref/Aly genes mRNAs is efficient and specific. The level of the residual transcripts following dsRNA injections was examined by RT–PCR with specific primers, as described previously (Longman et al. 2000) CeRef 1, 2, and 3 mRNAs are specifically depleted in RNAi treated animals (lanes 6,7,8, respectively) compared with wild-type animals (lanes 2,3,4, respectively), whereas the level of a control mRNA corresponding to CeNXF1/Tap1 gene is unaffected (lanes 1,5). The figure shows a negative of an ethidium bromide-stained agarose gel. (M) 100-bp ladder DNA size marker.
FIGURE 5.
Simultaneous suppression of all three C. elegans Ref/Aly genes does not result in nuclear accumulation of poly(A)+ mRNA. Injected animals and control N2 animals were fixed and subjected to RNA in situ hybridization as described. In each panel, the intestine is the predominant tissue characterized by the presence of large nuclei. No accumulation of poly(A)-containing RNAs was observed in nuclei of intestines of RNAi-treated animals (indicated by arrows in b and d). (a,c) The corresponding DAPI-stained nuclei. Poly(A)+ RNA distribution in intestines of wild-type, untreated worms are shown in f and h (nuclei are indicated by arrowheads), and corresponding DAPI- stained nuclei are shown in e and g. Bar, 25 μm.
Interestingly, two human nucleoctyoplasmic shuttling SR proteins, 9G8 and SRp20, promote mRNA export, suggesting a role for SR proteins as mRNA export adaptors (Huang and Steitz 2001; Huang et al. 2003). However, here we show that RNAi of all three C. elegans Ref genes in combination with individual SR proteins, or with the SR-related protein SRm160, failed to give a phenotype distinct from the phenotype caused by simultaneous depletion of all three Ref genes alone, indicating that REF proteins are not functionally interacting with individual SR proteins or with SRm160 (Table 2). It remains possible that CeSR proteins contribute toward mRNA export with a degree of functional redundancy (see Discussion).
Altogether, these results strongly suggest the existence of adaptors, other than the REF/Aly proteins, that facilitate the recruitment of the mRNA export factor NXF1/TAP to different mRNAs.
DISCUSSION
We have used C. elegans as a model system to functionally characterize genes involved in mRNA export, as well as components of the exon–exon junction complex that links pre-mRNA splicing with mRNA export and NMD.
The requirement for the TAP/p15 heterodimer for mRNA export has been clearly established in Drosophila. Small bristles, the Drosophila ortholog of NXF1, is essential for mRNA export throughout development (Wilkie et al. 2001). Furthermore, RNAi of NXF1/TAP and NXT1/p15 in Drosophila cultured cells leads to nuclear accumulation of poly(A)+ RNA (Herold et al. 2001; Wiegand et al. 2002). By RNAi, we have demonstrated that CeNXT1/p15 is an essential protein that is required early in development of C. elegans, and has a direct role in mRNA export.
We also show here that Y14, a component of the exon–exon junction complex, is essential for C. elegans viability, yet it does not affect bulk mRNA export. A role for Y14 in NMD was suggested upon the fact that hUpf3, a key factor in NMD, interacts with Y14 and is enriched in Y14-containing mRNP complexes (Kim et al. 2001). The strict requirement of Y14 for normal development in C. elegans is most likely unrelated to NMD, as this is a nonessential process in yeast and worms (Hodgkin et al. 1989; Culbertson 1999). Therefore, Y14 is likely to be involved in some other essential process that is neither related to mRNA export or NMD.
Whereas the cellular export factor NXF/TAP interacts with the CTE element to promote export of retroviral RNAs, adaptor proteins are required to promote its recruitment to cellular mRNAs (for review, see Izaurralde 2002; Reed and Hurt 2002). Several lines of evidence suggest the role of members of the REF/Aly family of proteins in mediating this task both in yeast and in higher eukaryotes, linking pre-mRNA splicing with mRNA export. However, recent results in Drosophila cultured cells showed that REF/Aly and also protein components of the EJC complex were dispensable for mRNA export. Only when REF and RNPS1 were codepleted or when all EJC proteins were depleted simultaneously, partial nuclear accumulation of poly(A)+ RNA was observed (Gatfield and Izaurralde 2002). In this study, we show that simultaneous depletion of all three C. elegans REF proteins does not compromise viability and only causes a minor phenotype. Moreover, depletion of all three REF proteins does not affect the nuclear export of bulk mRNAs from the nucleus. The fact that UAP56 is an essential protein required for mRNA export in C. elegans, whereas the REF proteins are dispensable, strongly suggests that UAP56 must perform some other additional function/s apart from recruiting REF/Aly to mRNAs.
Our results suggest the existence of alternative strategies to promote mRNA export, some of which may be REF independent. Interestingly, two nucleocytoplasmic shuttling SR proteins, SRp20 and 9G8, act to promote the export of intronless RNAs and also function as adaptors for TAP-dependent mRNA export (Huang and Steitz 2001; Huang et al. 2003). However, we observed that simultaneous depletion of all three Ref/Aly genes, together with individual SR proteins, did not affect the phenotype observed when depleting the three Ref/Aly genes alone. This suggests the existence of additional mRNA export adaptors other than CeRef/Aly and CeSR proteins. Alternatively, CeSR proteins may be acting as mRNA export adaptors with a degree of functional redundancy. In agreement, we have shown previously that depletion of individual SR proteins (with the exception of CeSF2) or certain combinations of SR proteins does not lead to any discernible phenotype in C. elegans, which is indicative of functional redundancy for this family of proteins in nematodes (Longman et al. 2000).
Together with the evidence described in Drosophila cells, our results strongly suggest that additional adaptor protein/s, with a certain degree of functional redundancy, may contribute to TAP-dependent mRNA export.
MATERIALS AND METHODS
dsRNA preparation and microinjection
Templates for RNA synthesis were generated by PCR from C. elegans genomic DNA using gene-specific primers with T3 and T7 promoter sequences added on to forward (F) and reverse (R) primers, respectively. Where possible, the region amplified corresponds to a large exon or an exon-rich part of the gene. Oligonucleotide primers were purchased from Genosys (Cambridge). PCR conditions using Vent DNA polymerase (New England Biolabs) were as follows: (1) 98°C, 5 min, 1×, (2) 98°C, 30 sec, 58°C, 50 sec, 72°C, 1 min, 30×, (3) 72°C, 10 min, 1×.
Primers used were as follows: T3 sequence: attaaccctcactaaagggaag; T7 sequence: taatac gactcactatagg; p15F1: tcagctatgcaacgagtcg; p15R1: attttccgtcttcaacgcc; not p15F: tgaac gagctacgagaagc; not p15R: atgaggaaggccaga atcc; Tap1F: ttctcatgcacgacaccac; Tap1R: cat tccactgaactggcac; ran-4F: acgaaagtgttgcaaaggc; ran-4R: gtgcaagtcaagtcggaag; Y14F: aacgcc gagagatcaagag; Y14R: tgacgaagcaccaatcgac; UAP56F: actgaaggaagccgagaag; UAP56R: atcgaagcggtcttgaacg; RNPS1F: atgcgaccatcac catcac; RNPS1R: tggagaacttctacgctgg; Ref1F: aacttgtcaaacttggccc; Ref1R: gcatccaactcttcgagtg; Ref2F: caaatgtcggaactccacg; Ref2R: aagggcaa ctccagcaaac; Ref3F: agctcgacgaggaaatacg; Ref3R: cgacaccgctaaacttctg.
PCR products were gel purified and used as templates for in vitro RNA synthesis with T3 and T7 RNA polymerase (Boehringer Mannheim) following instructions from the manufacturers. RNA was dissolved in sterile water with 0.4 U/μL RNase Inhibitor (Boehringer Mannheim) to reach a final concentration of 0.5μg/μL. Double-stranded RNA was assembled by mixing equal amounts of sense and antisense RNA followed by incubation at 68°C for 10 min, and then 37°C for 30 min. For each gene, 10–15 young adult hermaphrodites (Bristol strain N2) were injected with dsRNA into the gut or gonad. RNAi interferes with maternal mRNA, but it will not deplete maternal protein present in the mother at the time of RNAi. To minimize the contribution of maternal protein in RNAi-treated embryos, injected worms were left to recover and egg lay for 16 h. Then, injected animals were transferred onto individual plates and the phenotype was observed in the F1 progeny on days 3, 4, and 5 after injection. F1 progeny were scored for embryonic lethality, slow progression through larval stages, size of adults, abnormal organ development in adults, abnormalities in feeding and movements, and sterility. The affected progeny were examined using DIC microscopy.
RT–PCR
Total RNA from embryos was prepared as follows. Approximately 20 gravid hermaphrodites, either wild type or injected previously with dsRNA, were dissolved in 1:10 solution of bleach in 1 M NaOH. Embryos were collected and washed twice in 1 mL of PBS, pelleted, and resuspended in 200 μL 0.5% SDS, 5% β-mercaptoethanol, 10 mM EDTA, 10 mM Tris-HCl (pH 7.5), and 0.5 mg/mL proteinase K. Samples were incubated at 55°C for 1 h, and further processed using Total RNA Isolation Reagent (Advanced Biotechnologies Ltd.) following manufacturer’s instructions. Total RNA from whole worms or larvae was prepared as described above with exception of the bleaching step. RT–PCR was performed using SuperScript One-Step RT–PCR System (GIBCO BRL) following the manufacturer’s instructions. To test the efficiency of RNAi treatment by RT–PCR, 100 μL (200 μL for CeRefs RNAi RNA) reactions were prepared for wild-type and RNAi-treated samples. These reactions were split into identical fractions and RT–PCR analysis was performed to compare RNA levels corresponding to either a control gene or the gene(s) targeted in the RNAi experiment. After 23–28 cycles of amplification, RT–PCR products were loaded on ethidium bromide-stained agarose gel. Primers used for RT–PCR analysis were the same as the ones used for preparation of dsRNA fragments without the T3 and T7 sequences.
RNA in situ hybridization
On day 3, after dsRNA injection-treated worms or control wild-type worms were placed on polylysine-coated slides in 4 μL PBS with 0.25 mM levamisole, and cut at midsection with a needle. A total of 4 μL of fix solution [3.7% formaldehyde, 100 mM HEPES (pH6.9), 2 mM MgSO4, 1 mM EGTA in PBS] were added, coverslips were placed on a sample with a gentle pressure, and slides were put on a metal rack on dry ice. After 10 min, coverslips were cracked off, and slides were immediately placed in ice-cold methanol for 10 min. Slides were air dried briefly, fixed at room temperature for 30 min, then incubated in methanol for 5 min and in ethanol 2× for 5 min. At this point, the slides could be stored at −20°C for a few days. Slides were then incubated in 50% xylene/50% ethanol for 5 min, in xylene for 1 h, and in 50% xylene/50% ethanol for 5 min. Subsequently, slides were washed 3× in ethanol for 5 min, in methanol for 5 min, briefly air dried, post-fixed for 20 min, and washed 4× in PBS. Slides were then washed 2× in hybridization buffer without glycogen or salmon sperm DNA, and then prehybridized in 50 μL of hybridization buffer under a parafilm strip at 55°C for 1 h (hybridization buffer: 50% deionized formamide, 5× SSC, 1 mg/mL glycogen, 100 μg/mL salmon sperm DNA, 0.1% Tween-20). Slides were hybridized in 50 μL of hybridization buffer with 1 μL of cy3-labeled oligo(dT)30 probe (100 ng/μL stock) under a parafilm strip in a humidified sealed chamber at 55°C for 20 h. Then, slides were washed 5× for 20 min in hybridization buffer at 55°C and 4× for 10 min in PBS at room temperature. Slides were mounted in 0.5 μg/mL DAPI in Vectashield mounting medium (Vector Laboratories). DAPI and Cy3 signals were observed using Zeiss Axioplan2 fluorescence microscope under 63× magnification objective. Slide preparation was as follows: slides were wiped clean and dipped for a few seconds in the solution of 0.2% gelatine, 1mg/mL polylysine. Slides were drained, dried overnight, and used immediately, or stored at 4°C for several days.
Acknowledgments
We thank Elisa Izaurralde (EMBL) and Gavin Wilkie (MRC HGU) for critical reading of the manuscript. We acknowledge Betsy Goodwin for sharing protocols, Elsebet Lund for discussions, and Peg MacMorris and Tom Blumenthal for protocols and for communicating unpublished results. J.F.C. and D.L. were supported by the Medical Research Council (MRC), I.L.J. was supported by a MRC Senior Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5420503.
REFERENCES
- Bachi, A., Braun, I.C., Rodrigues, J.P., Pante, N., Ribbeck, K., von Kobbe, C., Kutay, U., Wilm, M., Gorlich, D., Carmo-Fonseca, M., et al. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6: 136–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear, J., Tan, W., Zolotukhin, A.S., Tabernero, C., Hudson, E.A., and Felber, B.K. 1999. Identification of novel import and export signals of human TAP, the protein that binds to the constitutive transport element of the type D retrovirus mRNAs. Mol. Cell. Biol. 19: 6306–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe, B.J., Issner, R., Nickerson, J.A., and Sharp, P.A. 1998. A coactivator of pre-mRNA splicing. Genes & Dev. 12: 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, I.C., Herold, A., Rode, M., Conti, E., and Izaurralde, E. 2001. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276: 20536–20543. [DOI] [PubMed] [Google Scholar]
- Bray, M., Prasad, S., Dubay, J.W., Hunter, E., Jeang, K.T., Rekosh, D., and Hammarskjold, M.L. 1994. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. 91: 1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalla, D., Zhu, J., Manche, L., Huber, E., Krainer, A.R., and Caceres, J.F. 2002. Nuclear export and retention signals in the RS domain of SR proteins. Mol. Cell. Biol. 22: 6871–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson, M.R. 1999. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 15: 74–80. [DOI] [PubMed] [Google Scholar]
- Cullen, B.R. 2000a. Connections between the processing and nuclear export of mRNA: Evidence for an export license? Proc. Natl. Acad. Sci. 97: 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000b. Nuclear RNA export pathways. Mol. Cell. Biol. 20: 4181–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. Nuclear RNA export. J. Cell Sci. 116: 587–597. [DOI] [PubMed] [Google Scholar]
- Dreyfuss, G., Kim, V.N., and Kataoka, N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 3: 195–205. [DOI] [PubMed] [Google Scholar]
- Espinet, C., de la Torre, M.A., Aldea, M., and Herrero, E. 1995. An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast 11: 25–32. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Fleckner, J., Zhang, M., Valcarcel, J., and Green, M.R. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes & Dev. 11: 1864–1872. [DOI] [PubMed] [Google Scholar]
- Fribourg, S., Braun, I.C., Izaurralde, E., and Conti, E. 2001. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell 8: 645–656. [DOI] [PubMed] [Google Scholar]
- Gatfield, D. and Izaurralde, E. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell. Biol. 159: 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield, D., Le Hir, H., Schmitt, C., Braun, I.C., Kocher, T., Wilm, M., and Izaurralde, E. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11: 1716–1721. [DOI] [PubMed] [Google Scholar]
- Gruter, P., Tabernero, C., von Kobbe, C., Schmitt, C., Saavedra, C., Bachi, A., Wilm, M., Felber, B.K., and Izaurralde, E. 1998. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1: 649–659. [DOI] [PubMed] [Google Scholar]
- Guzik, B.W., Levesque, L., Prasad, S., Bor, Y.C., Black, B.E., Paschal, B.M., Rekosh, D., and Hammarskjold, M.L. 2001. NXT1 (p15) is a crucial cellular cofactor in TAP-dependent export of intron-containing RNA in mammalian cells. Mol. Cell. Biol. 21: 2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet, O. and Ephrussi, A. 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11: 1666–1674. [DOI] [PubMed] [Google Scholar]
- Herold, A., Suyama, M., Rodrigues, J.P., Braun, I.C., Kutay, U., Carmo-Fonseca, M., Bork, P., and Izaurralde, E. 2000. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol. Cell Biol. 20: 8996–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold, A., Klymenko, T., and Izaurralde, E. 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7: 1768–1780. [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., Papp, A., Pulak, R., Ambros, V., and Anderson, P. 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. and Steitz, J.A. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7: 899–905. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Gattoni, R., Stevenin, J., and Steitz, J.A. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11: 837–843. [DOI] [PubMed] [Google Scholar]
- Hurt, E., Strasser, K., Segref, A., Bailer, S., Schlaich, N., Presutti, C., Tollervey, D., and Jansen, R. 2000. Mex67p mediates nuclear export of a variety of RNA polymerase II transcripts. J. Biol. Chem. 275: 8361–8368. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E. 2002. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur. J. Cell Biol. 81: 577–584. [DOI] [PubMed] [Google Scholar]
- Jensen, T.H., Boulay, J., Rosbash, M., and Libri, D. 2001. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11: 1711–1715. [DOI] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser, A.G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kang, Y. and Cullen, B.R. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes & Dev. 13: 1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira, J., Strasser, K., Podtelejnikov, A., Mann, M., Jung, J.U., and Hurt, E. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18: 2593–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, N., Yong, J., Kim, V.N., Velazquez, F., Perkinson, R.A., Wang, F., and Dreyfuss, G. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6: 673–682. [DOI] [PubMed] [Google Scholar]
- Kiesler, E., Miralles, F., and Visa, N. 2002. HEL/UAP56 binds cotranscriptionally to the balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr. Biol. 12: 859–862. [DOI] [PubMed] [Google Scholar]
- Kim, V.N., Kataoka, N., and Dreyfuss, G. 2001. Role of the nonsense-mediated decay factor hUpf3 in the pplicing-dependent exon-exon junction complex. Science 293: 1832–1836. [DOI] [PubMed] [Google Scholar]
- Koffa, M.D., Clements, J.B., Izaurralde, E., Wadd, S., Wilson, S.A., Mattaj, I.W., and Kuersten, S. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20: 5769–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., and Moore, M.J. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19: 6860–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Braun, I.C., Forler, D., and Izaurralde, E. 2001a. The protein Mago provides a link between splicing and mRNA localization. EMBO Rep. 2: 1119–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Izaurralde, E., and Moore, M.J. 2001b. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.S., Henry, M., and Silver, P.A. 1996. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes & Dev. 10: 1233–1246. [DOI] [PubMed] [Google Scholar]
- Lei, E.P. and Silver, P.A. 2002. Protein and RNA export from the nucleus. Dev. Cell 2: 261–272. [DOI] [PubMed] [Google Scholar]
- Li, W., Boswell, R., and Wood, W.B. 2000. mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev. Biol. 218: 172–182. [DOI] [PubMed] [Google Scholar]
- Libri, D., Graziani, N., Saguez, C., and Boulay, J. 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes & Dev. 15: 36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman, D., Johnstone, I.L., and Caceres, J.F. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19: 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman, D., McGarvey, T., McCracken, S., Johnstone, I.L., Blencowe, B.J., and Caceres, J.F. 2001. Multiple interactions between SRm160 and SR family proteins in enhancer-dependent splicing and development of C. elegans. Curr. Biol. 11: 1923–1933. [DOI] [PubMed] [Google Scholar]
- Lu, S., and Cullen, B.R. 2003. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA 9: 618–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M.J. and Reed, R. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. 96: 14937– 14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M.L., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M., and Reed, R. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., and Steitz, J.A. 2001. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836–1839. [DOI] [PubMed] [Google Scholar]
- MacMorris, M.A., Brocker, C., and Blumenthal, T. 2003. UAP56 levels affect viability and mRNA export in C. elegans. RNA (this issue). [DOI] [PMC free article] [PubMed]
- Mayeda, A., Badolato, J., Kobayashi, R., Zhang, M.Q., Gardiner, E.M., and Krainer, A.R. 1999. Purification and characterization of human RNPS1: A general activator of pre-mRNA splicing. EMBO J. 18: 4560–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey, T., Rosonina, E., McCracken, S., Li, Q., Arnaout, R., Mientjes, E., Nickerson, J.A., Awrey, D., Greenblatt, J., Grosveld, G., et al. 2000. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J. Cell. Biol. 150: 309–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3: 737–747. [DOI] [PubMed] [Google Scholar]
- Murphy, R., Watkins, J.L., and Wente, S.R. 1996. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol. Biol. Cell 7: 1921–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny, S. and Dreyfuss, G. 1996. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell. Biol. 134: 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmark, P.A., Mohr, S.E., Gong, L., and Boswell, R.E. 1997. mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila. Development 124: 3197–3207. [DOI] [PubMed] [Google Scholar]
- Nott, A., Meislin, S.H., and Moore, M.J. 2003. A quantitative analysis of intron effects on mammalian gene expression. RNA 9: 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal, B.M. and Gerace, L. 1995. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J. Cell. Biol. 129: 925–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli, A.E., Ernst, R.K., Lund, E., Grimm, C., Zapp, M.L., Rekosh, D., Hammarskjold, M.L., and Dahlberg, J.E. 1997. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 16: 7500–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portman, D.S., O’Connor, J.P., and Dreyfuss, G. 1997. YRA1, an essential Saccharomyces cerevisiae gene, encodes a novel nuclear protein with RNA annealing activity. RNA 3: 527–537. [PMC free article] [PubMed] [Google Scholar]
- Preker, P.J., Kim, K.S., and Guthrie, C. 2002. Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism. RNA 8: 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R. and Hurt, E. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108: 523–531. [DOI] [PubMed] [Google Scholar]
- Rodrigues, J.P., Rode, M., Gatfield, D., Blencowe, B.J., Carmo-Fonseca, M., and Izaurralde, E. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. 98: 1030–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra, C., Felber, B., and Izaurralde, E. 1997. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr. Biol. 7: 619–628. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., Moreno, H., Simos, G., Segref, A., Fahrenkrog, B., Pante, N., and Hurt, E. 1998. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol. Cell Biol. 18: 6826–6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref, A., Sharma, K., Doye, V., Hellwig, A., Huber, J., Luhrmann, R., and Hurt, E. 1997. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16: 3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge, C.A., Colot, H.V., Goldstein, A.L., and Cole, C.N. 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 17: 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser, K. and Hurt, E. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19: 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413: 648–652. [DOI] [PubMed] [Google Scholar]
- Stutz, F. and Rosbash, M. 1998. Nuclear RNA export. Genes & Dev. 12: 3303–3319. [DOI] [PubMed] [Google Scholar]
- Stutz, F., Bachi, A., Doerks, T., Braun, I.C., Seraphin, B., Wilm, M., Bork, P., and Izaurralde, E. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6: 638–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., Grishok, A., and Mello, C.C. 1998. RNAi in C. elegans: Soaking in the genome sequence. Science 282: 430–431. [DOI] [PubMed] [Google Scholar]
- Tan, W., Zolotukhin, A.S., Bear, J., Patenaude, D.J., and Felber, B.K. 2000. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA 6: 1762–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons, L. and Fire, A. 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- Tseng, S.S., Weaver, P.L., Liu, Y., Hitomi, M., Tartakoff, A.M., and Chang, T.H. 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 17: 2651–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, J.L., Murphy, R., Emtage, J.L., and Wente, S.R. 1998. The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+ RNA export. Proc. Natl. Acad. Sci. 95: 6779–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand, H.L., Coburn, G.A., Zeng, Y., Kang, Y., Bogerd, H.P., and Cullen, B.R. 2002. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 22: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie, G.S., Zimyanin, V., Kirby, R., Korey, C., Francis-Lang, H., Van Vactor, D., and Davis, I. 2001. Small bristles, the Drosophila ortholog of NXF-1, is essential for mRNA export throughout development. RNA 7: 1781–1792. [PMC free article] [PubMed] [Google Scholar]
- Zenklusen, D. and Stutz, F. 2001. Nuclear export of mRNA. FEBS Lett. 498: 150–156. [DOI] [PubMed] [Google Scholar]
- Zhao, X.F., Nowak, N.J., Shows, T.B., and Aplan, P.D. 2000. MAGOH interacts with a novel RNA-binding protein. Genomics 63: 145–148. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Luo, M.J., Straesser, K., Katahira, J., Hurt, E., and Reed, R. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407: 401–405. [DOI] [PubMed] [Google Scholar]