Abstract
Removal of introns from pre-messenger RNAs in eukaryotes is carried out by the spliceosome, an assembly of a large number of proteins and five small nuclear RNAs (snRNAs). We showed previously that an in vitro transcribed and assembled base-paired complex of U2 and U6 snRNA segments catalyzes a reaction that resembles the first step of splicing. Upon incubation with a short RNA oligonucleotide containing the consensus sequence of the pre-mRNA branch site, the U2/U6 complex catalyzed a reaction between the 2′ OH of a bulged adenosine and a phosphate in the catalytically important AGC triad of U6, leading to the formation of an X-shaped product, RNA X, apparently linked by an unusual phosphotriester bond. Here we characterize this splicing-related reaction further, showing that RNA X formation is an equilibrium reaction, and that the low yield of the reaction likely reflects an unfavorable equilibrium coefficient. Consistent with a phosphotriester linkage, RNA X is highly alkali-sensitive, but only mildly acid-sensitive. We also show that mutations in the AGC sequence of U6 can have significant effects on RNA X formation, further extending the similarities between splicing and RNA X formation. We also demonstrate that pseudouridylation of U2 enhances RNA X formation, and that U6 snRNA purified from nuclear extracts is capable of forming RNA X. Our data suggest that the ability to form RNA X might be an intrinsic property of spliceosomal snRNAs.
Keywords: U6 snRNA, U2 snRNA, ribozyme, spliceosome, phosphotriester, splicing
INTRODUCTION
The molecular machine responsible for removal of introns from mRNA precursors, the spliceosome, has an indispensable role in eukaryotic gene expression. Given the complexity and importance of pre-mRNA splicing, it is not surprising that the spliceosome is one of the most complicated cellular machines known, comprising five small nuclear RNA molecules (snRNAs) and a hundred or more proteins (e.g., Rappsilber et al. 2002; Zhou et al. 2002). The spliceosome appears to assemble de novo on each pre-mRNA in a stepwise fashion, with spliceosomal components joining the assembly either singly or in pre-formed complexes. Complementation studies, in vivo and in vitro functional assays, and cross-linking data have provided evidence for the existence of an extensive network of RNA–RNA and RNA–protein interactions in the spliceosome (for review, see Moore et al. 1993; Nilsen 1998; Brow 2002). Through these interactions, the 5′ and 3′ splice sites, and a conserved region of the intron close to the 3′ splice site called the branch site, are recognized in a number of steps and at the same time, interactions between snRNAs provide a scaffold for the assembly of the active site components.
In the fully assembled spliceosome, two of the snRNAs, U2 and U6, are extensively base-paired to each other through a number of helices (Datta and Weiner 1991; Wu and Manley 1991; Madhani and Guthrie 1992, 1994; Sun and Manley 1995). At the same time, these two snRNAs help juxtapose the reacting groups of the first step of splicing, the branch site and the 5′ splice site, by base-pairing to these regions of the intron (for review, see Nilsen 1998). The base-pairing interaction between U2 and the branch site bulges a specific adenosine residue, the branch site A, out of the RNA duplex (e.g., Query et al. 1994). The 2′ OH of this bulged adenosine then launches a nucleophilic attack on the 5′ splice site, resulting in the formation of a 2′–5′ linkage between this A and the first nucleotide of the intron, and release of the 5′ exon. The newly released exon is held in the active site via interactions with U5 snRNA and associated proteins, which also interact with the 3′ exon and the branch site (for review, see Nilsen 1998; Collins and Guthrie 2000). The second step involves a nucleophilic attack by the 3′ OH group of the 5′ exon on the phosphodiester bond at the 3′ splice site. After the second step of splicing the spliceosome dissembles, in preparation for the next round of splicing (for review, see Moore et al. 1993; Nilsen 1998; Brow 2002).
As with any other enzymatic system, a crucial step in understanding the spliceosome is elucidation of its catalytic mechanism and the organization and identity of the groups that form the active site of the splicing reactions. Although definitive evidence for the make up of the spliceosomal active site is still lacking, the majority of available data point to the possibility that the spliceosome, like the ribosome, is an RNA enzyme, with two of the spliceosomal snRNAs, U2 and U6, having major roles in splicing catalysis. Of the five spliceosomal snRNAs, U1 and U4 leave the spliceosome before the catalytic steps occur, and U5 snRNA seems to be largely dispensable at least for the first step of splicing (for review, see Nilsen 1998; Brow 2002). U2 and U6 snRNAs are both highly conserved and in addition to their role in positioning the 5′ splice site and the branch site, mutagenesis and phosphorothioate substitution studies suggest a crucial role for two invariant regions in U6, the ACAGAGA and the AGC domains, in catalysis (for review, see Nilsen 1998; Brow 2002). Taken together, the current data provide evidence for participation of U6 and U2 in both steps of splicing.
The possibility of an RNA catalytic site in the spliceosome is strengthened further by the intriguing mechanistic and structural similarities between the spliceosome and the self-splicing group II introns, ribozymes found in bacteria, protists, and organelles of higher eukaryotes (for reviews, see Jacquier 1996; Pyle 1996; Newman 1997; see also Sontheimer et al. 1997, 1999; Gordon et al. 2000; Yean et al. 2000; Shukla and Padgett 2002). It has been proposed that the spliceosome is an evolutionary descendent of self-splicing introns, such as group II introns, with the spliceosomal snRNAs being remnants of the ancestral ribozyme (Sharp 1985; Cech 1986; Newman 1997). However, despite a host of indirect data suggesting RNA-mediated catalysis in the spliceosome, direct evidence for the competence of the spliceosomal snRNAs to form the active site of the splicing reactions has been lacking.
Recent data has provided evidence for the catalytic potential of purified snRNAs. First, protein-free, in vitro-transcribed central domains of human U2 and U6 snRNAs were shown to form a base-paired complex that resembles the one thought to exist in the active spliceosome (Valadkhan and Manley 2000; Fig. 1 ▶). This work included the demonstration of a long-range tertiary interaction in the U2/U6 complex that closely resembles a functionally important association that had been deduced previously in yeast by genetic analysis (Madhani and Guthrie 1994). Importantly, this interaction has the potential to juxtapose the catalytically crucial ACAGAGA and AGC domains of U6, and likely contributes to the assembly of the active site of the spliceosome. Extending these results, Valadkhan and Manley (2001) found that upon incubation of the U2/U6 complex with a short RNA containing the branch site consensus sequence (designated Br; see Fig. 1 ▶), the 2′ OH of the bulged adenosine in Br was activated by the U2/U6 complex and attacked the phosphodiester backbone of U6 RNA in the catalytically crucial AGC triad (Valadkhan and Manley 2001). This reaction, which bears many similarities to the first step of splicing, including metal ion requirements and susceptibility of several key residues in both U6 and Br to mutagenesis, led to formation of an X-shaped product (RNA X) apparently containing an unusual phosphotriester linkage between U6 and the branch site substrate. The yield of product, however, was low and because of the unusual nature of the linkage, additional characterization of the reaction and the product is important (Villa et al. 2002).
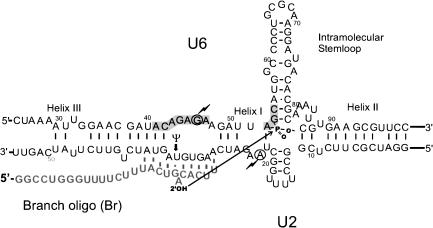
FIGURE 1.
Base-pairing interactions in the in vitro-assembled complex of U2–U6 and the branch oligonucleotide (Br). The shaded boxes mark the invariant regions in U6, and the three previously established base-paired helices between U2 and U6 and the intramolecular stemloop of U6 are indicated. Dashed lines enclose psoralen-crosslinked residues. The chemical groups involved in RNA X formation in U6 and Br are shown. Arrow indicates the reaction that results in formation of RNA X. Bold arrow points to the site of a pseudouridine modification in U2. The circled residues marked with thunderbolts can be joined together in a UV crosslink described previously. Numbers indicate nucleotide positions from the 5′ end.
Here we present further characterization of this snRNA-catalyzed reaction. We show that RNA X formation is an equilibrium-state reaction, rapidly reversed upon removal of unreacted Br. We also show that the unreacted U6 and Br remaining at the end of the reaction, when purified and reused, are completely active for RNA X formation. The low yield of the reaction therefore likely reflects an unfavorable equilibrium constant, which in turn reflects the unusual chemistry of the reaction. Alkali and acid sensitivity provide further support that RNA X is a phosphotriester. We also provide evidence that RNA X formation can be strongly affected by mutation of the catalytically important AGC triad in U6, further supporting its similarity with splicing. Significantly, we also show that U6 snRNA purified from human nuclear extracts is active in RNA X formation, and that pseudouridylation of a key residue in U2 enhances RNA X formation. Our data provide further evidence that RNA X formation is a splicing-related reaction catalyzed by spliceosomal snRNAs in the absence of the 5′ splice site. We discuss these results with respect to authentic pre-mRNA splicing, and also in the context of previous work analyzing the formation and properties of RNA phosphotriesters.
RESULTS
RNA X formation shows characteristics of an equilibrium reaction
A significant feature of the RNA X formation reaction is its low yield, with ~0.15% of the input converted to product at the maximum extent of reaction (Valadkhan and Manley 2001). This could reflect a number of different reasons. For example, only a small fraction of U2/U6 complex or Br oligonucleotide might be active for RNA X formation, which in turn could be attributable to misfolding of the majority of the molecules in the reaction. Alternatively, RNA X formation might require a certain modified nucleotide, perhaps arising artifactually during synthesis or purification, which would be present in trace quantities in the starting pool of RNAs. Another explanation for the low yield could be the nature of the reaction, which simply reaches its maximum extent after 0.15% of the input has been converted to product.
Characterization of the covalent bond formed in RNA X is consistent with a phosphotriester linkage formed between the 2′ OH of a bulged adenosine in Br and the phosphate between the A and G in the AGC triad of U6 (Valadkhan and Manley 2001; Fig. 1 ▶). This unusual chemistry can at least partially explain the low yield, since to form the triester, one of the nonbridging oxygens on the phosphate becomes the leaving group, and this reaction is known to have a slow rate (see Discussion). Also, the 2′ OH of the adenosine located 5′ to the phosphotriester, upon deprotonation, can attack the phosphotriester and lead to reversal of the reaction. Therefore, it would be expected that RNA X formation should be an equilibrium reaction with a low yield.
To determine if this is indeed the case, we first tested whether RNA X formation is in fact an equilibrium reaction. We reasoned that if RNA X was in equilibrium with the reactants (U6 and Br), removal of unreacted Br would result in reversal of the reaction and therefore a decrease in the amount of RNA X. To this end, typical RNA X formation reactions containing 2 μM U2/U6 complex and 50 nM Br were allowed to proceed until the reaction had reached its maximum extent (after ~20 h at room temperature). An excess amount of a DNA oligonucleotide complementary to Br (αBr; at a final concentration of 10 μM) was then added to the reactions and after 10 min to 2 h at 30°C, reactions were terminated by addition of urea loading buffer and analyzed by denaturing PAGE. Significantly, the amount of RNA X rapidly decreased after the addition of αBr, with a rate constant of 0.02 min−1 at 30°C (Fig. 2A ▶). To demonstrate that the rate of αBr-induced reversal of RNA X formation is not limited by the rate of exchange between the Br bound to the U2/U6 complex and the unbound Br, we further characterized the binding of Br to the U2/U6 complex. First, using a range of U2/U6 concentrations, we determined that Br binds to U2/U6 with an apparent Kd of 50 nM (see Materials and Methods). We then determined the rate of exchange between bound and unbound Br by first incubating Br with an excess of U2/U6, followed by the addition of a large excess of unlabeled Br and further incubation at room temperature (see Materials and Methods). After the addition of a 100-fold excess of unlabeled Br, exchange with the unbound Br was observed (koff = 0.025 min−1 at RT). Therefore, it is likely that chemistry is the rate limiting factor in αBr-induced reversion of RNA X.
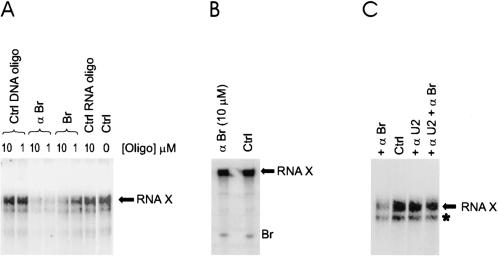
FIGURE 2.
RNA X formation is a reversible reaction. (A) RNA X formation can be reversed. Identity and concentration of the RNA or DNA oligonucleotide added to each reaction is shown above each lane. RNA products were analyzed by denaturing PAGE and detected by a PhosphorImager. The position of RNA X is indicated. No oligonucleotide was added to the reaction marked Ctrl. (B) Purified RNA X is resistant to reversal. The positions of RNA X and Br are shown. (Ctrl) No DNA or RNA oligonucleotide added. Identity and concentration of the DNA oligonucleotide added is shown above the left lane. (C) αBr-mediated RNA X reversal requires U2. The identity of the DNA oligonucloetides added to each lane is indicated. No oligonucleotide was added to the sample in the lane marked Ctrl. Position of RNA X is shown to the right. The band marked by an asterisk is a degradation product of RNA X.
If the decrease in RNA X induced by αBr indeed reflects the equilibrium state of the reaction, it would be expected that addition of excess unlabeled Br to reactions will have the same effect as αBr. To test this, we set up typical RNA X formation reactions, with radio-labeled Br as the only source of Br, and allowed them to proceed to the maximum extent of reaction. An excess amount of unlabeled Br (1 and 10 μM) was then added and products analyzed after 2 h of incubation at 30°C. Again, the amount of radioactive RNA X significantly decreased (Fig. 2A ▶). Addition of DNA or RNA oligonucleotides with no similarity or complementarity to Br did not affect the amount of RNA X formed (Fig. 2A ▶).
To show that this reversion was indeed a catalyzed reaction, we tested whether purified RNA X could be reversed by incubation with αBr. Addition of even the highest concentration of αBr (10 μM) did not result in reversal of RNA X (Fig. 2B ▶), which rules out the possibility of nonspecific hydrolysis after αBr addition. To provide further evidence that the αBr-mediated disappearance of RNA X is catalyzed, and therefore requires U2, we first allowed typical reactions to proceed for 20 h, after which an excess of an oligonucleotide complementary to U2 (αU2) was added and the reactions were incubated for an additional 2 h at 37°C. This treatment, which results in removal of U2 from the U2/U6 complex, stabilized RNA X from αBr-mediated reversal (Fig. 2C ▶). Therefore, the disappearance of RNA X after the addition of αBr is a specific, catalyzed reaction reflecting the equilibrium nature of RNA X formation. We also tested whether adding U2 snRNA back to purified RNA X would result in reversion, but were unable to detect any difference in RNA X levels (data now shown). This result, however, was not unexpected as the linkage of Br to U6 in RNA X interferes with the formation and correct refolding required to generate the active U2/U6 complex. This is consistent with our earlier finding that the U2/U6 complex must be formed prior to addition of Br for RNA X formation to occur (Valadkhan and Manley 2001).
The low yield is an intrinsic property of RNA X formation
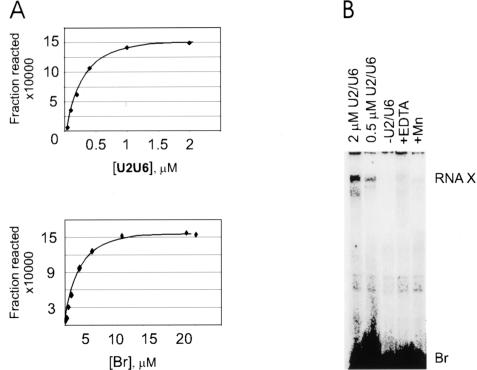
If RNA X formation is indeed in equilibrium, the final extent of the reaction should not change by switching the relative concentration of the two reactants, as the final amount of product will be determined by the equilibrium constant of the reaction that depends on the product of the concentrations of the reactants and not on individual concentrations. Therefore, to provide additional evidence that the low yield of RNA X reflects the equilibrium constant of the reaction, we determined the maximum extent of RNA X formation in reactions that contained an excess of unlabeled Br (at a final concentration of 0.1–20 μM), a trace amount of labeled U6 (50–100 nM), and unlabeled U2 at twofold excess to U6 (Fig. 3A ▶, bottom panel). We compared the final extent of these reactions to typical RNA X formation reactions, which contained an excess of unlabeled U2/U6 complex (0.1–2 μM) and a trace amount of labeled Br (50 nM; Fig. 3A, ▶top panel). Both sets of reactions showed the same maximum extent of RNA X formation, with 0.15% of the trace RNA species appearing in RNA X in each case (Fig. 3A ▶), further supporting the existence of an equilibrium state and the catalyzed nature of the reaction.
FIGURE 3.
RNA X formation shows characteristics of an equilibrium reaction. (A) Response of RNA X formation to various concentrations of Br and U2/U6. The graphs plot the amount of RNA X formed in various concentrations of U2/U6 (top panel) or Br (bottom panel). (B) RNA X formation by a U2/U6 chimera. The concentration of the U2/U6 chimera is noted above each lane. The reaction analyzed in the lane marked −U2/U6 contained only Br and reaction buffer. The reaction in lane marked +Mn contained Mn++ instead of Mg++, whereas that in the lane marked +EDTA had received 50 mM EDTA after the addition of reaction buffer (see Materials and Methods). The location of RNA X and Br are shown to the right.
We also tested the ability of a chimeric U2–U6 construct in which U6 and U2 were joined by a stable tetraloop between the 3′ end of U6 and 5′ end of U2 to form RNA X. Use of this construct essentially eliminates the possibility of inefficient or incomplete complex formation, which can occur when using separate U2 and U6 RNAs, and in addition, leads to a more uniform folding among the U2/U6 complexes. RNA X formation reactions using this construct were identical to those using separate U2 and U6 RNAs, with respect to, for example, the final extent of the reaction, Mg++ requirement, and reaction kinetics (Fig. 3B ▶ and data not shown). These data show that the low yield of RNA X cannot be due to inefficient formation or misfolding of the U2/U6 complex.
Unreacted U6 and Br are fully active in RNA X formation
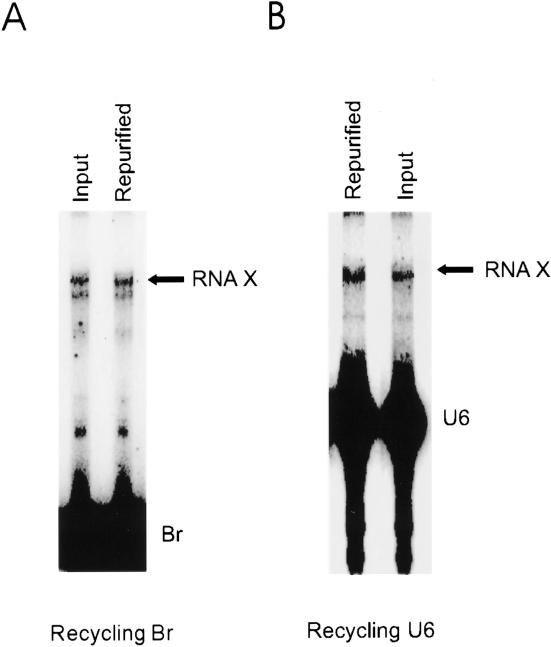
To provide additional evidence that the low yield of the reaction results from an unfavorable equilibrium constant and not an inherent defect in the unreacted material, we tested the ability of repurified, unreacted U6 and Br remaining after an initial RNA X formation reaction to form RNA X. In one set of reactions, Br was the labeled, trace species and the U2/U6 complex was in excess. In other reactions, Br was in excess, and U6 was the labeled species, with U2/U6 in trace concentration. After 24 h, the reactions were subjected to denaturing PAGE and the unreacted, labeled U6 or Br was purified. After elution and precipitation, the repurified U6 or Br was used as the sole source of U6 or Br in new reactions. In each case, the RNA species that was provided in trace concentration was recycled to ensure that all the molecules that could possibly react had reacted. Parallel reactions were set up using labeled U6 or Br that had not been used previously in RNA X formation. After 24 h, the reactions were terminated and analyzed. Significantly, the efficiency of RNA X formation was identical between reactions with repurified Br or U6 and those with Br and U6 not used previously for RNA X formation (Fig. 4A,B ▶). These results rule out the possibility that RNA X formation involves hypothetical chemically modified RNA molecules present in trace amounts in the original starting material, and show that the low yield of the reaction is an inherent property of RNA X formation.
FIGURE 4.
Unreacted Br and U6 are active for RNA X formation. The lanes marked Input display products of reactions that contained Br (panel A) or U6 (panel B) that was newly synthesized. The reactions analyzed in lanes marked Repurified contained the purified, unreacted RNAs (Br or U6) from previous RNA X formation reactions. The locations of RNA X and Br or U6 are shown to the right.
Consistent with a phosphotriester core, RNA X is extremely alkali-sensitive
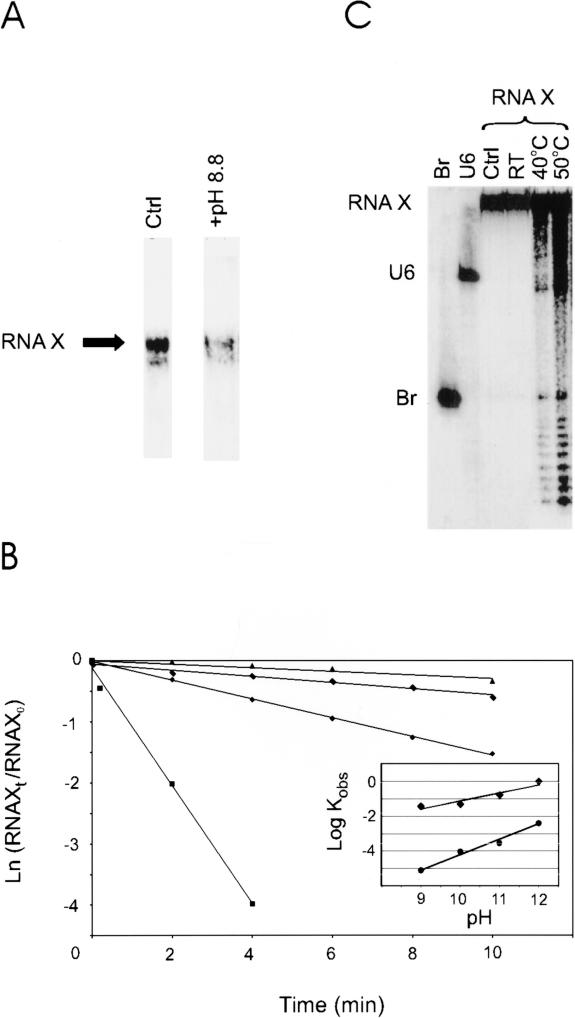
Phosphotriesters similar to the one we have proposed to exist at the core of RNA X, located 3′ to a ribose group, have been shown to be extremely sensitive to high pH, whereas they are significantly less sensitive to acidic pHs (e.g., see Kosonen et al. 1998; see also Discussion). We showed previously that RNA X is in fact sensitive to alkali, as incubation of RNA X at pH 12 for 20 min at room temperature resulted in ~50% dissociation (Valadkhan and Manley 2001). To extend this result, we investigated the stability of RNA X at a more modest pH (pH 8.8). Strikingly, this treatment resulted in a similar reversal of RNA X after 10 min (Fig. 5A ▶), indicating that RNA X is indeed remarkably alkali-sensitive. Consistent with this, RNA X formation is greatly reduced at pH values greater than 8.2, and undetectable at pH 8.8 (results not shown). We also determined the stability of RNA X at a range of pH values (pHs 9–12, Fig. 5B ▶). As a control, the stability of a natural RNA phosphodiester bond in Br was measured under identical conditions. Compared to the control, RNA X was dramatically more alkali-sensitive (between 250–5000-fold; Fig. 5B ▶), with rate constants similar to those observed in model phosphotriesters (e.g., t½of 45 sec for RNA X at pH 12, 50°C versus t½of 30 sec–11 min at pH 12, RT for various known phosphotriesters; Kosonen et al. 1998). In addition, we also examined the acid sensitivity of RNA X, by incubation of RNA X in a range of acidic pHs. RNA X was resistant to mild acid treatment (pHs 2–3, 10 min; data not shown) and to stronger treatment (pH 1, at RT, for 10 min; Fig. 5C ▶). At elevated temperatures (pH 1, 40°C and 50°C for 10 min.), very limited breakage of both the phosphodiester backbone linkages and the linkage between U6 and Br in RNA X was observed (Fig. 5C ▶). These results are not only consistent with a phosphotriester, but also rule out a number of alternative chemical linkages between U6 and Br in RNA X, such as O- or N-glycosidic bonds, which would have resulted in extreme and moderate acid sensitivity, respectively (see Discussion).
FIGURE 5.
Effects of alkali and acidic pH on RNA X. (A) RNA X is extremely alkali-sensitive. The reaction mixture analyzed in the lane on the left (Ctrl) was incubated for 10 min at pH 7.2, whereas the pH of the sample on the right was raised to pH 8.8. The position of RNA X is shown to the left. (B) Quantitative analysis of alkali-sensitivity of RNA X. The reactions are done at pH 9 (triangles), pH 10 (rectangles), pH 11 (diamonds), and pH 12 (squares). (Inset) Comparison of alkali-sensitivity of RNA X (diamonds) versus a phosphodiester bond (circles). (C) RNA X is relatively stable in acidic pHs. Samples were brought to pH 1.0 by addition of HCl to a final concentration of 0.2 N and reactions incubated for 10 min. The temperature of incubation is indicated above each lane. (Ctrl) Sample incubated at pH 7.2. Lanes labeled Br and U6 display input Br and U6 RNAs. The positions of U6, Br, and RNA X are shown to the left.
Mutations in the AGC triad of U6 can strongly affect the rate of RNA X formation
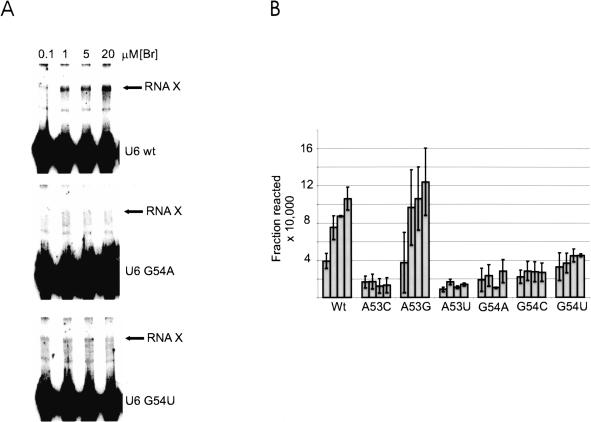
Our data support the conclusion that RNA X contains a phosphotriester linkage between the 2′ OH of the bulged A in Br and the phosphate located between the A and G of the AGC triad of U6. We showed previously that, although mutations in the other invariant region of U6, the ACAGAGA sequence, reduced or eliminated RNA X formation, mutations in the AGC sequence seemed to be well tolerated (Valadkhan and Manley 2001). It is possible, however, that the effect of these mutations on RNA X formation was masked because a step other than catalysis, such as binding or a conformational change, was rate-limiting under the conditions used. To determine if this is indeed the case, we tested whether mutations in the AGC sequence affect RNA X formation when the U2/U6 complex is the trace species in reactions, and Br is in excess, so that the binding of Br by U2/U6 would be eliminated as a possible rate-limiting step. Reactions containing U6 as the trace, labeled species were set up and the reactivity of these mutants was assayed by adding various concentrations of unlabeled Br. Interestingly, all point mutations of the G in the AGC sequence significantly decreased the rate of RNA X formation (Fig. 6A,B ▶). Two of the three mutations of the A also showed a strong reduction in the amount of RNA X formed (Fig. 6B ▶), showing that although not absolutely essential, the AGC triad has an important role in RNA X formation. Therefore, our data show that both invariant regions of U6 are involved in RNA X formation, strengthening the similarity with the authentic splicing reaction.
FIGURE 6.
Mutations in the AGC triad can block RNA X formation. (A) The effect of mutation of U6 G54 to A and U on RNA X formation. The position of RNA X and the U6 species used in each reaction is shown to the right. The concentration of Br in each lane is indicated above the panel. (B) Summary of the results of point mutations in the AGC triad. The U6 species used in each set of reactions is indicated below each series. The concentrations of Br tested in each series were 0.1, 1, 5, and 20 μM, and are graphed from left to right.
Authentic human U6 snRNA is active in RNA X formation
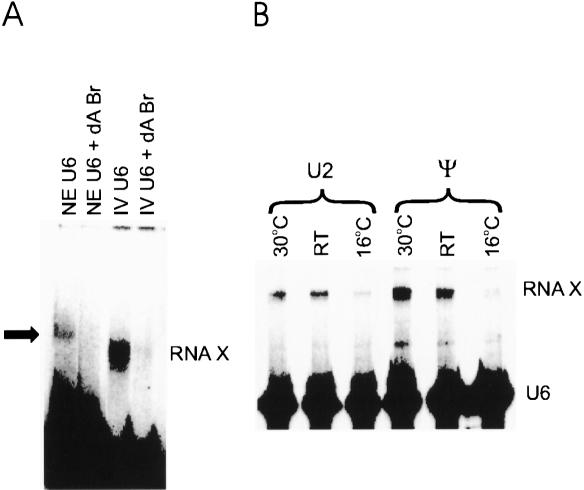
U6 and U2 snRNAs contain a large number of modified nucleotides (for review, see Massenet et al. 1998). Although most of these changes seem not to be absolutely necessary for the function of these snRNAs, as proved by reconstitution of U2- and U6-depleted nuclear extracts with in vitro synthesized U2 and U6 snRNAs (Fabrizio et al. 1989; McPheeters et al. 1989; McPheeters and Abelson 1992; Wolff and Bindereif 1992), in some cases they do seem to have significant roles, possibly by facilitating RNA–protein interactions or formation of a specific RNA structure (Yu et al. 1998; Newby and Greenbaum 2002). Interestingly, residues that flank the phosphate attacked by Br during RNA X formation, A53 and G54 in the AGC triad, are both 2′ O-methylated (Massenet et al. 1998). To test the possible effect of post-transcriptional modifications in U6 on the formation of RNA X, and more importantly, to test the ability of authentic U6 snRNA to catalyze RNA X formation, we purified U6 snRNA from HeLa nuclear extracts. As the presence of the 5′ domain of U6 leads to the formation of stable intramolecular interactions in U6 and reduced formation of the U2/U6 complex (results not shown), the first 25 nucleotides of U6 were removed (see Materials and Methods), which left only those nucleotides present in the in vitro-transcribed U6 normally used in RNA X formation, plus seven extra nucleotides at the 3′ end (nucleotides 100–106). The extract-purified U6 snRNA fragment was 5′ labeled and tested for RNA X formation. Figure 7A ▶ shows that the human U6 snRNA indeed formed RNA X after incubation with Br, albeit at reduced quantities (see Materials and Methods). Incubation of the purified U6 with a Br oligonucleotide that had deoxy substitutions at the bulged adenosine and the preceding nucleotide did not result in formation of any product (Fig. 7A ▶), indicating that, similar to RNA X formation with in vitro transcribed U2 and U6, this reaction also requires the 2′ OH of the bulged adenosine.
FIGURE 7.
The effect of post-transcriptional modifications in U2 and U6 on RNA X formation. (A) Authentic human U6 snRNA is active in RNA X formation. (IV U6) U6 made by in vitro transcription; (NE U6) U6 purified from nuclear extracts. Lanes marked NE U6 + dA Br or IV U6 + dA Br display RNAs from reactions that received a Br containing deoxy substitutions at A21 and G20. The position of RNA X formed with IV U6 is indicated to the right. The position of RNA X formed with NE U6 is marked by an arrow. The size difference between the two RNA X molecules is attributable to the larger size of NE U6 compared with IV U6 (seven nucleotides). (B) A conserved pseudouridine in U2 enhances formation of RNA X. (U2) U2 snRNA without any modifications was used in RNA X formation; (Ψ) U2 snRNA containing a pseudouridine at position 34. The positions of RNA X and U6 are indicated to the right. Reaction temperatures are indicated above each set of reactions.
A conserved pseudouridine in U2 snRNA increases RNA X formation
The solution structure of the branch-binding region of U2 in complex with a short RNA oligonucleotide that contains the branch consensus sequence was determined recently (Newby and Greenbaum 2002). A conserved pseudouridine in the branch-binding region of U2 (nucleotide 34) was found to have a major role in keeping the branch site A in the bulged conformation in these protein-free RNAs. To determine the effect of this modification on RNA X formation, a synthetic U2 snRNA fragment containing a pseudo U at this position (Fig. 1 ▶) was prepared and used in RNA X formation. Strikingly, significant increases in RNA X formation were detected with this U2 RNA, at both RT and 30°C (Fig. 7B ▶). When reaction rates at 30°C were measured, the forward rate of RNA X formation was found to be significantly enhanced in the presence of pseudouridine in U2, whereas the reverse rate was only very modestly increased (data not shown), resulting in the observed net increase in RNA X formation. Interestingly, the stimulatory effect was absent when reactions were performed at lower temperatures (e.g., 16°C; Fig. 7B ▶), presumably because of intrahelical stacking of the unpaired adenosine at lower temperatures despite the presence of the pseudouridine. This result is in agreement with the importance of a bulged conformation for the branch site A in RNA X formation and furthers the similarity between the splicing reaction and RNA X formation.
DISCUSSION
We have further characterized a splicing-related reaction catalyzed by an in vitro-assembled complex of protein-free U2 and U6 snRNAs with the aims, first, to prove that it is indeed a genuine RNA-catalyzed reaction, second, to explore further its relationship to the role played by snRNAs during authentic splicing, and finally, to provide additional data consistent with the hypothesis that the product of the reaction, RNA X, contains a phosphotriester linkage. We have provided new evidence that RNA X formation is a catalyzed reaction in equilibrium and strengthened both our previous conclusion that the linkage in RNA X is a phosphotriester and the relationship between RNA X formation and the authentic splicing reaction that occurs within the spliceosome.
The low yield of RNA X formation likely reflects the unusual chemistry of the reaction
Prior to the experiments described here, several explanations for the low yield of RNA X could be put forth. One was that the low yield reflects the involvement of a hypothetical, unknown chemically modified nucleotide as the reacting group in either U6 or Br, which would lead to termination of the reaction once all such rare, modified molecules in the reaction were used up (Villa et al. 2002). If so, it would be expected that the unreacted material should be inactive in RNA X formation, or at least show significantly reduced activity. Our results, which indicate that the unreacted RNAs have the same reactivity as they did originally, rules out this possibility, and also proves that the reactive group for RNA X formation should be one of the chemical groups found in natural RNA.
Because the low yield of RNA X does not result from a requirement for hypothetical modified nucleotides, it can be attributed either to structural constraints, such as misfolding of the majority of the U2/U6 complex into an inactive conformation, or to the intrinsic nature of the actual chemical reaction, or both. The small size of the U2/U6 complex (130 nucleotides total) could result in a high degree of conformational freedom, as lack of extensive stabilizing tertiary interactions makes the complex structurally flexible and potentially amenable to a high level of misfolding (Narlikar and Herschlag 1997; Burkard et al. 1999). The high efficiency of the long-range UV crosslink in the U2/U6 complex (Valadkhan and Manley 2000), however, is suggestive of a more or less uniform folding. In addition, if folding was rate-limiting in RNA X formation, different folding conditions or use of different, more stable constructs (such as the chimeric U2/U6 RNA) should have had detectable effects on RNA X formation, but none of the changes tested had any significant effect. The enhancement in RNA X formation observed with the use of pseudouridine-substituted U2 suggests that nonproductive binding (see Fersht 1985) of Br to the U2/U6 complex might be partially responsible for the low yield of the reaction, since based on structural studies (Newby and Greenbaum 2002), pseudouridine substitution in U2 helps keep the branch A in the bulged conformation necessary for its activity. The magnitude of the observed enhancement, however, suggests that nonproductive binding of Br makes only a relatively minor contribution to the low yield. Therefore, U2/U6 folding and Br binding are unlikely to be the major determinants of the yield of RNA X.
Alternatively, the low yield can reflect the chemical nature of the linkage in RNA X. The data presented here and previously suggest that RNA X contains a phosphotriester at its core. Formation of such a phosphotriester requires extraction of a molecule of water by direct or indirect protonation of one of the nonbridging phosphate oxygens. It is likely that formation of such a linkage is slow, since, although phosphate oxygens do exchange with solvent, this exchange is slow at neutral pH (Bunton et al. 1961, see below). The rate of phosphate oxygen exchange, however, increased in more acidic pHs, showing that as the solvent pH approaches the pKa of the nonbridging phosphate oxygens, they become better leaving groups. This same effect can be achieved by perturbing the pKa of the phosphate oxygens and increasing it to levels close to the pH of solvent, and it is plausible that the U2/U6 complex is employing this strategy to catalyze RNA X formation (see below). Nonetheless, the low yield of RNA X is entirely consistent with the expected properties of a phosphotriester linkage.
Phosphotriesters frequently occur in nucleic acids
Phosphotriesters have long been known to occur in both DNA and RNA as a result of exposure to certain toxins and chemicals (e.g., see Bannon and Verly 1972; Singer et al. 1975). Formation of a phosphotriester similar to the type proposed to exist in RNA X was observed to occur as a side reaction during chemical synthesis of DNA (Weimann and Khorana 1962). The reaction is thought to involve initial extraction of a water molecule from the phosphate, leading to the formation of an anhydride, in this case, between two adjacent phosphates. In the next step, the anhydride reacts with an incoming group, the 5′ OH of a nucleotide, leading to the formation of a phosphotriester with three nucleotide ligands. It is conceivable that formation of a phosphotriester in RNA X might follow a similar path, involving multiple steps and unstable intermediates. At any rate, the spontaneous formation of a phosphotriester as a side reaction proves that such compounds can form and accumulate in vitro, and because they form in low quantities, phosphotriesters might in fact occur more commonly than generally thought.
A seemingly ubiquitous family of enzymes that display phosphotriesterase activity has been described in a large variety of organisms, from bacteria to mammals (Raushel and Holden 2000; Sogorb and Vilanova 2002). It has been shown that such enzymes catalyze cleavage of phosphotriesters by positioning and activating a hydroxyl radical from the solvent for attack on the phosphate at the core of the phosphotriester. Because phosphotriesterase is capable of catalyzing the cleavage of phosphotriesters, according to enzymatic microreversibility principles, the enzyme should also be able to catalyze the formation of phosphotriesters, by subtracting a hydroxyl group from a phosphodiester and joining a third ligand to the phosphate. Therefore, an active site similar to that of phosphotriesterase should be capable of phosphotriester formation, although the yield, similar to the yield observed with RNA X formation, is expected to be very low (see below).
To form a phosphotriester such as seems to exist in RNA X, in the simplest scenario, one of the nonbridging phosphate oxygens should at some point become the leaving group. It has been shown that even without the involvement of an enzyme, phosphate oxygens do exchange with water, especially in mildly acidic pHs, but the rate of this exchange is slow (Bunton et al. 1961). However, a number of protein enzymes are capable of catalyzing this reaction at much faster rates (e.g., see Bock and Cohn 1978; Van Etten and Risley 1978). To be able to catalyze departure of a nonbridging oxygen, the pKa of this oxygen needs to be perturbed significantly. Such perturbations in pKa are common features of the active site of many protein and RNA enzymes (e.g., Connell and Yarus 1994; Legault and Pardi 1994; Narlikar and Herschlag 1997). Therefore, it is conceivable that an RNA enzyme can form an active site capable of increasing the pKa of the nonbridging oxygen on the phosphate and assisting its departure by protonating it. Because the AGC triad is close to a four-way junction in the U2/U6 complex, it is possible that the elaborate structural organization in the surrounding region could help form an active site with enough complexity to catalyze the unusual chemistry that seems to occur during RNA X formation.
Phosphotriesters in nucleic acids can be very stable
Studies on RNA phosphotriesters at the level of nucleotides have shown that depending on the nature of the third ligand, such linkages can range in stability from very stable (with a half-life of many days) to rather labile (t½ ~ 60 min) even in neutral pH (e.g., see Conrad et al. 1986; Kosonen et al. 1998). They can be quite stable, however, in the context of a large RNA molecule. For example, it has been observed that treatment of RNA with alkylating agents results in formation of molecules containing multiple phosphotriesters throughout their length. These RNAs can be subjected to shearing forces and other stresses, for example through multiple rounds of centrifugation and extended incubations at room temperature, without backbone breakage (Singer et al. 1975). Therefore, the relative stability of RNA X does not preclude the presence of a phosphotriester linkage.
The data suggesting that phosphotriesters display greater stability in the context of larger RNA molecules are in accordance with our results from the analysis of products obtained after complete digestion of purified RNA X with nucleases P1 and T2 (S. Valadkhan and J.L. Manley, unpubl.). Although RNA X can be subjected to limited digestion with nucleases, complete digestion apparently resulted in destabilization of the linkage between U6 and Br. This data is again consistent with a phosphotriester linkage, as exposure of the phosphotriester core to the solvent by removal of the surrounding nucleotides would likely result in occasional, solvent-mediated deprotonation of the 2′ OH located 5′ to the phosphotriester, which in turn would result in cleavage of the phosphotriester by the attack of this deprotonated 2′ OH. Our observations from hydroxyl radical and RNase fingerprinting experiments also support this conclusion, as in the folded U2/U6 complex, the area in the vicinity of the AGC triad seems to be strongly protected from the solvent (S. Valadkhan and J.L. Manley, unpubl.), which offers an explanation for why RNA X doesn’t naturally hydrolyze without the removal of most of the nucleotides surrounding the phosphotriester core.
The extreme sensitivity of RNA X to alkali pH, and its relative stability to acidic pHs, imposes further limits on the chemical identity of the linkage between U6 and Br in RNA X. For example, an alternative linkage between U6 and Br in RNA X could be via a reaction between the 2′ OH of Br and a group on a purine base in U6 (e.g., on A53 or G54), which would have resulted in a C-, N-, or O-glycosidic bond. Whereas C-glycosides show neither acid- nor alkali-sensitivity, an N- or O-glycosidic linkage in RNA X would have resulted in significant acid-sensitivity (Shabarova and Bogdanov 1994). Therefore, all three possible glycosidic linkages are ruled out based on the acid- and alkali-sensitivity profile of RNA X. (This was also suggested by our previous analysis of the iodoethanol cleavage pattern of thio-substituted RNA X; Valadkhan and Manley 2001).
Human U6 snRNA is active in RNA X formation
One of the most significant aspects of RNA X formation is that one of the reactive groups, the 2′ OH of Br, is also a reactive group in the first step of splicing. And the other reactive group, the phosphate in the AGC sequence, is part of a catalytically crucial domain that is thought to form part of the active site of the spliceosome. Therefore, RNA X formation requires that the U2/U6 complex position the reactive group of the first step of splicing in what would be the spliceosomal active site. In this regard, the linkage formed between Br and U6 in RNA X is reminiscent of the covalent attachment of suicide inhibitors (Fersht 1985) to the active site of enzymes. In our case, the natural substrate has, by mistake, attacked the active site of the enzyme, resulting in the formation of a covalent linkage (the phosphotriester). An alternative interesting possibility is that RNA X might reflect an intermediate in the splicing reaction, which has accumulated because of the inability of the system to complete catalysis of the first step, perhaps because of the lack of a 5′ splice site. This would suggest that the spliceosome employs a covalent catalysis strategy, by first making a covalent linkage to the 2′ OH of the branch A, which would then be positioned to attack the 5′ splice site to complete the reaction. (Note that this pathway allows the reaction to proceed with inversion of configuration at the phosphate center, as has been demonstrated; Maschhoff and Padgett 1993; Moore and Sharp 1993.) Formation of a transitory covalent linkage between substrates and enzyme active sites is a common feature in many enzymatic reactions (Jencks 1987), and it is possible that RNA X formation might reflect the occurrence of covalent catalysis during the authentic splicing reaction. Although there is currently no evidence supporting this intriguing possibility, there is also no data that rule it out, and our experiments suggest that investigation of possible covalent catalysis during pre-mRNA splicing is warranted.
Despite similarities in the sequence requirements and possibly active site arrangement between RNA X formation and splicing reactions, the two products of course differ in their chemistry. The ability of authentic human U6 snRNA to catalyze RNA X formation, however, points to the possibility that RNA X formation, similar to other “acknowledge” side reactions performed by the spliceosome (e.g., Yu et al. 1993; also see Tani and Ohshima 1991), reflects a legitimate activity of the spliceosome that can occur in the absence of a properly positioned 5′ splice site acceptor. RNA X formation might therefore be part of the catalytic repertoire of the spliceosomal snRNAs.
MATERIALS AND METHODS
RNA X formation reactions
Central domains of human U2 and U6 snRNAs were transcribed as described (Valadkhan and Manley 2000) and RNAs were diluted to a concentration of 10 μM by the addition of H20 before use. For each reaction, 1 μL of 10 μM U2, 1 μL of 10 μM U6, and 0.5 μL of an annealing buffer (400 mM Tris pH 7.2, 200 mM MgCl2) and 4.5 μL of H20 were added. Annealing mixtures were cooled slowly from 70°C to 28°C over the course of 1 h. The mixture, which was reduced in volume to about 4 μL because of evaporation, was then transferred to a fresh tube. One-half microliter of reaction buffer (100 mM Tris pH 7.2, 200 mM MgCl2) and 0.5 μL of 5′-labeled Br (60 K cpm, for a final concentration of about 50 nM) were added, followed by mixing by pipeting up and down. Reaction mixtures were left at room temperature for up to 24 h, after which 5 μL of loading buffer (9.8 M urea, 10 mM EDTA) was added and the sample loaded onto a 12%–20% denaturing PAGE.
For reactions that contained labeled U6, the annealing mixture contained 60 K cpm (for a final concentration of 50–100 nM) of 5′-labeled U6, 0.5 μL of 2 μM U2, 0.5 μL of the annealing buffer (400 mM Tris pH 7.2, 200 mM MgCl2) in a total volume of 7 μL. The mixture was slowly cooled from 70°C to 28°C as described above for typical reactions. Reaction mixtures were assembled as described above, except that instead of end-labeled Br, unlabeled Br was added at the final concentration of 5 μM (or the desired concentration). For assaying the effect of pseudoU-containing U2 (U2Ψ) on RNA X formation, the reactions were set up as described above for reactions with 5′-labeled U6, except U2Ψ was used instead of Wt U2.
Reversal of RNA X reactions
RNA X formation reactions were set up as above. After 20 h 0.5 μL of 10 or 100 μM αBr or unlabeled Br was added to the reactions and mixtures were incubated at 30°C or 37°C for 10 min to 2 h, followed by the addition of loading buffer and analysis on 12% denaturing PAGE. No DNA or RNA oligonucleotide was added to the control reactions.
Kinetic analysis of RNA X reversal
RNA X reactions were set up as described above with trace, labeled U6, 200 nM U2 wild type or U2 34Ψ and 10 μΜ Br. After 20 h of reaction at 30°C, αBr was added at a final concentration of 100 μΜ and the reactions were incubated at 30°C for 10 min to 2 h. No αBr was added to the control reactions, which were either left on ice for the duration of the reaction, or were incubated at 30°C for 2 h to ensure that the RNA X reaction has indeed reached its end point. The reactions were quenched by the addition of equal volume of a loading buffer containing 10 M urea and were loaded onto a 16% gel, followed by exposure to phosphorImager screens. The amount of remaining RNA X at each time point (RNAXt) was quantitated. The amount of RNA X in the two control reactions (RNAX0) was similarly quantitated and was found to be the same in both sets of control reactions. The rate constant for reversal of RNA X was calculated as the slope of the line obtained from plotting Ln(RNAXt/RNAX0) versus time.
Analysis of binding of Br to U2/U6
U2/U6 complexes were formed as described above, at a concentration of 2 μΜ. Before the addition of Br, U2/U6 was diluted to 100, 200, 500 nM and 1 μΜ concentrations by the addition of a buffer containing 50 mM Tris pH 7.2 and 40 mM MgCl2. 5′-labeled Br was added at a final concentration of 50 nM followed by 1 h of incubation at room temperature. The reactions were loaded onto an 8% native PAGE that contained 10 mM MgCl2 and was run in the cold room for 8 h at 150 V. The gel was exposed to PhosphorImager screens and the amount of bound Br was quantitated. The ratio of bound Br to total Br (bound + unbound) was plotted against the concentration of U2/U6, and Kd of Br binding to U2/U6 was calculated as [U2/U6 concentration at midpoint] − (1/2 [Brtotal]). The midpoint was defined as the point where [Brbound] = [Brunbound].
To calculate the koff of Br binding to U2/U6, reactions were set up as above with the final U2/U6 concentration of 1 μΜ and Br concentration of 50 nM, followed by 1 h of incubation at RT. Next, unlabeled Br at a final concentration of 10 μΜ was added, followed by incubation at RT for 10 min–1.5 h. A control reaction did not receive any unlabeled Br. After each time point was reached, the reactions received 1/10 volume of 50% glycerol loading buffer and were immediately loaded onto an 8% native PAGE. After exposure to the phosphorImager screens, the amount of bound Br at each time point (BrBdt) and the amount of bound Br in the control lane (BrBd0) were quantitated. The natural log of BrBdt/BrBd0 was plotted against time, and the slope of the line (−koff) was calculated.
Recycling of unreacted Br and U6
To recycle Br, reactions were set up as above and were allowed to proceed for 24 h, after which they were loaded onto 12% denaturing PAGE and the unreacted, radiolabeled Br was purified. For recycling U6, reactions were set up that contained a trace concentration of U2/U6 complex and an excess of Br (50 nM radiolabeled U6, 100 nM of U2, and 5 μΜ Br). After 24 h, reactions were loaded onto denaturing PAGE and unreacted U6 was purified from the gel. After elution, extraction with phenol and chloroform and precipitation, the repurified U6 or Br was used in new reactions. Identical reactions were set up with U6 and Br not previously used in RNA X formation reactions.
Kinetic analysis of alkali sensitivity of RNA X
RNA X reactions were set up as described above with final concentrations of 2 μΜ for U2/U6 and 10 μΜ for Br. A trace amount of 5′-labeled Br was added to aid in purification. After 24 h of reaction, the reactions were loaded onto a 12% PAGE and RNA X was purified. The purified RNA X was 5′ labeled followed by a second round of gel purification. To determine the alkali-sensitivity rate of RNA X, reactions were set up that contained phosphate buffers with pHs of 9, 10, 11, or 12, and 6 K cpm of the 5′-labeled RNA X, at an ionic strength of 0.25 M. Identical reactions were set up for 5′-labeled Br. The reactions were incubated for 2–10 min at 50°C, followed by quenching on ice. For each series, a control reaction was set up and was kept on ice for the duration of the reaction. The reactions were loaded on 12% gels and exposed to phosphorImager screens. The amount of RNA X remaining at each time point (RNAXt) and in the control lane (RNAX0) was calculated and the natural log of (RNAXt/RNAX0) was plotted against time. The slope of the line obtained in the graph was calculated as kobs. An identical analysis was performed on the Br reactions. As the rate of cleavage of all phosphodiester bonds in Br was roughly equal, the rate of cleavage of each phosphodiester bond would be 1/26 of the kobs obtained from the disappearance of Br (Br is 26 nucleotides long). Log10 of the first order reaction rates were plotted against pH for both RNA X and Br.
Acid treatment of RNA X
The reactions contained 3 K cpm of purified RNA X, 0.1 μg/ μL tRNA, 100 mM NaCl, and 0.2, 0.02, or 0.002 N HCl. Reactions were performed at room temperature, 40°C, and 50°C for 10 min. No HCl was added to control reactions.
Assaying the effect of mutations in the AGC sequence
Reactions were set up as described above for reactions with 5′-labeled U6, except instead of wild-type U6, the desired mutant U6 was used. The concentrations of unlabeled Br tested with each mutant were 0.1, 1, 5, and 20 μM. Each reaction was repeated at least three times. The results were quantitated using Molecular Dynamics PhosphorImager ImageQuant utility. The amount of RNA X formed in each case was normalized to the amount of U6 input and the average of the three trials was plotted in the graph, with two standard deviations as error bars.
RNA X formation with U6 snRNA from nuclear extracts
One-milliliter aliquots of HeLa nuclear extracts (Dignam et al. 1983) were treated with Proteinase K in reaction mixtures that contained 30 μL extract, 50 mM Tris pH 7.5, 10 mM EDTA, 100 mM NaCl, 5 μg tRNA, 10 μg Proteinase K, and 1% SDS in a total volume of 200 μL. After 30 min at 30°C, samples were extracted with phenol and chloroform, precipitated, and loaded onto a 12% denaturing PAGE along with a size marker for U6 snRNA. U6 was visualized by ethidium bromide staining and was purified. After elution, the first 25 nucleotides were removed by a 10–23 DNAzyme (Santoro and Joyce 1997) and after removal of the DNAzyme by DNase I digestion and two ethanol precipitations (to remove the dNTPs), U6 25–106 fragment was labeled at the 5′ end as described (Valadkhan and Manley 2000) and purified from a gel. RNA X formation reactions were performed as described above. Yields of U6 25–106 from extract were much lower than when synthesized in vitro, and it is possible that this contributed to the apparent lower efficiency of RNA X formation.
Acknowledgments
We thank Tim Nilsen, Reinhard Luhrmann, Harri Lonnberg, Dan Herschlag, Magda Konarska, Geoffrey Zubay, and Joe Piccirilli for helpful suggestions, discussions, and insight. We also thank Nima Shah for technical assistance. This work is supported by National Institutes of Health grant R37 GM48259.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5440303.
REFERENCES
- Bannon, P. and Verly, W. 1972. Alkylation of phosphates and stability of phosphate triesters in DNA. Euro. J. Biochem. 31: 103–111. [DOI] [PubMed] [Google Scholar]
- Bock, J.L. and Cohn, M. 1978. Metal dependence of the phosphate oxygen-water exchange reaction of Escherichia coli alkaline phosphatase. Kinetics followed by 31P(18O) NMR. J. Biol. Chem. 253: 4082–4085. [PubMed] [Google Scholar]
- Brow, D.A. 2002. Allosteric cascade of spliceosome activation. Annu. Rev. Genet. 36: 333–360. [DOI] [PubMed] [Google Scholar]
- Bunton, C.A., Llewellyn, D.R., Vernon, C.A., and Welch, V.A. 1961. The reactions of organic phosphates. Part IV. Oxygen exchange between water and orthophosphoric acid. J. Chem. Soc. 1961: 1636–1640. [Google Scholar]
- Burkard, M.E., Turner, D.H., and Tinoco, I. 1999. The interactions that shape RNA structure. In The RNA world II. (eds. R.F. Gesteland et al.), pp. 233–264. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Cech, T.R. 1986. The generality of self-splicing RNA: Relationship to nuclear mRNA splicing. Cell 44: 207–210. [DOI] [PubMed] [Google Scholar]
- Collins, C.A. and Guthrie, C. 2000. The question remains: Is the spliceosome a ribozyme? Nat. Struct. Biol. 7: 850–854. [DOI] [PubMed] [Google Scholar]
- Connell, G.J. and Yarus, M. 1994. RNAs with dual specificity and dual RNAs with similar specificity. Science 264: 1137–1141. [DOI] [PubMed] [Google Scholar]
- Conrad, J., Muller, N., and Eisenbrand, G. 1986. Studies on the stability of trialkyl phosphates and di-(2′-deoxythymidine) phosphotriesters in alkaline and neutral solution. A model study for hydrolysis of phosphotriesters in DNA and on the influence of a β hydroxyethyl ester group. Chem. Biol. Interact. 60: 57–65. [DOI] [PubMed] [Google Scholar]
- Datta, B. and Weiner, A.M. 1991. Genetic evidence for base pairing between U2 and U6 snRNA in mammalian mRNA splicing. Nature. 352: 821–824. [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., and Roeder, R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P., McPheeters, D.S., and Abelson, J. 1989. In vitro assembly of yeast U6 snRNP: A functional assay. Genes Dev. 3: 2137–2150. [DOI] [PubMed] [Google Scholar]
- Fersht, A. 1985. Enzyme structure and mechanism. W. H. Freeman and Company, New York, NY.
- Gordon, P.M., Sontheimer, E.J., and Piccirilli, J.A. 2000. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: Extending the parallels between the spliceosome and group II introns. RNA 6: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier, A. 1996. Group II introns: Elaborate ribozymes. Biochimie 78: 474–487. [DOI] [PubMed] [Google Scholar]
- Jencks, W.P. 1987. Covalent catalysis. In Catalysis in chemistry and enzymology. pp. 42–162. Dover Publications, Inc., Mineola, NY.
- Kosonen, M., Hakala, K., and Lonnberg, H. 1998. Hydrolysis and intramolecular transesterification of ribonucleoside 3′-phosphotriesters: The effect of alkyl groups on the general and specific acid-base-catalyzed reactions of 5′-O-pivaloyluridin-3′-yl dialkyl phosphates. J. Chem. Soc. Perkin Trans. 2: 663–670. [Google Scholar]
- Legault, P. and Pardi, A. 1994. 31P chemical shift as a probe of structural motifs in RNA. J. Magn. Reson. B. 103: 82–86. [DOI] [PubMed] [Google Scholar]
- Madhani, H.D. and Guthrie, C. 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817. [DOI] [PubMed] [Google Scholar]
- ———. 1994. Randomization-selection analysis of snRNAs in vivo: Evidence for a tertiary interaction in the spliceosome. Genes Dev. 8: 1071–1086. [DOI] [PubMed] [Google Scholar]
- Maschhoff, K.L. and Padgett, R.A. 1993. The stereochemical course of the first step of pre-mRNA splicing. Nucleic Acids Res. 21: 5456–5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet, S., Mougin, A., and Branlant, C. 1998. Posttranscriptional modifications in the U small nuclear RNAs. In The modification and editing of RNA (eds. H. Grosjean and R. Benne), pp. 201–227. ASM Press, Washington, DC.
- McPheeters, D.S. and Abelson, J. 1992. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell 71: 819–831. [DOI] [PubMed] [Google Scholar]
- McPheeters, D.S., Fabrizio, P. and Abelson, J. 1989. In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev. 3: 2124–2136. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. and Sharp, P.A. 1993. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 365: 364–368. [DOI] [PubMed] [Google Scholar]
- Moore, M.J., Query, C.C., and Sharp, P.A. 1993. Splicing of precursors to messenger RNAs by the spliceosome. In The RNA world (eds. R.F. Gesteland and J.F. Atkins), pp. 303–357. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Narlikar, G.J. and Herschlag, D. 1997. Mechanistic aspects of enzymatic catalysis: Lessons from comparison of RNA and protein enzymes. Annu. Rev. Biochem. 66: 19–59. [DOI] [PubMed] [Google Scholar]
- Newby, M.I. and Greenbaum, N.L. 2002. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat. Struct. Biol. 9: 958–965. [DOI] [PubMed] [Google Scholar]
- Newman, A. 1997. RNA splicing: Out of the loop. Curr. Biol. 7: R418–R420. [DOI] [PubMed] [Google Scholar]
- Nilsen, T.W. 1998. RNA-RNA interactions in nuclear pre-mRNA splicing. In RNA structure and function (eds. R. Simon and M. Grunberg-Manago), pp. 279–307. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- Pyle, A.M. 1996. Catalytic reaction mechanisms and structural features of group II intron ribozymes. Nucleic Acids Mol. Biol. 10: 75–107. [Google Scholar]
- Query, C.C., Moore, M.J., and Sharp, P.A. 1994. Branch nucleophile selection in pre-mRNA splicing: Evidence for the bulged duplex model. Genes Dev. 8: 587–597. [DOI] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., and Mann, M. 2002. Large-scale proteomic analysis of the human spliceosome. Genome Res. 12: 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raushel, F.M. and Holden, H.M. 2000. Phosphotriesterase: An enzyme in search of its natural substrate. Adv. Enzymol. Relat. Areas Mol. Biol. 74: 51–93. [DOI] [PubMed] [Google Scholar]
- Santoro, S.W. and Joyce, G.F. 1997. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. 94: 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabarova, Z. and Bogdanov, A. 1994. Advanced organic chemistry of nucleic acids. pp. 60–70. VCH publications, Weinheim, Germany.
- Shukla, G.C. and Padgett, R.A. 2002. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol. Cell. 9: 1145–1150. [DOI] [PubMed] [Google Scholar]
- Singer, B., Sun, L., and Fraenkel-Conrat, H. 1975. Effects of alkylation of phosphodiesters and of bases of infectivity and stability of tobacco mosaic virus RNA. Proc. Natl. Acad. Sci. 72: 2232–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogorb, M.A. and Vilanova, E. 2002. Enzymes involved in the detoxification of organophosphorus, carbamate and pyrethroid insecticides through hydrolysis. Toxicol. Lett. 128: 215–228. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J., Sun, S., and Piccirilli, J.A. 1997. Metal ion catalysis during splicing of premessenger RNA. Nature 388: 801–805. [DOI] [PubMed] [Google Scholar]
- Sontheimer, E.J., Gordon, P.M., and Piccirilli, J.A. 1999. Metal ion catalysis during group II intron self-splicing: parallels with the spliceosome. Genes & Dev. 13: 1729–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J.S. and Manley, J.L. 1995. A novel U2-U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev. 9: 843–854. [DOI] [PubMed] [Google Scholar]
- Tani, T. and Ohshima, Y. 1991. mRNA-type introns in U6 small nuclear RNA genes: Implications for the catalysis in pre-mRNA splicing. Genes Dev. 5: 1022–1031. [DOI] [PubMed] [Google Scholar]
- Valadkhan, S. and Manley, J.L. 2000. A tertiary interaction detected in a human U2-U6 snRNA complex assembled in vitro resembles a genetically proven interaction in yeast. RNA 6: 206–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Splicing-related catalysis by protein-free snRNAs. Nature 413: 701–707. [DOI] [PubMed] [Google Scholar]
- Van Etten, R.L. and Risley, J.M. 1978. Phosphate oxygen-water exchange reaction catalyzed by human prostatic acid phosphatase. Proc. Natl. Acad. Sci. 75: 4784–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa, T., Pleiss, J.A., and Guthrie, C. 2002. Spliceosomal snRNAs: Mg(2+)-dependent chemistry at the catalytic core? Cell 109: 149–152. [DOI] [PubMed] [Google Scholar]
- Weimann, G. and Khorana, H.G. 1962. Studies on polynucleotides. XIII. Stepwise synthesis of deoxyribo-oligonucleotides. An alternative general approach and the synthesis of Thymidine di-and tri- and tetranucleotides bearing 3′-phosphomonoester end groups. J. Am. Chem. Soc. 84: 419–430. [Google Scholar]
- Wolff, T. and Bindereif, A. 1992. Reconstituted mammalian U4/U6 snRNP complements splicing: a mutational analysis. EMBO J. 11: 345–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.A. and Manley, J.L. 1991. Base pairing between U2 and U6 snRNAs is necessary for splicing of a mammalian pre-mRNA. Nature 352: 818–821. [DOI] [PubMed] [Google Scholar]
- Yean, S.L., Wuenschell, G., Termini, J., and Lin, R.J. 2000. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y.T., Maroney, P.A., and Nilsen, T.W. 1993. Functional reconstitution of U6 snRNA in nematode cis- and trans-splicing: U6 can serve as both a branch acceptor and a 5′ exon. Cell 75: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Yu, Y.T., Shu, M.D., and Steitz, J.A. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 17: 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185. [DOI] [PubMed] [Google Scholar]