Abstract
Peptide bond formation on the ribosome is catalyzed by RNA. Kinetic studies using Escherichia coli ribosomes have shown that catalysis (>105-fold overall acceleration) is due to a large part to substrate positioning. However, peptide bond formation is inhibited ~100-fold by protonation of a ribosomal group with pKa=7.5, indicating either a contribution of general acid-base catalysis or inhibition by a pH-dependent conformational change within the active site. The function of a general base has been attributed to A2451 of 23S rRNA, and a charge relay system involving G2447 has been postulated to bring about the extensive pKa shift of A2451 implied in the model. Using a rapid kinetic assay, we found that the G2447A mutation, which has essentially no effect on cell growth, lowers the rate of peptide bond formation about 10-fold and does not affect the ionization of the ribosomal group with pKa=7.5 taking part in the reaction. This result does not support the proposed charge relay mechanism involving G2447 and the role of A2451 as general base in the catalysis of peptide bond formation.
Keywords: Ribosome, peptidyl transfer, rapid kinetics, puromycin, rRNA
INTRODUCTION
During ribosomal protein synthesis, peptide bond formation is catalyzed by the ribosome. The active site, the peptidyl transferase (PT) center, is located on the large subunit of the ribosome, called 50S in bacteria. High-resolution crystal structures of the large ribosomal subunit from the Archaeon Haloarcula marismortui (Ban et al. 2000; Hansen et al. 2002; Schmeing et al. 2002) and from the bacterium Deinococcus radiodurans (Harms et al. 2001; Bashan et al. 2003) revealed that the PT center is built up of highly conserved residues of 23S rRNA and does not contain protein, implying that peptide bond formation is catalyzed by RNA. As revealed by rapid kinetic analysis, a large part of the overall catalysis is contributed by substrate positioning (Katunin et al. 2002), in line with previous suggestions based on biochemical data (Krayevsky and Kukhanova 1979; Nierhaus et al. 1980). Several base pairs between the CCA sequences of substrate or product analogs and bases in the P and A loops of the active site were revealed by the 50S crystal structures (Ban et al. 2000; Hansen et al. 2002; Schmeing et al. 2002). In the P site, C74 and C75 are paired to G2252 and G2251 (Escherichia coli numbering) of the P loop, and in the A site C75 is paired to G2553 in the A loop; these base pairs are essential for the PT reaction (Samaha et al. 1995; Kim and Green 1999). The functional relevance of the model complex structures was suggested by the demonstration of PT activity of 50S crystals (Schmeing et al. 2002). The comparison with the structure of 70S ribosomes with bound tRNAs suggests that the CCA ends of full-size tRNA substrates are arranged in the active site in a similar fashion (Yusupov et al. 2001).
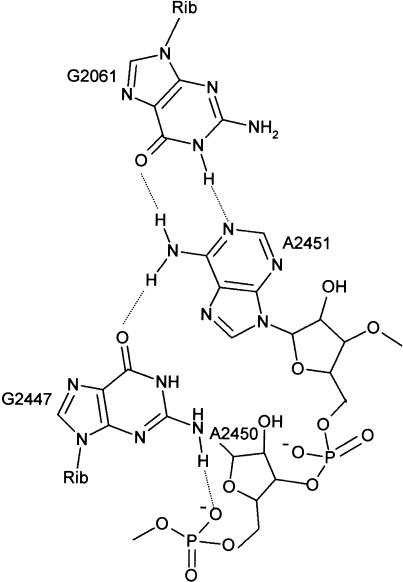
The PT reaction is strongly pH-dependent (Fahnestock et al. 1970) and is inhibited 100-fold by protonation of a single ribosomal residue with pKa=7.5 (Katunin et al. 2002). This suggested that there might be a contribution of general acid-base catalysis in addition to positional catalysis. In the crystal structures, there was no evidence for the presence of catalytic metal ions, suggesting that 23S rRNA bases function as catalytic residues. Based on the crystal structure of the 50S subunit from H. marismortui, Nissen et al. (2000) proposed a model in which bases located within the PT center take part in the chemical mechanism of peptide bond formation. In this model, A2451 was proposed to function as a general base that abstracts a proton from the nucleophilic amino group during the formation of the tetrahedral intermediate. The suggestion was based on the observation that in the crystal structure of the 50S complex with CCdAp-Pm (Pm, puromycin) mimicking the tetrahedral intermediate bound to the active site, N3 of A2451 is in hydrogen-bonding distance to the nitrogen of the tetrahedral intermediate (Nissen et al. 2000). Similarly, the α-NH2 group of Pm coupled to a minihelix bound in the A site is hydrogen-bonded to N3 of A2451, which may have a function in positioning the amino group for the nucleophilic attack (Nissen et al. 2000; Hansen et al. 2002). A general-base function of A2451 requires that the pKa of N3 is shifted from a very low unperturbed value towards neutrality, and it was proposed that groups in the vicinity of A2451, most importantly G2447, form a charge relay system that brings about the pKa shift (Fig. 1 ▶; Nissen et al. 2000). In its central function in forming hydrogen bonds to A2451 and to the phosphate of A2450, G2447 cannot be replaced by adenine or other bases. Therefore, if the postulated general-base function of A2451 were valid, a G2447 to A mutation would be expected to eliminate acid-base catalysis and the protonation at pKa=7.5.
FIGURE 1.
Residues of 23S rRNA at the peptidyl transferase site. Residues are numbered according to the E. coli 23S rRNA sequence.
Mutant ribosomes with a G2447A base exchange were studied both in vivo and in vitro, and small or insignificant effects on cell growth and peptide bond formation were reported (Polacek et al. 2001; Thompson et al. 2001). However, rates of peptide bond formation in both studies were rather low (minutes), precluding direct conclusions regarding the chemistry step of the reaction. A larger effect of the G2447 mutation on Vmax of the puromycin reaction was reported recently (Bobkova et al. 2003); however, it was not shown which step of the reaction measured in turnover was actually affected. In order to obtain quantitative information about the effect of the G2447A mutation, we have used a rapid kinetic assay which measures the rate of peptide bond formation between P site-bound peptidyl-tRNA and A site-bound Pm under conditions where the chemistry step is rate-limiting (Katunin et al. 2002). The same assay was used to determine the pH dependence of peptide bond formation on G2447A ribosomes.
RESULTS AND DISCUSSION
Effect of the G2447A mutation on cell growth
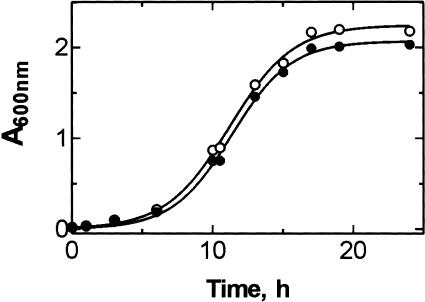
We compared the growth curves of wild-type and mutant strains of E. coli. The growth rate of Δ7 prrn cells (strain MC250) transformed with plasmid pLK35 carrying the rrnB operon with wild-type sequence was decreased compared to E. coli strain DH5α, which contains all seven rDNA operons (data not shown). However, the G2447A mutation had no significant effect on the growth of MC250 cells (Fig. 2 ▶), indicating that the G2447A mutation did not impair overall translation, in line with previous reports (Polacek et al. 2001; Thompson et al. 2001).
FIGURE 2.
Growth curves of E. coli MC250 strain transformed with either plasmid pLK35(wt) (open circles) or pLK35(G2447A) (closed circles).
Kinetics of peptide bond formation on mutant G2447A ribosomes
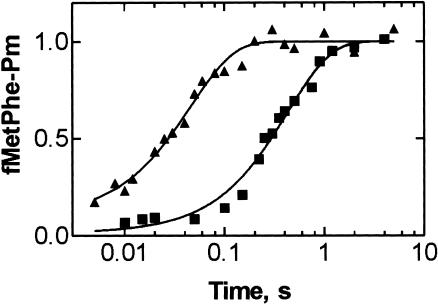
To measure the rate of peptide bond formation, we performed single turnover experiments monitoring the reaction of Pm with fMetPhe-tRNAPhe in the P site. Posttranslocation complexes were prepared as described previously (Katunin et al. 2002) with minor changes (see Materials and Methods). At saturating Pm concentration (10 mM) and pH 7.2, G2447A mutant posttranslocation complexes produced fMetPhe-Pm with a rate constant kpep of 2.3±0.2 s−1 (Fig. 3 ▶). This rate constant is nearly ten times smaller than that determined for wild-type ribosomes from either pLK35-transformed Δ7 prrn cells (kpep = 21 s−1; Fig. 3 ▶) or E. coli MRE600 (kpep = 18 s−1; Katunin et al. 2002).
FIGURE 3.
Effect of the G2447A mutation on the peptidyl transferase reaction. Posttranslocation complexes of G2447A (squares) or wild-type (triangles) ribosomes were reacted with Pm at 37°C (Materials and Methods). Values are normalized by setting to 1.0 the maximum extent of fMetPhe-Pm formed per posttranslocation complex present. Actual values were 0.8–0.9 for wild-type and 0.5–0.6 for mutant ribosomes.
pH dependence of peptide bond formation on G2447A mutant ribosomes
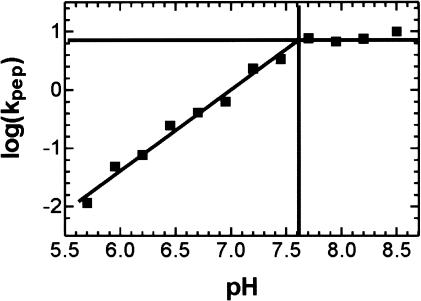
The rate of peptide bond formation between Pm and f[3H]Met[14C]Phe-tRNAPhe on G2447A mutant ribosomes was measured at pH values ranging from 5.7 to 8.5. The plot of log(kpep) versus pH (Fig. 4 ▶) was similar to the one previously obtained with wild-type ribosomes, except that the final level of kpep was about 7 s−1, seven times lower than the maximum rate previously observed with wild-type ribosomes, kpep = 50 s−1 (Katunin et al. 2002). The slope of the plot was 1.4, and the highest pKa of the titrated groups was 7.6 ± 0.1, practically the same values as those determined for wild-type ribosomes.
FIGURE 4.
The pH dependence of the PT reaction with G2447A ribosomes. log(kpep), determined as in Figure 3 ▶, is plotted against pH. The highest titrable pKa is 7.6±0.1, and the slope is 1.4.
The ionization at pKa=7.5±0.1 was assigned to a ribosomal group, and the second pKa involved in the reaction was shown to be due to the protonation of the α-NH2 group of Pm (pKa=6.9; Katunin et al. 2002). The results in Figure 4 ▶ show that the G2447A mutation does not eliminate the ionization of the ribosomal group and its effect on the rate of peptide bond formation. The finding that the pKa values of the ionizing groups in mutant and wild-type ribosomes are the same strongly suggests that the same group is titrated in both types of ribosomes. Thus, the present results are inconsistent with the idea that G2447 has an important function in promoting a pKa shift of A2451 (Nissen et al. 2000), making it unlikely that A2451 exhibits the pKa around 7.5 observed in the titrations. The tenfold inhibition of the PT reaction caused by the G2447A mutation can be explained by the loss of the hydrogen bond to NH2 of A2451 (Fig. 1 ▶) that may result in a different arrangement of A2451 and interfere with positioning the nucleophilic amino group of the A-site substrate by hydrogen bonding to N3 of A2451 (Hansen et al. 2002). It should be noted that lowering the rate of peptide bond formation by a factor of ten is compatible with the unchanged growth rate of cells containing G2447A ribosomes, because the elongation rate is limited by the binding of the substrate, aminoacyl-tRNA, rather than by peptide bond formation (Pape et al. 1999).
Our results leave unresolved the question of whether there is a contribution of general acid-base catalysis to the chemical mechanism of peptide bond formation and, if so, which ribosomal group(s) may be involved, or whether the pH effect is due to a conformational change for which evidence has been reported (Muth et al. 2001; Xiong et al. 2001). We have suggested that the ionizing group in the PT center may be A2450 (Katunin et al. 2002), which in the protonated state can form a noncanonical base pair with C2063 (Muth et al. 2001), and that proton uptake by A2450 may promote the formation of the tetrahedral intermediate of peptide bond formation. At present, this model remains a possibility that needs substantiation by further mutational and structural analysis.
MATERIALS AND METHODS
E. coli strains and plasmids
Plasmid pLK35 containing one rDNA operon coding for either wild-type or G2447A mutant 23S rRNA (Thompson et al. 2001) was transformed into the Δ7 prrn E. coli strain MC250 (Asai et al. 1999). Transformed cells were a gift from A. Dahlberg, Brown University. Mutant cells were grown in LB medium at 37°C in the presence of both ampicillin (125 μg/mL) and chloramphenicol (34 μg/mL). The wild-type E. coli strain DH5α was grown in LB medium at 37°C.
Preparation of ribosome complexes
Materials were prepared as described (Rodnina and Wintermeyer 1995; Rodnina et al. 1999). Ribosomes with 23S rRNA transcribed from the pLK35 plasmid (wild-type or G2447A) were isolated from MC250 cells and purified as described (Rodnina and Wintermeyer 1995). During G2447A mutant ribosome preparation, an aliquot of the culture in the log phase was taken, and the mutation on plasmid pLK35 was confirmed by DNA sequencing. The 70S initiation complex with f[3H]Met-tRNAfMet in the P site was prepared in buffer A (50 mM Tris-HCl, 30 mM KCl, 70 mM NH4Cl, and 7 mM MgCl2, pH 7.2) by incubating 1 μM ribosomes with MFT-mRNA (4 μM; coding for fMetPheThr) and f[3H]Met-tRNAfMet (2 μM) in the presence of initiation factors 1, 2, and 3 (1.7 μM each) for 15 min at 37°C. Binding of EF-Tu•GTP•[14C]Phe-tRNAPhe (1.5 μM) and peptide bond formation resulted in ribosomes carrying deacylated tRNAfMet in the P site and f[3H]Met[14C]Phe-tRNAPhe in the A site. Upon incubation (5 min) with EF-G (0.4 μM) and GTP (1 mM), f[3H]Met[14C]Phe-tRNAPhe was translocated to the P site. Complexes were purified and concentrated by centrifugation through 1.1 M sucrose cushions at 295,000g for 1.5 h (Sorvall M120GX). Ribosome pellets were redissolved in buffer B (50 mM Tris-HCl, 20 mM Bis-Tris-HCl, pH 7.2, 30 mM KCl, 70 mM NH4Cl, and 7 mM MgCl2) to about 4 μM concentration, frozen in liquid nitrogen, and stored at −80°C.
Kinetic measurements
Buffer B was used to cover the pH range from 5.7 to 8.5. For the kinetic assays, ribosomal posttranslocation complexes and Pm stocks were diluted 1:10 into the respective buffer, and the pH (37°C) of both ribosome and Pm solutions was controlled using a small-size pH electrode. Quench-flow assays were performed at 37°C in a KinTek apparatus, mixing equal volumes (12 μL) each of ribosome and Pm solution. Final concentrations were 0.2 μM posttranslocation complex and 10 mM Pm. Reactions were quenched with 25% formic acid. f[3H]Met[14C]Phe-Pm was extracted in 750 μL ethyl acetate in the presence of 500 μL 1.5 M sodium acetate saturated with MgSO4 at pH 4.5. Next, 500 μL of the organic phase was analyzed by double-label radioactivity counting. Results were fitted to a single exponential curve using GraphPad Prism software.
Acknowledgments
We thank A. Dahlberg for generously providing E. coli strains for the preparation of mutant ribosomes, V.I. Katunin and Y.P. Semenkov for tRNA preparations, and Astrid Böhm, Simone Möbitz, Petra Striebeck, and Carmen Schillings for expert technical assistance. The work was supported by the Deutsche Forschungsgemeinschaft, the Alfried Krupp von Bohlen und Halbach-Stiftung, the EC-Program Quality of Life (Contract QLK2-CT-2002-00892), and the Fonds der Chemischen Industrie.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
Pm, puromycin
PT, peptidyl transferase
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5600503.
REFERENCES
- Asai, T., Condon, C., Voulgaris, J., Zaporojets, D., Shen, B., Al-Omar, M., Squires, C., and Squires, C.L. 1999. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 181: 3803–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Bashan, A., Agmon, I., Zarivach, R., Schluenzen, F., Harms, J., Berisio, R., Bartels, H., Franceschi, F., Auerbach, T., Hansen, H.A., et al. 2003. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell 11: 91–102. [DOI] [PubMed] [Google Scholar]
- Bobkova, E.V., Yan, Y.P., Jordan, D.B., Kurilla, M.G., and Pompliano, D.L. 2003. Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones. J. Biol. Chem. 278: 9802–9807. [DOI] [PubMed] [Google Scholar]
- Fahnestock, S., Neumann, H., Shashoua, V., and Rich, A. 1970. Ribosome-catalyzed ester formation. Biochemistry 9: 2477–2483. [DOI] [PubMed] [Google Scholar]
- Hansen, J.L., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2002. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. 99: 11670–11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688. [DOI] [PubMed] [Google Scholar]
- Katunin, V.I., Muth, G.W., Strobel, S.A., Wintermeyer, W., and Rodnina, M.V. 2002. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol. Cell 10: 339–346. [DOI] [PubMed] [Google Scholar]
- Kim, D.F. and Green, R. 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell 4: 859–864. [DOI] [PubMed] [Google Scholar]
- Krayevsky, A.A. and Kukhanova, M.K. 1979. The peptidyltransferase center of ribosomes. Prog. Nucleic Acid. Res. Mol. Biol. 23: 1–51. [DOI] [PubMed] [Google Scholar]
- Muth, G.W., Chen, L., Kosek, A.B., and Strobel, S.A. 2001. pH-dependent conformational flexibility within the ribosomal peptidyl transferase center. RNA 7: 1403–1415. [PMC free article] [PubMed] [Google Scholar]
- Nierhaus, K.H., Schulze, H., and Cooperman, B.S. 1980. Molecular mechanisms of the ribosomal peptidyl transferase center. Biochem. Int. 1: 185–192. [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., and Rodnina, M. 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 18: 3800–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacek, N., Gaynor, M., Yassin, A., and Mankin, A.S. 2001. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature 411: 498–501. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V. and Wintermeyer, W. 1995. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl. Acad. Sci. 92: 1945–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina, M.V., Savelsbergh, A., Matassova, N.B., Katunin, V.I., Semenkov, Y.P., and Wintermeyer, W. 1999. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl. Acad. Sci. 96: 9586–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha, R.R., Green, R., and Noller, H.F. 1995. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature 377: 309–314. [DOI] [PubMed] [Google Scholar]
- Schmeing, T.M., Seila, A.C., Hansen, J.L., Freeborn, B., Soukup, J.K., Scaringe, S.A., Strobel, S.A., Moore, P.B., and Steitz, T.A. 2002. A pretranslocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9: 225–230. [DOI] [PubMed] [Google Scholar]
- Thompson, J., Kim, D.F., O’Connor, M., Lieberman, K.R., Bayfield, M.A., Gregory, S.T., Green, R., Noller, H.F., and Dahlberg, A.E. 2001. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc. Natl. Acad. Sci. 98: 9002–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Polacek, N., Sander, P., Bottger, E.C., and Mankin, A. 2001. pKa of adenine 2451 in the ribosomal peptidyl transferase center remains elusive. RNA 7: 1365–1369. [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]