Abstract
CA dinucleotide repeat sequences are very common in the human genome. We have recently demonstrated that the polymorphic CA repeats in intron 13 of the human endothelial nitric oxide synthase (eNOS) gene function as an unusual, length-dependent splicing enhancer. The CA repeat enhancer requires for its activity specific binding of hnRNP L. Here we show that in the absence of bound hnRNP L, the pre-mRNA is cleaved directly upstream of the CA repeats. The addition of recombinant hnRNP L restores RNA stability. CA repeats are both necessary and sufficient for this specific cleavage in the 5′ adjacent RNA sequence. We conclude that—in addition to its role as a splicing activator—hnRNP L can act in vitro as a sequence-specific RNA protection factor. Based on the wide abundance of CA repetitive sequences in the human genome, this may represent a novel, generally important role of this abundant hnRNP protein.
Keywords: Repetitive sequence, hnRNP L, RNA stability, splicing
INTRODUCTION
CA dinucleotide repetitive sequences represent the most common fraction in the microsatellite DNA of the human genome (approximately 0.25%; Lander et al. 2001). Very often these are polymorphic sequence elements, and their function has remained largely unclear. In this study, we focus on the human gene for the endothelial isoform of nitric oxide synthase (eNOS), which plays an important physiological role in vascular homeostasis. This gene carries a well-characterized CA repeat polymorphism in its intron 13 (4.1 kb), varying between 14 and 44 CA repeats and mapping only 80 nt downstream of the 5′ splice site of exon 13 (Nadaud et al. 1994). Interestingly, a large statistical evaluation revealed that the CA repeat number can be considered an independent risk factor for coronary artery disease, in particular at higher CA repeat numbers (Stangl et al. 2000). To address the potential mechanistic basis for this correlation, we have recently demonstrated that the CA repeats act as an intronic splicing-regulatory element (Bilbao and Valcárcel 2003; Hui et al. 2003). In addition, we found that the heterogenous nuclear ribonucleoprotein L (hnRNP L)—through specific binding to the CA repeat enhancer—activates eNOS splicing, establishing for the first time a splicing-regulatory role of this abundant nuclear protein.
hnRNP L had been identified as a major nuclear protein of 64–68 kD (pI = 7.4–7.7) containing three RNA recognition motifs (RRMs) as well as several glycine- and proline-rich regions (Piñol-Roma et al. 1989). Its protein sequence is most similar to hnRNP I/polypyrimidine tract binding protein (PTB: 27% identity; Ghetti et al. 1992). In addition to some general nucleoplasmic staining, hnRNP L preferentially localizes to the landmark giant loops of the newt lampbrush chromosomes, which are rich in transcribed repetitive sequence elements (Piñol-Roma et al. 1989, and references therein).
Regarding the function of hnRNP L, there are several reports on specific RNA binding: First, hnRNP L could be UV-crosslinked to a cis-acting RNA element of the herpes simplex virus thymidine kinase (HSV-TK) gene, which is necessary for intron-independent cytoplasmic mRNA accumulation (Liu and Mertz 1995). Second, hnRNP L was found to bind to the 3′ border region of the hepatitis C virus (HCV) internal ribosome entry site (IRES; Hahm et al. 1998). Third, an hnRNP L-binding site was mapped within the 3′ UTR of the human vascular endothelial growth factor (VEGF) mRNA, functionally important for regulated mRNA stability under conditions of low oxygen supply (Shih and Claffey 1999). Close inspection revealed that in each of these three cases, the binding site contains A/C-rich sequence clusters (see references above and data not shown). Because these studies and our own work include both nuclear (mRNA splicing enhancement, mRNA export) and cytoplasmic functions (mRNA translation and stability), hnRNP L—like the well-studied hnRNP A1 (Piñol-Roma and Dreyfuss 1992)—may also shuttle between nucleus and cytoplasm.
Other members of the hnRNP family participate in many functions during gene expression, ranging from transcription, hnRNA packaging, splicing regulation, and polyadenylation to mRNA export and stability (Krecic and Swanson 1999). This diversity is generated by differential RNA-binding specificities and multiple protein–protein interactions.
In this study, we demonstrate that hnRNP L—through binding with high affinity to CA repeats—functions in vitro as an RNA-stabilizing factor. CA repeats are sufficient for rendering the adjacent upstream RNA sequence particularly sensitive to cleavage. Because CA repeats are widely distributed in the human genome, the RNA protection function of hnRNP L—in addition to splicing activation—probably constitutes a general role of this abundant nuclear hnRNP protein.
RESULTS AND DISCUSSION
Destabilization of CA-repeat containing RNA by (CA)32 competitor RNA
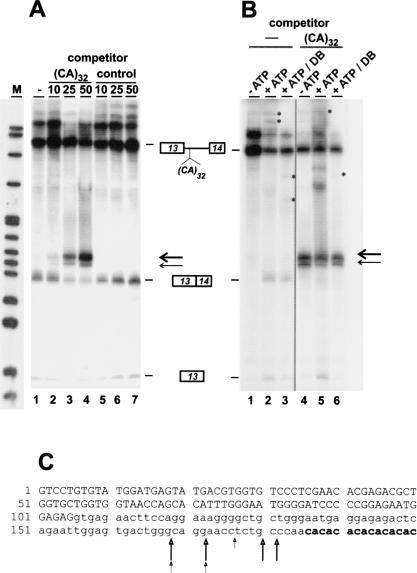
We have recently identified hnRNP L as the major protein binding to the CA repeat splicing enhancer, using affinity purification on an RNA oligonucleotide consisting of 32 CA dinucleotides, (CA)32 (Hui et al. 2003). To test for competition effects of the (CA)32 RNA, we performed in vitro splicing of 32P-labeled pGEM-CA32, an eNOS minigene pre-mRNA that consists of eNOS exons 13 and 14 interrupted by a shortened version of intron 13 with 32 CA repeats (Hui et al. 2003). When we added (CA)32 RNA as a competitor during in vitro splicing of this minigene pre-mRNA, we were surprised to observe—in addition to the expected splicing products—a major additional fragment of ca. 195 nt; furthermore, a minor, slightly shorter fragment of ca. 190 nt was resolved (Fig. 1A ▶, see bands marked by arrows). These two bands appeared only in the presence of the (CA)32 RNA and strongly increased in abundance as the molar excess of (CA)32 RNA was changed from 10- to 25- to 50-fold (lanes 1–4). Furthermore they were specific for (CA)32 RNA, as they were absent when instead of (CA)32 a control RNA was used as competitor (lanes 5–7). In subsequent experiments, we observed these bands also in the absence of ATP (Fig. 1B ▶, cf. lanes 1/2 and 4/5). After a debranching treatment, both bands remained intact, in contrast to the lariat intermediate and excised lariat bands, which were linearized (see bands marked by asterisks on lanes 2/3 and 5/6). Taken together, these results indicate that the RNA fragments observed represent specific, splicing-independent cleavage products of the eNOS CA32 pre-mRNA. Moreover they suggest that (CA)32 RNA competes for a specific protein factor, resulting in RNA destabilization.
FIGURE 1.
Competition by (CA)32 RNA destabilizes CA-repeat containing RNA. (A) 32P-labeled eNOS pre-mRNA with 32 CA repeats was incubated under splicing conditions for 90 min in the absence of competitor (lane 1) or with a 10-, 25-, or 50-fold molar excess of unlabeled (CA)32 RNA (lanes 2–4) or a 64-nt control RNA (lanes 5–7). After incubation, the RNAs were separated on an 8% denaturing polyacrylamide gel, followed by autoradiography. The positions of pre-mRNA, spliced product, and first exon are shown on the right (the strong band above the pre-mRNA appeared already without incubation and varied between different extract preparations). Thick and thin arrows mark the positions of the major and minor degradation products, respectively. (M) pBR322/HpaII marker fragments. (B) 32P-labeled eNOS pre-mRNA with 32 CA repeats was incubated for 90 min without (lanes 1,4) or with ATP (lanes 2,3,5,6), and without (lanes 1–3) or with a 50-fold molar excess of (CA)32 RNA (lanes 4–6). In addition, the RNAs shown in lanes 3 and 6 had been treated with HeLa cell S100 extract for debranching (DB). The putative lariat intermediate and excised lariat bands as well as their debranching products are indicated by asterisks. (C) The cleavage sites were precisely mapped within intron 13 of the eNOS gene, as described in Materials and Methods (eNOS exon 13 sequence in capital letters; intron 13 sequence in small letters; CA repeats in bold). Thick and thin arrows indicate cleavage sites derived from the major and minor degradation products, respectively.
To identify these RNA cleavage products, oligonucleotide-directed RNase H cleavage reactions were done, targeting different regions of the pre-mRNA; this revealed that the fragments contained exon 13 and adjacent intron sequences, but not exon 14 (data not shown). We subsequently mapped precisely the cleavage sites by gel purifying both the major and minor fragments, followed by 3′ end-tailing with poly(A) polymerase, cDNA cloning, and sequence analysis (see Materials and Methods). The results are summarized in Figure 1C ▶. Both the major and minor fragments contain the complete exon 13 and extend into intron 13 up to the region immediately upstream of the CA repeats [after positions −18 to −5 relative to (CA)32]. Surprisingly, we have never detected the remaining 3′ part of the pre-mRNA. In sum, these data demonstrate that competition by (CA)32 RNA induces specific cleavage within the intronic region upstream of the CA repeats, resulting in a stable 5′ terminal RNA fragment.
hnRNP L stabilizes CA-repeat-containing RNA
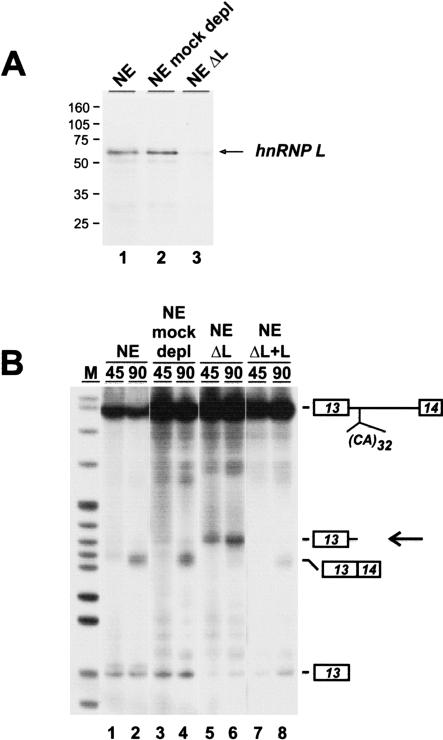
Because hnRNP L represents the major protein binding to CA repeats (Hui et al. 2003), we next tested for RNA degradation in extract specifically depleted of hnRNP L (Fig. 2 ▶). Nuclear extract was pretreated with a 5′-biotinylated (CA)32 RNA oligonucleotide and streptavidin agarose, resulting in depletion of more than 95% of hnRNP L, as detected by Western blot analysis (Fig. 2A ▶). As a control, nuclear extract was treated in the same way in the absence of oligonucleotide, resulting in mock-depleted extract. 32P-labeled eNOS minigene pre-mRNA with 32 CA repeats was incubated under splicing conditions for 45 and 90 min in untreated nuclear extract (NE; Fig. 2B ▶, lanes 1,2), mock-depleted extract (NE mock-depl; lanes 3,4), hnRNP-depleted extract (NEΔL; lanes 5,6), and hnRNP-depleted extract complemented by baculovirus-expressed recombinant hnRNP L (NEΔL + L; lanes 7,8).
FIGURE 2.
HnRNP L stabilizes CA-repeat containing RNA. (A) Western blot analysis of untreated nuclear extract (lane 1), mock-depleted nuclear extract (lane 2), and hnRNP L-depleted nuclear extract (lane 3). The sizes of marker proteins are given in kilodaltons. (B) 32P-labeled eNOS pre-mRNA with 32 CA repeats was incubated under splicing conditions for 45 and 90 min in untreated extract (lanes 1,2), mock-depleted extract (lanes 3,4), hnRNP L-depleted extract (lanes 5,6), and in depleted extract complemented with recombinant hnRNP L (600 ng per 25 μL reaction). RNAs were isolated and analyzed by denaturing polyacrylamide gel electrophoresis (8%) and autoradiography. The positions of pre-mRNA, spliced product, first exon, and the degradation product (arrow) are marked on the right. (M) pBR322/HpaII marker fragments.
In both normal and mock-depleted nuclear extract, the eNOS pre-mRNA was spliced (lanes 1–4), whereas in hnRNP L-depleted extract, only very little spliced product could be detected. Instead, the 5′ cleavage fragment appeared already after 45 min of incubation (lanes 5,6). After complementation by recombinant hnRNP L (600 ng per 25 μL reaction) no degradation occurred, and splicing activity was restored to a significant level. This demonstrated directly that, first, hnRNP L depletion is responsible for the destabilization effect and the appearance of the specific 5′ terminal cleavage products. Second, hnRNP L is required for protecting CA-repeat-containing RNA from degradation.
CA repeats are necessary and sufficient for destabilizing RNA
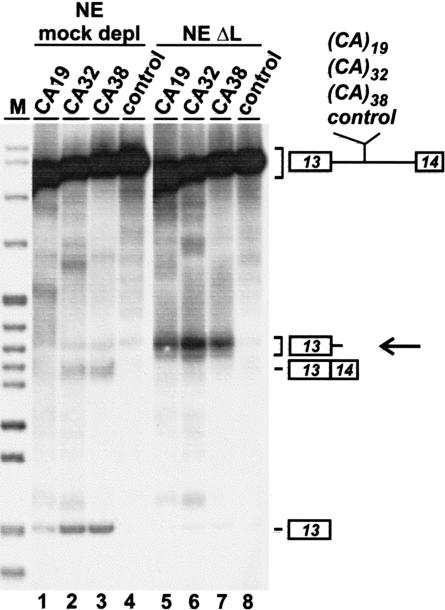
We next asked whether specific cleavage depends on the CA repeats and their length (Fig. 3 ▶). Four different 32P-labeled eNOS minigene pre-mRNAs (Hui et al. 2003) were tested that are composed of exons 13 and 14, interrupted by a shortened version of intron 13 and containing at the normal intron position of the polymorphic CA repeats either 19, 32, or 38 CA repeat copies (CA19, CA32, and CA38), or a 64-nt nonspecific sequence (control). Each of these 32P-labeled substrates was incubated under splicing conditions for 90 min in mock-depleted (lanes 1–4) or hnRNP L-depleted nuclear extract (lanes 5–8), followed by direct RNA analysis. Low splicing activity could be detected in mock-depleted extract for the CA19 construct (lane 1, see exon 13 band) and higher activities for the CA32 and CA38 constructs (lanes 2,3); in contrast, there was no detectable activity for the control pre-mRNA (lane 4), consistent with our recent finding that splicing activity correlates with the CA repeat number (Hui et al. 2003). In hnRNP L-depleted extract, we could observe the cleavage products for each of the substrates with 19, 32, and 38 CA repeats, but not for the control pre-mRNA (lanes 5–8, see band marked by the arrow). In conclusion, the specific cleavage depended on the CA repeat sequence, although there was no apparent strong correlation with the CA repeat length.
FIGURE 3.
CA repeats are necessary for RNA destabilization. The following 32P-labeled eNOS pre-mRNAs were incubated under splicing conditions for 90 min in mock-depleted extract (lanes 1–4) and hnRNP L-depleted extract (lanes 5–8): eNOS with 19 CA repeats (CA19; lanes 1,5), 32 CA repeats (CA32; lanes 2,6), 38 CA repeats (CA38; lanes 3,7), and a 64-nt control sequence (control; lanes 4,8). RNAs were isolated and analyzed by denaturing polyacrylamide gel electrophoresis (8%) and autoradiography. The positions of pre-mRNA, spliced product, first exon, and the degradation products (arrow) are marked on the right. (M) pBR322/HpaII marker fragments.
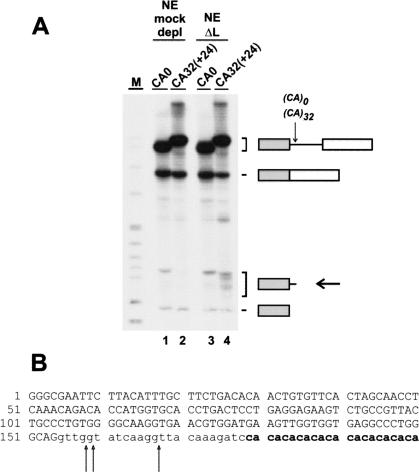
To investigate whether CA repeats are sufficient for the observed destabilization, we also tested a heterologous CA repeat-containing construct. A human β-globin construct was made that contains the first two exons and 32 CA repeats inserted after intron position +24 [CA32(+24); Fig. 4A ▶]. β-Globin pre-mRNAs derived from the starting construct without CA repeats (CA0) and from the (CA)32-containing construct [CA32(+24)] were incubated in either mock- or hnRNP L-depleted nuclear extract under splicing conditions for 90 min, followed by direct RNA analysis. As a result, in mock-depleted extract, each of these two substrates was spliced at approximately the same, high efficiency (lanes 1,2). This indicates that in the context of the β-globin pre-mRNA, which is spliced in vitro relatively efficiently, no further stimulation can be achieved by the CA repeat splicing enhancer. When incubated in hnRNP L-depleted extract, normal β-globin pre-mRNA gave the same product pattern as in mock-depleted extract (cf. lanes 1 and 3). However, the β-globin CA32(+24) pre-mRNA with intronic CA repeats produced a series of additional fragments ranging in size from 160 to 180 nt. As done for the eNOS minigene pre-mRNA (see Fig. 1 ▶), we characterized these degradation products by poly(A) tailing, followed by cDNA cloning and sequence analysis of individual clones. As a result, cleavage sites were precisely mapped at three different positions upstream of the CA repeats (after positions −21, −20, and −12 relative to the CA repeats; see Fig. 4B ▶). Finally, another β-globin derivative, CA32(+85) pre-mRNA, in which 32 CA repeats were inserted in intron 1 after position +85, was incubated in hnRNP L-depleted extract; cleavage products were observed in the size range expected from cleavage upstream of this new CA repeat location (data not shown).
FIGURE 4.
CA repeats are sufficient for RNA destabilization. (A) 32P-labeled β-globin pre-mRNAs without CA repeats (CA0; lanes 1,3) and with 32 CA repeats after position +24 in the intron [CA32(+24); lanes 2,4] were incubated under splicing conditions for 90 min in mock-depleted (lanes 1,2) or hnRNP L-depleted nuclear extract (lanes 3,4), followed by direct RNA analysis on an 8% denaturing polyacrylamide gel and autoradiography. The positions of pre-mRNA, spliced product, first exon, and the degradation products (arrow) are shown on the right. (M) pBR322/HpaII marker fragments. (B) The cleavage sites (arrows) were precisely mapped within the β-globin intron, as described in Materials and Methods (β-globin first exon sequence in capital letters; intron sequence in small letters; CA repeats in bold).
Taken together, the data from Figures 3 ▶ and 4 ▶ demonstrate that CA repeats are both necessary and sufficient for specifying—in the absence of hnRNP L—RNA cleavage within the adjacent upstream region.
Combining the mapping data from the eNOS and β-globin degradation products (Figs. 1C ▶ and 4B ▶), we have identified several cleavage sites within approximately 20 nt upstream of the CA repeats, with a preference for cleavage after guanosine or cytidine nucleotides (however, we cannot be certain that all sites mapped reflect primary cleavage by the responsible RNase). Because surprisingly in both cases only the 5′ terminal cleavage product appeared as a stable product and we were unable to detect the corresponding 3′ terminal fragment, the latter one is most likely rapidly degraded after the initial cleavage. Several attempts to detect specific cleavage products of the eNOS pre-mRNA in vivo—after knock-down of hnRNP expression by RNA interference—so far have been unsuccessful, which may be due to the instability of the degradation intermediates in vivo (data not shown). Is the hnRNP-dependent stabilization of the eNOS pre-mRNA (this study) related to the RNA stability function of hnRNP L reported for the human VEGF mRNA, where a C/A-rich element in the 3′-UTR binds hnRNP L and mediates mRNA stability under hypoxic growth conditions (Shih and Claffey 1999)? The latter case of hnRNP L-dependent regulation was studied in a cell culture system, operates at the level of mRNA stability in the cytoplasm, and was not characterized further in its molecular mechanism. In contrast, we have uncovered here a CA-repeat-dependent RNA stabilizing role of hnRNP L that should function at the nuclear pre-mRNA level. Therefore it will be interesting to investigate further whether these two RNA stability effects are based on the same principle, that is, recognition of C/A-rich regulatory RNA elements by hnRNP L. Regarding the mechanism of hnRNP L-mediated protection, an attractive model would involve competition between an endonuclease and a specific RNA-binding protein such as hnRNP L (see Beelman and Parker 1995, and references therein). Because of the high abundance of CA dinucleotide repeats in the human genome, we propose that hnRNP L may provide an RNA-stabilizing function in many cases, not only for regular CA repeat RNA (this study), but also for other C/A-rich RNA elements (see references in Introduction). Consistent with such a general RNA protection function, a large fraction of hnRNP L had originally been detected in micrococcal-nuclease sensitive complexes (Piñol-Roma et al. 1989).
MATERIALS AND METHODS
DNA oligonucleotides
B1: 5′-GATC(CA)32-3′; BB2: 5′-GATC(TG)32-3′; DSX1: 5′-CTA GA(CA)32A-3′; DSX2: 5′-AGCTT(TG)32T-3′; DSX3: 5′-GAAAA ATTCCGCTATCCTTG-3′; DSX4: 5′-CACATACGATTTAGGTG ACA-3′; EN1: 5′-TTAAGAATTCGTCCTGTGTATGGATGAGTA-3′; oligo (dT)18.
Plasmids
pGEM-CA19, -CA32, -CA38, and -control were used for T7 transcription and have been previously described (Hui et al. 2003). pComp-CA32 and pComp-control: DNA fragments containing 32 CA repeats or a 64-nt control sequence were generated by annealing oligonucleotides DSX1 and DSX2 or by amplifying from pGEM5Zf(+) using oligonucleotides DSX3 and DSX4, respectively. These two fragments were cloned into pGEM-3Z between the XbaI and HindIII sites. To move the inserted sequences close to the T7 transcription initiation site, the new clones were cleaved by EcoRI and XbaI and religated after filling in by Klenow enzyme to generate pComp-CA32 and pComp-control. For in vitro transcription, pComp-CA32 and pComp-control were linearized by HindIII. Transcribed RNAs were used as competitors in splicing competition assays. pHβG-CA0 and pHβG-CA32(+24): pHβG-CA0 was made by introducing through PCR a BglII restriction site at position +24 of β-globin intron 1 in pT7-Hβ (Reed et al. 1988), which contains exon 1, intron 1, and exon 2 of the human β-globin gene. For pHβG-CA32(+24), a DNA fragment generated by annealing the two oligonucleotides BB1 and BB2 was cloned into the BglII site of pHβG-CA0, resulting in the insertion of 32 CA repeats after position +24 of β-globin intron 1.
In vitro transcription and splicing
In vitro transcription and splicing were performed as previously described (Hui et al. 2003). For splicing competition assays, a 10-, 25-, or 50-fold molar excess of unlabeled short RNAs containing 32 CA repeats or a 64-nt random sequence (transcribed from pComp-CA32 and pComp-control, respectively) was added to the in vitro splicing reaction. Debranching analysis after splicing was carried out in HeLa cell S100 extract (Ruskin and Green 1985). Purified RNAs from splicing or debranching reactions were separated on 8% denaturing polyacrylamide gels.
HnRNP L depletion and complementation in HeLa nuclear extract
Mock or affinity depletions of hnRNP L in HeLa nuclear extract were performed as previously described (Hui et al. 2003). HnRNP L depletion was assessed through Western blotting using anti-hnRNP L monoclonal antibody 4D11 (Piñol-Roma et al. 1989) as described. For complementation of depleted extract, 600 ng of purified recombinant hnRNP L were added to a 1× splicing reaction (25 μL). Recombinant hnRNP L was expressed in the baculovirus system and purified as described (Hui et al. 2003).
Identification of RNA cleavage products
The 32P-labeled RNA cleavage products generated after a 90-min incubation in hnRNP L-depleted nuclear extract were gel-purified, followed by poly(A) tailing for 2 h at 37°C in a 20 μL reaction containing 40 mM Tris (pH 8.0), 200 mM NaCl, 10 mM MgCl2, 2 mM of MnCl2, 50 μg/mL BSA, 3 mM ATP, 40 U RNasin, and 1.4 U poly(A) polymerase (GIBCO Life Technology). The poly(A)-tailed RNAs were then reverse transcribed, using oligo (dT)18 primer, followed by a PCR reaction with eNOS exon 13-specific primer EN1 and oligo (dT)18. The amplified cDNA fragments were subsequently cloned into pCR2.1-TOPO (Invitrogen) for sequencing.
Acknowledgments
We thank Gideon Dreyfuss for monoclonal anti-hnRNP L antibody 4D11, Silke Schreiner for technical assistance, and Zsofia Palfi for comments on the manuscript. This work was supported by the Deutsche Forschungsgemeinschaft.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5660803.
REFERENCES
- Beelman, C.A. and Parker, R. 1995. Degradation of mRNA in eukaryotes. Cell 81: 179–183. [DOI] [PubMed] [Google Scholar]
- Bilbao, D. and Valcárcel, J. 2003. Getting to the heart of a splicing enhancer. Nat. Struct. Biol. 10: 6–7. [DOI] [PubMed] [Google Scholar]
- Ghetti, A., Piñol-Roma, S., Michael, W.M., Morandi, C., and Dreyfuss, G. 1992. hnRNP I, the polypyrimidine tract-binding protein: Distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 20: 3671–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm, B., Kim, Y.K., Kim, J.H., Kim, T.Y., and Jang, S.K. 1998. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 72: 8782–8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, J., Stangl, K., Lane, W.S., and Bindereif, A. 2003. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat. Struct. Biol. 10: 33–37. [DOI] [PubMed] [Google Scholar]
- Krecic, A.M. and Swanson, M.S. 1999. hnRNP complexes: Composition, structure, and function. Curr. Opin. Cell Biol. 11: 363–371. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nussbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Liu, X. and Mertz, J.E. 1995. HnRNP L binds a cis-acting RNA sequence element that enables intron-independent gene expression. Genes & Dev. 9: 1766–1780. [DOI] [PubMed] [Google Scholar]
- Nadaud, S., Bonnardeaux, A., Lathrop, M., and Soubrier, F. 1994. Gene structure, polymorphism and mapping of the human endothelial nitric oxide synthase gene. Biochem. Biophys. Res. Comm. 198: 1027–1033. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma, S. and Dreyfuss, G. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355: 730–732. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma, S., Swanson, M.S., Gall, J.G., and Dreyfuss, G. 1989. A novel heterogeneous nuclear RNP protein with a unique distribution on nascent transcripts. J. Cell. Biol. 109: 2575–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R., Griffith, J., and Maniatis, T. 1988. Purification and visualization of native spliceosomes. Cell 53: 949–961. [DOI] [PubMed] [Google Scholar]
- Ruskin, B. and Green, M.R. 1985. An RNA processing activity that debranches RNA lariats. Science 229: 135–140. [DOI] [PubMed] [Google Scholar]
- Shih, S.C. and Claffey, K.P. 1999. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J. Biol. Chem. 274: 1359–1365. [DOI] [PubMed] [Google Scholar]
- Stangl, K., Cascorbi, I., Laule, M., Klein, T., Stangl, V., Rost, S., Wernecke, K.D., Felix, S.B., Bindereif, A., Baumann, G., and Roots, I. 2000. High CA repeat numbers in intron 13 of the endothelial nitric oxide synthase gene and increased risk of coronary artery disease. Pharmacogenetics 10: 133–140. [DOI] [PubMed] [Google Scholar]