Abstract
Cells infected with human cytomegalovirus (HCMV) after commencing DNA replication do not initiate viral immediate-early (IE) gene expression and divide before arresting. To determine the nature of this blockade, we examined cells that were infected 24 h after release from G0 using immunofluorescence, laser scanning cytometry, and fluorescence-activated cell sorting (FACS) analysis. Approximately 40 to 50% of the cells had 2N DNA content, became IE+ in the first 12 h, and arrested. Most but not all of the cells with >2N DNA content did not express IE antigens until after mitosis. To define the small population of IE+ cells that gradually accumulated within the S and G2/M compartments, cells were pulsed with bromodeoxyuridine (BrdU) just prior to S-phase infection and analyzed at 12 h postinfection for IE gene expression, BrdU positivity, and cell cycle position. Most of the BrdU+ cells were IE− and had progressed into G2/M or back to G1. The majority of the IE+ cells in S and G2/M were BrdU−. Only a few cells were IE+ BrdU+, and they resided in G2/M. Multipoint BrdU pulse-labeling revealed that, compared to cells actively synthesizing DNA at the beginning of the infection, a greater percentage of the cells that initiated DNA replication 4 h later could express IE antigens and proceed into S. Synchronization of the cells with aphidicolin also indicated that the blockade to the activation of IE gene expression was established in cells soon after initiation of DNA replication. It appears that a short-lived protein in S-phase cells may be required for IE gene expression, as it is partially restored by treatment with the proteasome inhibitor MG132.
Human cytomegalovirus (HCMV) is a widespread human pathogen that latently infects 50 to 90% of the world's population. It is the leading viral cause of birth defects, with a congenital infection rate of approximately 1%. Serious neurological defects are manifest at birth in 5 to 10% of these infants, and an additional 10 to 15% will later develop various levels of sensorineural hearing loss and/or learning disabilities (6, 9, 11, 21). HCMV infection is also a major medical problem in immunocompromised individuals (9) and may contribute to the development of atherosclerosis and restenosis following coronary angioplasty (54).
The significance of this virus as a human pathogen has highlighted the importance of developing an understanding of its interaction with its host on a cellular level. It is clear from earlier studies that initial contact of the virus with the plasma membrane generates a rapid physiological response similar to a second-messenger cascade (1). This is also accompanied by a rapid, albeit transient, induction of c-fos, c-jun, and c-myc mRNAs (5) and an induction of a subset of the mRNAs encoding genes induced by alpha interferon in uninfected cells (40, 55, 56). The ability of UV-inactivated virus to stimulate the initial signaling responses within the cell indicates that binding and passage of the virus through the cell membrane is sufficient for activation. In fact, work from the laboratory of Teresa Compton has shown that the majority of the signaling events in the early stages of infection can be triggered exclusively by the binding of the viral glycoprotein gB to its receptor on the cell surface (7).
In the past few years, a more complex story has unfolded concerning the interaction of the virus with the host cell cycle machinery. In order to observe the effects of the virus on a population of synchronous cells, most of these initial studies were performed in permissive fibroblast cells infected just after release from a G0 arrest (8, 15, 31). Under these conditions, there is marked dysregulation of the cell cycle, with induction of elevated levels of p53, active cyclin E and B complexes, and hyperphosphorylated Rb. Expression of cyclin A, however, is delayed until late times in the infection (31). Of primary importance to the viral program is the hyperphosphorylation of Rb, which releases the inhibition that this protein imposes upon the E2F/DP transactivator complexes. Key targets of these complexes are genes encoding proteins involved in DNA replication (16) and include DNA polymerase α, thymidine kinase, and dihydrofolate reductase, all of which are stimulated by HCMV infection (17, 27, 52). Two other regulatory proteins that are controlled by E2F and play an important role in the control of cellular DNA replication are cdc6 and Hsorc1 (24, 42). In addition, ornithine decarboxylase (30) and PCNA (15) are also stimulated by viral infection. It should be noted that although the virus encodes its own polymerase, processivity factor, single-stranded binding protein, and helicase/primase complex (43, 44), it is dependent upon the DNA replication machinery of the cell as well. Upregulation of the above-mentioned proteins ultimately leads to what has been termed a fully “activated” state for viral DNA replication.
Several reports have shown that cells infected in a quiescent state will arrest before dividing. Cellular DNA synthesis is blocked, and whether the observed inhibition is complete seems to depend on the cell synchronization methods used and timing of the infection after release (8, 15, 31, 35, 47). The absence of active cyclin A/Cdk complexes (which are essential for S-phase entry and exit) during early times of a G0 infection is likely responsible, at least in part, for the block in cellular DNA synthesis. Recent work has also suggested that UL69, a structural protein brought in with the virion, may be partially responsible for the G0/G1 arrest in these cells (25, 36).
Another protein potentially responsible for the observed cell cycle dysregulation is the viral immediate-early (IE) protein IE86 (53). Under G0 infection conditions, the IE proteins appear in the host cell nucleus by 1 to 2 h postinfection (hpi) (29; this paper). These IE proteins play a large role in initiating the viral replication cycle, as they are the major transactivator proteins for the early class of genes necessary for viral DNA replication. Studies from our laboratory and others have shown that the two major IE proteins can interact with many cellular and viral proteins in their roles as initiators of infection. The IE86 protein can interact not only with itself, but also with components of the cellular basal transcription machinery, the tumor suppressor proteins Rb and p53, and a number of transcription factors (for a review, see reference 20). The IE72 protein has also been found to form complexes with multiple regulatory proteins, including several TBP-associated factors, the transcription factor CTF-1, and the Rb-related protein p107 (26, 37, 46). Transient expression of IE86 alone has been shown to block cell division either by arresting the cells at G1/S or some time after the cells have committed to replication (10, 39, 53).
We have extended the analysis of viral interactions with the host cell by looking at the infection not only in a G0-arrested population, but also in cells that are released from confluence and then allowed to proceed into the ensuing cell cycle for 24 h before infection (S-phase infection). Previous work showed that at this point, approximately 50% of the cells are in S and G2/M and that cells actively replicating their DNA at the time of infection (as evidenced by bromodeoxyuridine [BrdU] incorporation) do not express viral IE gene products for the first 12 hpi (47). Additionally, unlike cells that are infected in G0 or G1, S-phase cells can traverse the cell cycle and divide once before becoming arrested in the following G1.
In this paper, we further define this blockade to expression of IE proteins during an infection in S phase. We found that most cells that were actively synthesizing their DNA at the beginning of the infection did not express the IE proteins until the cells underwent mitosis and entered the following G1. Through the use of aphidicolin synchronization, we also demonstrated that the blockade to activation of IE gene expression occurred very quickly after the initiation of DNA replication. When cells were pulsed with BrdU just prior to S-phase infection and harvested at 12 hpi, we found that a small population of IE+ cells gradually appeared in the S and G2/M compartments as the infection progressed. However, these cells were not actively synthesizing their DNA at the beginning of the infection, as evidenced by their BrdU− status. By pulse-labeling cells with BrdU at 4 hpi, we showed that at least some of these cells originated in G1 and were able to express IE antigens and traverse through S phase. Of particular interest is our finding that treatment of cells with the proteasome inhibitor MG132 partially relieved this block to IE gene expression in S phase. The significance of these findings with respect to infection of different cell types, the implications of partial reversal of the block through proteasome inhibition, and other possible mechanisms of repression are discussed.
MATERIALS AND METHODS
Cells and virus
Primary human foreskin fibroblasts were obtained from the University of California, San Diego, Medical Center and propagated in Earle's minimal essential medium (MEM) in incubators maintained at 37°C and 5% CO2. Medium was supplemented with 10% heat-inactivated fetal bovine serum (FBS), l-glutamine (2 mM), penicillin (200 U/ml), streptomycin (200 μg/ml), amphotericin B (1.5 μg/ml), and gentamicin sulfate (50 μg/ml). The Towne strain of HCMV was obtained from the American Type Culture Collection (no. VR977) and propagated as described earlier (50). All infections were performed at a multiplicity of 5.
Cell synchronization methods. (i) G0
Cells were grown to confluence and maintained at high density for 3 days. Cells were then trypsinized, reseeded at a density of 0.75 × 106 to 1.0 × 106 per 10-cm dish, and allowed to settle for 1 h before infection with virus- or mock-infected supernatant. Viral supernatant was removed 2 h later, and the cells were washed once and refed. Cells were then harvested at the indicated times postinfection (p.i.) and treated as described below for different assays.
(ii) S phase
S-phase infections were carried out as described earlier (19, 47). Briefly, cells were treated as in a G0 infection, but instead of being infected at 1 h postplating, cells were allowed to grow for 24 h before infection. Cells were harvested at indicated time points p.i.
(iii) Aphidicolin synchronization at G1/S boundary
Confluent cells were trypsinized and seeded at 5 × 105 cells per 10-cm plate. After 15 to 16 h, cells were placed into medium containing 2 μg of aphidicolin per ml for 24 h. Cells were then washed in fresh medium and infected at a multiplicity of infection of 5. The cells were again washed and refed after 2 h and harvested at the indicated time points.
Immunofluorescence. (i) Standard
Cells were seeded onto coverslips and infected as above for G0- or S-phase infections. At various times p.i., a set of mock- and virus-infected coverslips was harvested, washed twice with phosphate-buffered saline (PBS), and then simultaneously fixed and permeabilized in −20°C methanol for 10 min. Cells were again washed twice in PBS and stored at 4°C until the time course was complete. All coverslips were stained first with primary antibody (Ab) to IE72/86 (CH16.0; Goodwin Institute), followed by detection using fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin secondary Ab (Jackson Laboratories). FITC+ cells were then scored using a Zeiss Axiophot fluorescence microscope. At least 100 cells were counted for each time point.
(ii) BrdU pulse-labeling experiments
BrdU pulse-labeling experiments were carried out as described by Salvant et al. (47) with the following changes. Cells were seeded for S-phase infections. BrdU labeling was carried out for 30 min either 1 h prior to infection (time zero) or at 4 hpi. If labeled at time zero, cells were harvested at 4 or 12 hpi. If labeled at 4 hpi, cells were harvested at 5 or 12 hpi. Processing of coverslips was carried out as described to detect both BrdU incorporation and IE positivity.
Laser scanning cytometry
Cells were plated onto coverslips for S-phase infection as described above and pulsed with BrdU just prior to infection at 24 h postplating. Cells were then washed at 2 hpi with fresh medium to remove the virus inoculum. At 12 hpi and 18 hpi, the cells were harvested and simultaneously fixed and permeabilized in methanol as described above. They were subsequently treated in 4 N HCl (diluted in H2O) for 10 min to expose BrdU residues and washed twice in PBS.
Coverslips were then sequentially stained as follows (all incubations were at room temperature for 10 min, followed by PBS rinsing): normal goat serum (Jackson Laboratories); Ab to the IE72 and IE86 proteins (CH16.0; Goodwin Institute); donkey anti-mouse immunoglobulin secondary Ab conjugated with FITC (and cross adsorbed) (Jackson Laboratories); rat anti-BrdU Ab (Accurate Scientific); biotin-conjugated rabbit anti-rat immunoglobulin secondary Ab; and streptavidin conjugated with allophycocyanin (APC; Pharmingen). The coverslips were subsequently incubated in PBS containing 10 μg of propidium iodide and 100 μg of RNase per ml before mounting onto glass slides. Cells were then visualized and analyzed for all three fluors simultaneously using a CompuCyte Corporation laser scanning cytometer (analysis was generously performed by Jim Borree). It should be noted that with this method, gates are sometimes set so that apoptotic cells or doublets are not included. Using these gate settings, the total number of cells did not always add up to 100% for all cell populations. Percentages used in Fig. 3 and 4 were calculated from these raw data, using the percentages derived from the cytometer readings.
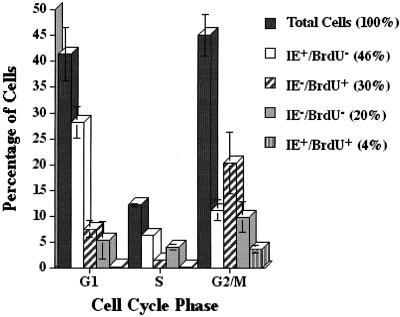
FIG. 3.
Blockade to IE gene expression occurred primarily in cells that were actively engaged in DNA synthesis at the beginning of the infection. Cells on coverslips were pulse labeled with BrdU just prior to infection at 24 h postplating. At 12 hpi, the cells were processed for laser scanning cytometry as described in Materials and Methods. Cells were simultaneously measured for the three fluors representing BrdU labeling (APC), IE positivity (FITC), and DNA content (propidium iodide). Percentages of cells in each cell cycle phase are given for each combination of IE and BrdU status compared to the distribution of the total cell population. Percentages are the averages of two trials, and error bars represent 1 standard deviation. IE+ cells accounted for an average of 50%, BrdU+ cells for 34%, and doubly positive cells for 4% of the total cells present in the two studies.
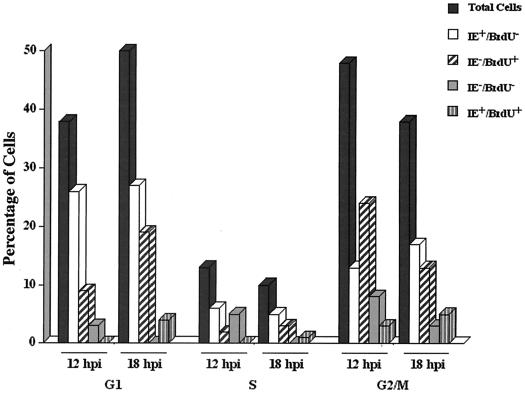
FIG. 4.
Laser scanning cytometry to track the movement of cells between 12 and 18 hpi. Cells on coverslips were pulse labeled with BrdU just prior to infection at 24 h postplating. At 12 and 18 hpi, the cells were processed for laser scanning cytometry as described in Materials and Methods. Cells were simultaneously measured for the three fluors representing BrdU labeling (APC), IE positivity (FITC), and DNA content (propidium iodide). Percentages of cells in each cell cycle phase are given for each combination of IE and BrdU status compared to the distribution of the total cell population. These data are from one of the experiments graphed in Fig. 3. IE+ cells accounted for 48 and 59% of the cells at 12 and 18 hpi, respectively, BrdU+ cells for 38 and 45%, respectively, and double-positive cells for 3 and 10% of the total cells present, respectively. The increase in the BrdU+ population can be accounted for by the BrdU+ cells that have undergone mitosis during the 6-h period.
FACS analysis of IE antigen (FITC) positivity versus cell cycle phase
At various times p.i., cells were harvested by trypsinization, pelleted, and resuspended in 600 μl of PBS. Ice-cold ethanol (1.4 ml) was then added dropwise while vortexing to achieve a final concentration of 70% ethanol. Cells were incubated overnight at 4°C before they were repelleted, rinsed, and resuspended in PBS. After samples were taken at all time points for a given experiment, the cells were again pelleted and stained for 30 min at room temperature in 50 μl of a mixture of primary Abs (CH16.0 and a mouse monoclonal Ab to IE72, provided by Bill Britt [University of Alabama, Birmingham]) in dilution buffer (PBS-0.5% bovine serum albumin [BSA]-0.5% Tween 20). Then 2 ml of wash buffer (PBS-0.5% BSA) was added, and the rinsed cells were repelleted. The pellets were then resuspended in 50 μl of FITC-conjugated goat anti-mouse immunoglobulin secondary Ab (in dilution buffer) and incubated for 30 min at room temperature in the dark. Cells were again rinsed in wash buffer and then in just PBS before the pelleted cells were resuspended in PBS containing propidium iodide (10 μg/ml) and RNase (100 μg/ml). The cells were analyzed for DNA content and FITC positivity on a FACScan Flow Cytometer.
Cycloheximide inhibition experiments
Cells were pulse labeled for 30 min with BrdU at 22 h postplating. At 23 h postplating, cells were incubated with cycloheximide (100 μg/ml) for 1 to 2 h prior to infection in the presence of the drug. Inoculum was removed after 2 to 4 h, and cells were refed with fresh medium without cycloheximide. Cells were fixed either 2 or 4 h after release from the cycloheximide block. Control S-phase infections in the absence of cycloheximide and G0/G1 infections in the presence of the drug were performed in parallel to assess limits of detection for IE proteins at 2 h post-cycloheximide release. Cells were stained as described above but treated with DNase instead of HCl to reveal incorporated BrdU residues.
Proteasome inhibition experiments
Cells were seeded for an S-phase experiment, and 1 h prior to infection (at 23 h postplating), cells were pulse labeled with BrdU as described above. At 24 h postplating, cells were mock or virus infected in the presence of 2.5 μM MG132 or an equivalent amount of dimethyl sulfoxide (DMSO) carrier. Viral supernatant containing the drug was left on the cells until they were harvested at 8 to 12 hpi and processed for IE and BrdU detection via immunofluorescence.
RESULTS
Differential patterns of IE72 and IE86 gene expression during G0- and S-phase infections.
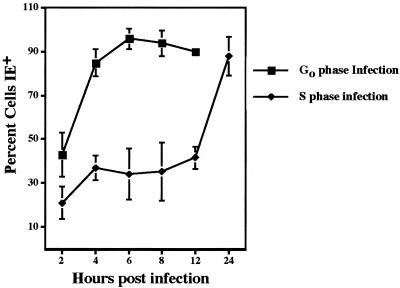
Earlier studies from our laboratory (47) indicated that the percentage of cells able to initiate IE gene expression immediately after infection is dependent upon the cell cycle phase at the time of infection. However, in these studies, the earliest time point examined was 12 hpi. To obtain a more complete picture of this disparity, we assayed IE gene expression using immunofluorescence at various times prior to and after 12 hpi. These results are shown in Fig. 1 and represent the averages of at least two trials. During G0 infection, IE expression was observed as early as 2 hpi, and by 4 to 8 hpi, at least 90% of the cells were IE+. However, when cells were infected in S phase, a bimodal distribution curve was observed, where a fraction of the cells (40 to 50%) became positive in the first 12 h, but expression of IE in the majority of the remaining cells was not observed until approximately 24 hpi.
FIG. 1.
Disparity between timing of IE gene expression during infection in G0 versus S phase. Cells were confluence synchronized, trypsinized, and reseeded onto coverslips for infection immediately (G0) or 24 h later (S). At the indicated times, coverslips were harvested and stained for IE protein expression with CH16.0 Ab. Points represent the averages of at least two trials, with error bars indicating 1 standard deviation.
Cells positive for IE at early times following S-phase infection have 2N DNA content.
In order to better understand the delay in IE gene expression, we proceeded to determine where in the cell cycle the IE+ cells resided during the course of the S-phase infection. Although our earlier data pointed to a block in S phase for IE expression, interpretation of the results was complicated by the complexity of the DNA profile at the beginning of the S-phase infections. Approximately 50% of the cells appeared to be in G1 while the other half of the cells were distributed through S and G2/M. To assess more directly how position in the cell cycle influenced the pattern of IE gene expression, we assayed the infected cells simultaneously for both their cellular DNA content and IE gene expression at various times p.i. Using FACS analysis of cells incubated both with propidium iodide (to label DNA) and with a combination of primary Abs against IE proteins detected with FITC-conjugated secondary Abs, we were able to examine the cell cycle profile of the FITC+ cells versus the whole population.
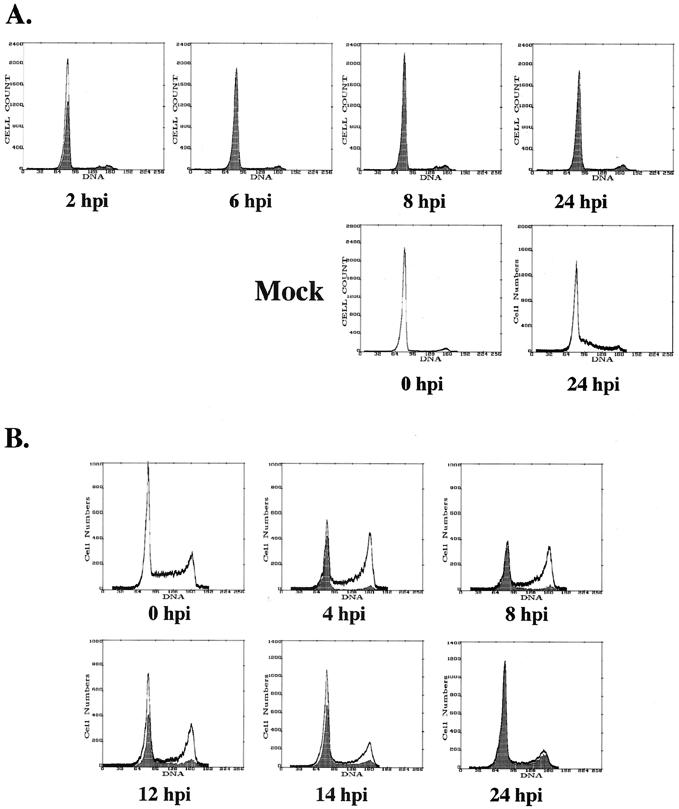
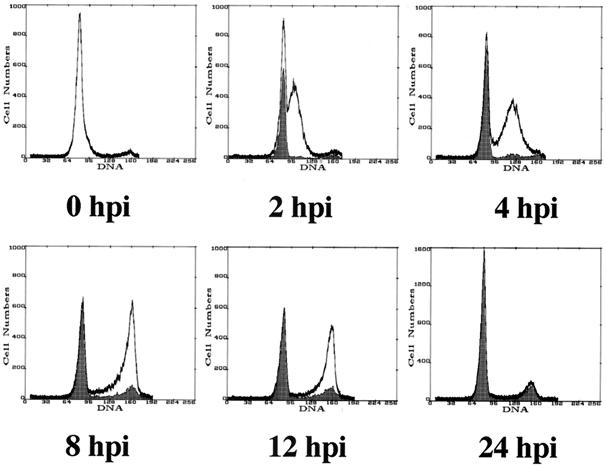
Representative results are presented in Fig. 2. The unshaded profiles represent the DNA content for the total population of cells at each time point, whereas the dark overlays represent the profiles for IE+ cells. As shown in Fig. 2A, the cells infected directly from confluence release (G0 infection) quickly became IE+ and then arrested with 2N DNA content without further progression through the cell cycle, while the corresponding mock-infected cells continued to cycle and divide. The profile of the IE+ cells during the S-phase infection is shown in Fig. 2B. Similar to the G0 infection, the cells with 2N DNA content quickly became IE+ and constituted the majority of positive cells for the first 12 h of the time course. As the cell population progressed through S to G2/M, a small population of IE+ cells gradually accumulated in both compartments, although it appeared that most cells cycled back to G1 before expressing detectable IE proteins. In the experiments described below, we set out to further define the origin of these IE+ cells and identify the boundaries of and potential mechanisms underlying the blockade to IE gene expression.
FIG. 2.
Cells positive for IE gene expression at early times p.i. are primarily in the G1 compartment. (A) Cells were virus- or mock-infected in G0 and harvested for FACS analysis at the indicated times p.i. as described in Materials and Methods. Cells were stained with a combination of Abs that recognized the IE gene products. Dark overlays represent IE+ cells. (B) Cells were virus- or mock-infected (data not shown) in S phase as described in Materials and Methods and harvested and stained as above at the indicated times. Profiles for mock-infected cells mirror those for virus-infected cells. Profiles in both panels of this figure are representative of at least two independent trials.
There is no inherent block to IE expression in S and G2/M for cells that were not actively synthesizing their DNA at the beginning of the infection.
In a previous publication (47), we described an experiment in which cells were labeled with BrdU just prior to infection to assess whether cells that were actively replicating their DNA at the time of infection were IE+. The data indicated that there was almost mutual exclusivity between BrdU+ and IE+ cells. However, beginning at about 12 hpi, we detected a small population of labeled BrdU+ cells that were also IE+. In these prior studies, we had not determined where in the cell cycle these dually positive cells first appeared and whether there was a requirement that the cells progress through mitosis to G1 before expression of IE.
To address this question, we used laser scanning cytometric analysis to assay cells simultaneously for active DNA replication at the time of infection, IE gene expression, and DNA content. Cells were plated onto coverslips for a standard S-phase infection. Just prior to infection at 24 h postplating, cells were pulse labeled with BrdU. Coverslips were then harvested at 12 hpi, fixed, and stained for BrdU incorporation (APC secondary label), IE gene expression (FITC secondary label), and cell cycle position (propidium iodide DNA staining). The cells were then analyzed for all three parameters simultaneously with a laser scanning cytometer. The results are shown in Fig. 3 and represent averages of two separate experiments.
On the basis of DNA content at 12 hpi, approximately 41% of the cells were in G1, 12% in S, and 45% in G2/M; 34% of the cells in the population were actively replicating their DNA at the time of infection (BrdU+). The majority of these cells at 12 hpi (30%) were still IE− and had cycled out of S phase and were distributed with approximately 8% in G1, 2% in S, and 20% in G2/M. Only a very small population of cells was positive for both BrdU and IE (4%), and these cells resided in G2/M. These results showed that a minor population of the cells that were infected while the cells were actively replicating their DNA could express the IE proteins in the G2/M phase of the cell cycle. The IE+ BrdU− cells (46% of the total population) were distributed throughout the cell cycle, with approximately 28% in G1, 7% in S, and 11% in G2/M. The IE− BrdU− cells constituted 20% of the population, with 6% in G1, 4% in S, and 10% in G2. Since over 50% of the BrdU− cells in the S and G2/M compartments were IE+, there does not appear to be an absolute block to IE gene expression in these two phases of the cell cycle for cells not actively replicating their DNA at the time of infection.
To further follow the progression of the infected cells, labeled cells from the same experiment were analyzed at both 12 hpi and 18 hpi. As shown in Fig. 4, between 12 and 18 hpi, approximately 10% of the cells in the G2/M compartment cycled back to G1. Most of these cells were IE− BrdU+. The number of cells positive for both BrdU and IE increased to 10%, with 4% in G1, 1% in S, and 5% in G2. These results indicate that although a small number of the BrdU+ cells became positive for IE in G2/M over time, the majority of the BrdU+ cells appeared to cycle back to G1 before commencing IE gene expression. Between 12 and 18 hpi, there was also a decrease in all compartments of the IE− BrdU− cells concomitant with a modest increase in the number of IE+ BrdU− cells in the G2/M compartment. The latter cells have either moved from the G1/S boundary or began in G2/M as IE− BrdU− cells and slowly converted to IE+ BrdU− before division.
Cells near the G1/S border at the time of infection express IE proteins and move into S.
In the experiments shown in Fig. 3, it is noteworthy that approximately 7% of the total population were IE+ BrdU− cells residing in S phase at 12 hpi. These likely had been infected in G1, became IE+, and moved to S by 12 hpi. An additional 11% of the total population were IE+ BrdU− and resided in the G2/M compartment. These IE+ cells were either infected in G1 and then moved to G2/M or infected in G2/M and remained there during the first 12 h of the infection. The results were consistent with the data shown in Fig. 2B and 4, where there was a gradual increase in IE+ cells in the S and G2/M compartments as the infection progressed.
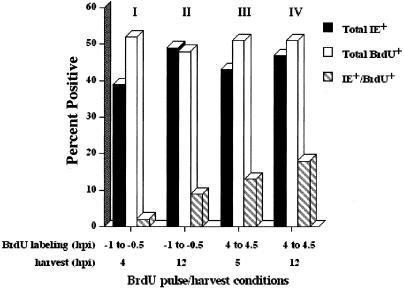
To more clearly define the mobile population of IE+ cells initiating at the G1/S boundary, we designed an experiment where cells were labeled with BrdU for 30 min beginning at 4 hpi. The cells were then harvested at 5 hpi (1 h after adding BrdU) and at 12 hpi, and the number of IE+ BrdU+ cells was determined. These results were then compared to those obtained when the cells were labeled with BrdU for 30 min just prior to normal S-phase infection and harvested at 4 hpi and 12 hpi. A representative set of data is shown in Fig. 5. In accord with the results shown in Fig. 3, pulsing the cells with BrdU just prior to infection at 24 h postplating resulted in only a small percentage (9%) of cells that were IE+ BrdU+ when the cells were examined at 12 hpi (group II), and at 4 hpi (group I) very few doubly positive cells (2%) could be detected.
FIG. 5.
Timing of the BrdU pulse determines percentages of doubly positive cells. S-phase infections were set up on coverslips. One set was pulsed at 23 h postplating for 30 min just prior to infection (−1 to −0.5 hpi) and harvested at either 4 hpi (group I) or 12 hpi (group II). The other set was pulsed for 30 min with BrdU at 4 hpi (4 to 4.5 hpi), with harvests at 5 hpi (group III) and 12 hpi (group IV). Cells were then stained for both BrdU and IE antigens as described in Materials and Methods. The data shown are representative of at least two trials.
However, by infecting the cells 4 h prior to pulse labeling with BrdU, we were able to detect the cells that had not yet entered S phase at the time of infection but had initiated cellular DNA synthesis during the next 4 h. Under these conditions, the results were quite different. Cells harvested at 5 hpi (1 h postpulse) consisted of 13% IE+ BrdU+ cells (group III), and at 12 hpi, 18% of the population were doubly positive (group IV). Thus, it appears that within the first 5 hpi, there is a population of cells at the G1/S boundary that can become IE+ and move into S phase, as is demonstrated in both Fig. 2B and 3. Moreover, by 12 hpi, there was a further increase in the number of BrdU+ cells with detectable IE gene expression, suggesting that for some of these BrdU+ cells, IE gene expression may be delayed. However, as demonstrated below, there appears to be a very narrow window of time during which the infected cells can both initiate viral gene expression and carry out cellular DNA synthesis.
Initiation of DNA replication alone blocks IE gene expression.
To determine the characteristics of the cells at the G1/S boundary, it was necessary to synchronize the cells so that they would cluster at this point in the cell cycle just prior to infection. Cells were confluence synchronized, trypsinized, and replated as before and 15 h later were placed into aphidicolin for 24 h in order to arrest them at the G1/S border. Aphidicolin allows the assembly of replication complexes and the initial firing of early origins, but not the elongation of newly initiated DNA strands (41). The cells were then infected at the time of drug release and followed for 24 h (Fig. 6). The majority of the cells were tightly synchronized with this treatment and quickly moved into S phase after release from the drug. However, the cells that moved rapidly into S phase did not become IE+ during this cell cycle, indicating that early events in S phase can block IE gene expression.
FIG. 6.
Cells released from aphidicolin arrest do not immediately express IE antigens. Cells were arrested with aphidicolin as described in Materials and Methods and then infected directly upon release from the drug. At the indicated time points, cells were harvested and stained for analysis of IE positivity by FACS analysis. The data are representative of at least two trials.
Interestingly, we again observed a small population of cells (presumably cells not already firing their origins, but residing at the G1/S boundary) that were able to express IE proteins and move into the S-phase compartment, in accord with the results described above. We could follow these cells as they moved through S and into G2/M. Since under these conditions there appears to be a population of IE+ cells that remain in the G2/M compartment as the infection progresses, it is possible that these cells arrested with 4N DNA content and did not go on to divide. This would be consistent with the data in Fig. 4 as well as with earlier data from our laboratory (31) and from Lu and Shenk (35), which indicate that cells infected with HCMV can arrest in the G2/M compartment.
Inhibition of proteasome function partially restores IE gene expression in replicating cells.
A key question evoked by the above results is the nature of the mechanism underlying the blockade to IE gene expression. To address this question, we treated the cells with several inhibitors of specific cellular functions. One possibility was that the presence of active cyclin A/Cdk2 complexes both at the G1/S transition and during replication might be responsible for inhibiting IE expression. To test this possibility, the cells were pulsed with BrdU for 1 h prior to S-phase infection in the presence of roscovitine, a selective inhibitor of Cdk1, -2, -5, and -7 (38). Incubation with 15 μM roscovitine (3, 12) for 1 or 2 h prior to and during S-phase infection had no effect on the percentage of IE+ BrdU+ cells observed at 12 hpi (data not shown).
A second possibility was that another inhibitor of IE expression was present during S phase. If this inhibitor had a short half-life, then infection in the presence of cycloheximide might allow the accumulation of IE transcripts that could then be translated into protein upon cycloheximide release and accumulation of the inhibitor. BrdU-labeled cells were incubated with cycloheximide for 1 or 2 h prior to infection in the presence of the drug. After 2 to 4 h, cells were refed with fresh medium to release the cycloheximide block and then fixed 2 to 4 h later. Under these conditions, IE proteins were detected in the BrdU− cells at a level similar to that in the nontreated, S-phase-infected controls (data not shown). There was, however, no increase in the number of IE+ BrdU+ cells. IE gene expression was easily detected under these conditions in a parallel experiment in which cells were infected in G0/G1 in the presence of cycloheximide (data not shown). These data indicated that IE expression in S phase was not inhibited by the presence of a short-lived inhibitor.
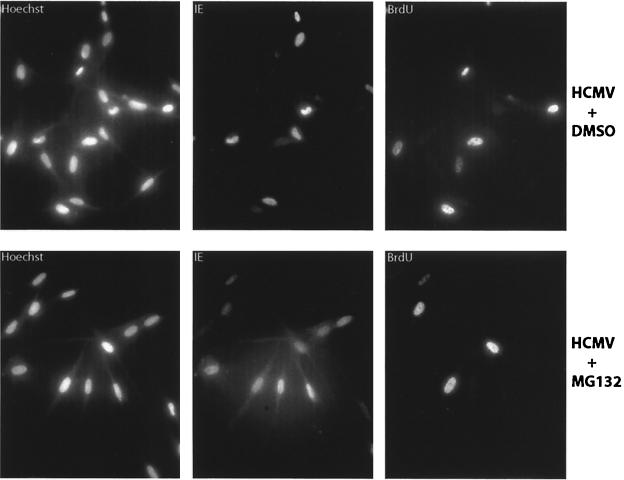
A third possibility was that some unstable protein in S phase was required for IE gene expression. To examine this possibility, the cells were infected and maintained in medium containing 2.5 μM of the proteasome inhibitor MG132 (34) and then harvested at 8 to 12 hpi. As shown in Fig. 7, the number of IE+ BrdU+ cells greatly increased following MG132 treatment. The data from four separate experiments with MG132 are presented in Table 1. These results suggest that degradation of a protein(s) required for expression of IE genes may be at least partially responsible for the S-phase blockade. Other potential explanations are discussed below.
FIG. 7.
Cells treated with MG132 display a higher percentage of IE+ BrdU+ cells at 8 to 12 hpi. S-phase infections were set up on coverslips. Cells were pulsed with BrdU at 23 h postplating for 30 min, followed by incubation without label for 30 min. At 24 h postplating, cells were mock or virus infected in the presence or absence of 2.5 μM MG132 (or an equivalent amount of DMSO carrier). At 8 to 12 h later, cells were harvested, fixed, and processed for immunofluorescence using Abs for both BrdU and IE antigens as described in Materials and Methods. These results are representative of the data shown in Table 1.
TABLE 1.
Addition of proteasome inhibitor MG132 increases the level of IE protein expression in S-phase-infected cellsa
| Expt | Infection and agent | % IE+ BrdU+ cells | Fold increase after MG132 treatment |
|---|---|---|---|
| 1 | Viral + DMSO | 2.8 | |
| Viral + MG132 | 30 | 10.7 | |
| 2 | Viral + DMSO | 2.8 | |
| Viral + MG132 | 12 | 4.3 | |
| 3 | Viral + DMSO | 14 | |
| Viral + MG132 | 31 | 2.2 | |
| 4 | Viral + DMSO | 6 | |
| Viral + MG132 | 26 | 4.3 |
Cells were pulsed with BrdU just prior to S-phase infection in the presence or absence of MG132 as described in Materials and Methods. Cells were then processed for immunofluorescent analysis of both IE and BrdU antigens between 8 and 12 hpi. The results of four separate experiments are reported.
DISCUSSION
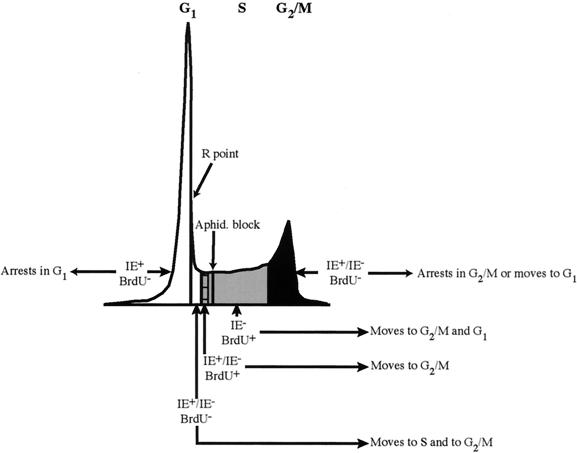
Previously we demonstrated that the cell cycle position of the host cell at the time of viral entry influenced the initiation of HCMV gene expression (47). Cells infected in S phase showed little cytopathic effect and lagged in expression of the IE antigens necessary to begin the viral life cycle. In this work, we have attempted to more fully define this disparity and the complexity of the interaction of HCMV with its host at different times in the cell cycle. Figure 8 presents a model of how expression of IE proteins and the ability of the cells to progress through the cell cycle may be related to the position of the cells at the time of the infection.
FIG. 8.
Model for IE gene expression in S-phase-infected cells. Cells that have 2N DNA content and have not reached the R point (BrdU−) are fully permissive for infection. They quickly express IE antigens and are arrested in G1. The small population of cells that have passed R but not yet begun S phase continue to cycle and have the potential to become IE+. Another population that have just entered S phase but are halted by aphidicolin also continue cycling, and some of these cells have the potential to become IE+. If either of these populations express IE antigens before dividing, they may arrest in G2/M. The vast majority of cells that have already begun cellular DNA synthesis (BrdU+) will not express IE antigens until after they have completed replication and passed through mitosis. Cells that begin in G2/M (BrdU−) also have the potential to express IE antigens; however, if they do, they may arrest prior to dividing.
As was observed in a G0 infection, some of the cells in the S-phase infections rapidly expressed IE antigens (Fig. 1). Figure 2B shows that these cells had 2N DNA content and became growth arrested in G0/G1. However, within the S and G2/M compartments, there gradually appeared a small population of cells that were IE+. This population was further defined by the pulse-labeling experiments described in Fig. 3 and 4. In these experiments, cells that were pulsed with BrdU just prior to infection in S and harvested 12 h later had populations of IE+ BrdU− cells in both S and G2/M. Since, in these compartments, over 50% of the BrdU− cells were IE+, there does not appear to be an absolute block to IE gene expression in these two phases of the cell cycle for cells that were not actively replicating their DNA at the beginning of the infection. In fact, over the next 6 h (to 18 hpi), there was an increase in the IE+ BrdU− population that resided in G2/M (Fig. 4).
Evidence that at least some of the IE+ cells in S phase resided in G1 at the time of the infection was provided by the experiment shown in Fig. 5. Here cells were either pulse labeled at the start of S-phase infection (24 h postrelease from confluence) or 4 h after S-phase infection. In contrast to cells that incorporate BrdU and are therefore actively replicating at the time of S-phase infection, cells that are 4 h behind these cells in the cycle are more likely to both express IE proteins and move into S phase, as evidenced by their incorporation of BrdU at 4 hpi (2% versus 13% double positive at 5 hpi). Although this is not a large population of cells, these results indicate the presence of a restriction point within G1. Before this point, IE antigens are rapidly expressed and the cells are arrested by HCMV. After this point, the cell cycle is not immediately blocked by HCMV, and some cells have the ability to move through S in the presence of newly expressed viral IE proteins.
From the results of the experiments involving arrest of the cells by aphidicolin (Fig. 6), it appears that there is another restriction point for cells that have initiated DNA replication but still have 2N DNA content. After this point, IE antigens are not expressed and cell cycle transit is not inhibited by HCMV until cells have progressed through mitosis and entered a new round of G1. It has been reported that the formation of prereplication complexes (pre-RCs) is not inhibited by aphidicolin (14, 41). Under normal conditions Orc is constitutively bound to all origins, and the pre-RCs are assembled during G1. The CDC6 protein binds to Orc and then loads the MCM2 to MCM7 proteins onto the complex at the origin throughout G1 (14, 51). During the progression from G1 to S, CDC7/Dbf4 phosphorylates MCM2, causing a conformational change that opens up the origin so that replication can be initiated. Aphidicolin does not prevent the firing of replication origins or the formation of short primers of 40 nucleotides or less, but does prevent elongation from these primers (41). Because the aphidicolin block did not alter the profile of IE expression, our results suggest that the block to IE gene expression occurs very early in S phase.
Taken together, these results raise the question of what events occurring between the restriction point in G1 and the point in S defined by the aphidicolin block might inhibit IE gene expression. One possibility that we considered was the accumulation of active cyclin A. Several reports in the literature note that during aphidicolin arrest, levels of cyclin A continue to accumulate within the cell, even though progression into S phase is inhibited (13, 23). Since cyclin A is the predominant S-phase cyclin (and is not immediately downregulated in S-phase-infected cells as it is in G0-infected cells), it seemed possible that the presence of active cyclin A was involved in the delayed expression of IE antigens in both aphidicolin-arrested and actively replicating cells.
Our results indicating a lack of effect of the Cdk inhibitor roscovitine appear to rule out the involvement of cyclin A complexes in the blockade. However, the fact that cyclin A is degraded in late G2 and that cyclin B levels increase as the cells progress toward mitosis may be important for understanding what defines the back edge of the block to IE protein expression. From the experiments in Fig. 3, it appears that the small percentage of IE+ BrdU+ cells observed at 12 hpi are located in G2/M. These results, coupled with the observation that over 50% of the BrdU− cells in G2/M at this time point are IE+, would suggest that there is no longer an inherent blockade to IE expression once cells have exited from S phase. Preliminary tests done on cells arrested in G2 and then infected upon release also showed that these cells can express IE antigens (data not shown). It should be noted, however, that some of these G2/M cells that do express IE antigens may arrest before they can divide (see Fig. 2B and 4).
Of particular interest is our finding that treatment of cells in S phase with the proteasome inhibitor MG132 resulted in a 3- to 10-fold increase in the number of BrdU+ cells that expressed the IE gene products. These results were obtained with both Abs that detect domains that are common to IE86 and IE72 and Abs specific for the C terminus of IE72. Thus, it seems unlikely that inhibition of the proteasome led to the unmasking of some epitope on the IE proteins. The simplest explanation for this result with the proteasome inhibitor is that IE gene expression requires some factor that has a short half-life in cells replicating their DNA. This factor could be a direct activator of IE gene expression or could counter the effects of some inhibitor. The inhibitor, however, would have to be relatively stable during S phase, since treatment of the cells with cycloheximide did not relieve the blockade upon removal of the drug (data not shown).
Although stabilization of some short-lived activator of IE expression is the most direct explanation, we cannot exclude the possibility that the newly described phenomenon of nuclear translation (28), which is more easily revealed in the presence of proteasome inhibitors, is involved. In this scenario, during a typical S phase, the newly synthesized IE mRNA would be rapidly translated in the nucleus, but the protein (and RNA) would be degraded. Proteasome inhibition would allow an accumulation of these IE proteins translated in the nucleus.
Since proteasome inhibition does not fully restore the expression of IE genes during S phase, additional factors would appear to be involved. One potential reason for nonexpression is that the input genomes may not be correctly localized. Ishov and colleagues (29) have shown that at 2 hpi, a certain proportion of the input HCMV DNA is associated with ND10 sites. When the early mRNA transcripts are observed, all are positioned at these ND10 sites (but not all ND10 sites have transcripts), indicating that only the genomes deposited at ND10 are engaged in early transcription. The number and distribution of ND10 structures change during the cell cycle, so that fewer sites might be available for genome deposition during S phase (18). In addition, the localization of the proteasome to ND10s may further influence the expression of the IE proteins during S phase (4). In any case, our results suggest that multiple factors are involved in inhibition of IE gene expression in cells actively replicating their DNA at the time of infection.
The repression of IE expression that occurs in S phase is reminiscent of the various levels of restriction to HCMV productive infection that are observed in different cell types. Spiller et al. (49) found that in HOG oligodendroglioma cells, which are relatively nonpermissive under normal growth conditions, IE mRNA synthesis is delayed and only small amounts of IE proteins are expressed. However, upon differentiation, these HOG cells express higher levels of IE antigens and also synthesize infectious virions, indicating that there is a switch to productive infection when the cells no longer divide. Both Poland et al. (45) and Jault et al. (32) also demonstrated that there is complete restriction to IE expression in the A172 glioblastoma cell line and that the glioblastoma line T98G allows only very limited expression of IE proteins. In addition, early studies on NT2D1 embryonal carcinoma cells showed that these cells were nonpermissive for HCMV in their undifferentiated/dividing state and had a complete block to IE gene expression. However, when these cells were made to differentiate with retinoic acid for 5 days, viral gene expression and replication were observed (22, 33). Again, this indicates that there is some inherent block to viral gene expression in dividing cells. The potential mechanisms underlying this inhibition are currently under investigation in our laboratory.
An important question yet to be answered is why cells infected with a 2N DNA content and presumably before the restriction point stop there and fail to replicate their DNA? Two viral gene products, IE86 and UL69, may play some role in this scenario, as each can at least partially block exit from G1 (25, 36, 53). Perhaps in combination, and with other viral genes, they are able to fully arrest the cell.
What is the significance of this blockade to viral protein synthesis and, more importantly, to the cells that may be able to first express IE antigens and then go on to replicate their DNA? This latter population may be important not so much in permissive cells, which are eventually killed by the virus, but in nonpermissive or semipermissive cells, where IE gene expression alone may lead to mutagenic effects (2, 48). The fact that these S-phase-infected cells maintain the ability to divide (at least once) is important in light of our most recent finding that approximately 10% of the mitotic cells in this first round of division sustain specific chromosomal damage at position 1q42 (19). This damage may prove important in the development of virus-induced sequelae, especially in cells that are semipermissive or nonpermissive for the virus, as viral protein expression is not required.
Acknowledgments
We thank Steven Hohmann for help in the initial phases of these experiments and the staff at the VA Medical Center FACS facility for help with data analysis and presentation. We also thank the members of the Spector laboratory for helpful discussions and critical reading of the manuscript.
This work was sponsored in part by March of Dimes grant 5-FY98-0727 to E.A.F. and NIH grants CA73490 and CA34729 to D.H.S. J.Y.Y. was supported by NIH training grant T32 GM07240.
REFERENCES
- 1.Albrecht, T., I. Boldogh, M. Fons, S. AbuBakar, and C. Z. Deng. 1990. Cell activation signals and the pathogenesis of human cytomegalovirus. Intervirology 31:68-75. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, T., M. P. Fons, C. Z. Deng, and I. Boldogh. 1997. Increased frequency of specific locus mutation following human cytomegalovirus infection. Virology 230:48-61. [DOI] [PubMed] [Google Scholar]
- 3.Alessi, F., S. Quarta, M. Savio, F. Riva, L. Rossi, L. A. Stivala, A. I. Scovassi, L. Meijer, and E. Prosperi. 1998. The cyclin-dependent kinase inhibitors olomoucine and roscovitine arrest human fibroblasts in G1 phase by specific inhibition of CDK2 kinase activity. Exp. Cell Res. 245:8-18. [DOI] [PubMed] [Google Scholar]
- 4.Anton, L. C., U. Schubert, I. Bacik, M. F. Princiotta, P. A. Wearsch, J. Gibbs, P. M. Day, C. Realini, M. C. Rechsteiner, J. R. Bennink, and J. W. Yewdell. 1999. Intracellular localization of proteasomal degradation of a viral antigen. J. Cell Biol. 146:113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boldogh, I., S. AbuBakar, C. Z. Deng, and T. Albrecht. 1991. Transcriptional activation of cellular oncogenes fos, jun, and myc by human cytomegalovirus. J. Virol. 65:1568-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boppana, S. B., K. B. Fowler, W. J. Britt, S. Stagno, and R. F. Pass. 1999. Symptomatic congenital cytomegalovirus infection in infants born to mothers with preexisting immunity to cytomegalovirus. Pediatrics 104:55-60. [DOI] [PubMed] [Google Scholar]
- 7.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 9.Britt, W., and C. Alford. 1996. Cytomegalovirus, p. 2493-2523. In P. M. Howley et al. (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 10.Castillo, J. P., A. D. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cinque, P., R. Marenzi, and D. Ceresa. 1997. Cytomegalovirus infections of the nervous system. Intervirology 40:85-97. [DOI] [PubMed] [Google Scholar]
- 12.David-Pfeuty, T. 1999. Potent inhibitors of cyclin-dependent kinase 2 induce nuclear accumulation of wild-type p53 and nucleolar fragmentation in human untransformed and tumor-derived cells. Oncogene 18:7409-7422. [DOI] [PubMed] [Google Scholar]
- 13.David-Pfeuty, T., Y. Nouvian-Dooghe, and D. Rouillard. 1997. Delaying the onset of M phase in NIH 3T3 cells blocked in early S phase occurs via accumulating cyclin B1 and tyrosine-phosphorylated p34cdc2 in the nucleus. Biol. Cell 89:179-197. [PubMed] [Google Scholar]
- 14.Dimitrova, D. S., I. T. Todorov, T. Melendy, and D. M. Gilbert. 1999. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146:709-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 17.Estes, J. E., and E.-S. Huang. 1977. Stimulation of cellular thymidine kinases by human cytomegalovirus. J. Virol. 24:13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., P. Lomonte, T. Sternsdorf, R. van Driel, and A. Orr. 1999. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112:4581-4588. [DOI] [PubMed] [Google Scholar]
- 19.Fortunato, E. A., M. L. Dell'Aquila, and D. H. Spector. 2000. Specific chromosome 1 breaks induced by human cytomegalovirus. Proc. Natl. Acad. Sci. USA 97:853-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortunato, E. A., and D. H. Spector. 1999. Regulation of human cytomegalovirus gene expression. Adv. Virus Res. 54:61-128. [DOI] [PubMed] [Google Scholar]
- 21.Fowler, K. B., F. P. McCollister, A. J. Dahle, S. Boppana, W. J. Britt, and R. F. Pass. 1997. Progressive and fluctuating sensorineural hearing loss in children with asymptomatic congenital cytomegalovirus infection. J. Pediatr. 130:624-630. [DOI] [PubMed] [Google Scholar]
- 22.Gonczol, E., P. W. Andrews, and S. A. Plotkin. 1984. Cytomegalovirus replicates in differentiated but not in undifferentiated human embryonal carcinoma cells. Science 224:159-161. [DOI] [PubMed] [Google Scholar]
- 23.Gong, J., F. Traganos, and Z. Darzynkiewicz. 1995. Growth imbalance and altered expression of cyclins B1, A, E, and D3 in MOLT-4 cells synchronized in the cell cycle by inhibitors of DNA replication. Cell Growth Differ. 6:1485-1493. [PubMed] [Google Scholar]
- 24.Hateboer, G., A. Wobst, B. O. Petersen, L. Le Cam, E. Vigo, C. Sardet, and K. Helin. 1998. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell. Biol. 18:6679-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi, M. L., C. Blankenship, and T. Shenk. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc. Natl. Acad. Sci. USA 97:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayhurst, G. P., L. A. Bryant, R. C. Caswell, S. M. Walker, and J. H. Sinclair. 1995. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J. Virol. 69:182-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirai, K., and Y. Watanabe. 1976. Induction of α-type DNA polymerases in human cytomegalovirus-infected WI-38 cells. Biochim. Biophys. Acta 447:328-339. [DOI] [PubMed] [Google Scholar]
- 28.Iborra, F. J., D. A. Jackson, and P. R. Cook. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293:1139-1142. [DOI] [PubMed] [Google Scholar]
- 29.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isom, H. C. 1979. Stimulation of ornithine decarboxylase by human cytomegalovirus. J. Gen. Virol. 42:265-278. [DOI] [PubMed] [Google Scholar]
- 31.Jault, F. M., J.-M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated RB, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jault, F. M., S. A. Spector, and D. H. Spector. 1994. The effects of cytomegalovirus on human immunodeficiency virus replication in brain-derived cells correlate with permissiveness of the cells for each virus. J. Virol. 68:959-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LaFemina, R., and G. S. Hayward. 1986. Constitutive and retinoic acid-inducible expression of cytomegalovirus immediate-early genes in human teratocarcinoma cells. J. Virol. 58:434-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 35.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, M., and T. Shenk. 1999. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J. Virol. 73:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukac, D. M., N. Y. Harel, N. Tanese, and J. C. Alwine. 1997. TAF-like functions of human cytomegalovirus immediate-early proteins. J. Virol. 71:7227-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meijer, L. 1996. Chemical inhibitors of cyclin-dependent kinases. Trends Cell Biol. 6:393-397. [DOI] [PubMed] [Google Scholar]
- 39.Murphy, E. A., D. N. Streblow, J. A. Nelson, and M. F. Stinski. 2000. The human cytomegalovirus IE86 protein can block cell cycle progression after inducing transition into the S phase of permissive cells. J. Virol. 74:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro, L., K. Mowen, S. Rodems, B. Weaver, N. Reich, D. Spector, and M. David. 1998. Cytomegalovirus activates interferon immediate-early response gene expression and an interferon regulatory factor 3-containing interferon-stimulated response element-binding complex. Mol. Cell. Biol. 18:3796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nethanel, T., and G. Kaufmann. 1990. Two DNA polymerases may be required for synthesis of the lagging DNA strand of simian virus 40. J. Virol. 64:5912-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohtani, K., J. DeGregori, G. Leone, D. R. Herendeen, T. J. Kelly, and J. R. Nevins. 1996. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol. Cell. Biol. 16:6977-6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poland, S. D., P. Costello, G. A. Dekaban, and G. P. Rice. 1990. Cytomegalovirus in the brain: in vitro infection of human brain-derived cells. J. Infect. Dis. 162:1252-1262. [DOI] [PubMed] [Google Scholar]
- 46.Poma, E. E., T. F. Kowalik, L. Zhu, J. H. Sinclair, and E. S. Huang. 1996. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J. Virol. 70:7867-7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen, Y., H. Zhu, and T. Shenk. 1997. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc. Natl. Acad. Sci. USA 94:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spiller, O. B., L. K. Borysiewicz, and B. P. Morgan. 1997. Development of a model for cytomegalovirus infection of oligodendrocytes. J. Gen. Virol. 78:3349-3356. [DOI] [PubMed] [Google Scholar]
- 50.Tamashiro, J. C., L. J. Hock, and D. H. Spector. 1982. Construction of a cloned library of the EcoRI fragments from the human cytomegalovirus genome (strain AD169). J. Virol. 42:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tye, B. K. 1999. MCM proteins in DNA replication. Annu. Rev. Biochem. 68:649-686. [DOI] [PubMed] [Google Scholar]
- 52.Wade, M., T. F. Kowalik, M. Mudryj, E. S. Huang, and J. C. Azizkhan. 1992. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol. Cell. Biol. 12:4364-4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 73:9274-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, Y. F., M. B. Leon, M. A. Waclawiw, J. J. Popma, Z. X. Yu, T. Finkel, and S. E. Epstein. 1996. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N. Engl. J. Med. 335:624-630. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, H., J. P. Cong, G. Mamtora, T. Gingeras, and T. Shenk. 1998. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:14470-14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]