Abstract
It is widely believed that the reason proteins dominate biological catalysis is because polypeptides have greater chemical complexity compared with nucleic acids, and thus should have greater enzymatic power. Consistent with this hypothesis is the fact that protein enzymes typically exhibit chemical rate enhancements that are far more substantial than those achieved by natural and engineered ribozymes. To investigate the true catalytic power of nucleic acids, we determined the kinetic characteristics of 14 classes of engineered ribozymes and deoxyribozymes that accelerate RNA cleavage by internal phosphoester transfer. Half approach a maximum rate constant of ~1 min−1, whereas ribonuclease A catalyzes the same reaction ~80,000-fold faster. Additional biochemical analyses indicate that this commonly encountered ribozyme “speed limit” coincides with the theoretical maximum rate enhancement for an enzyme that uses only two specific catalytic strategies. These results indicate that ribozymes using additional catalytic strategies could be made that promote RNA cleavage with rate enhancements that equal those of proteins.
Keywords: Ribozyme, deoxyribozyme, RNA transesterification, RNA cleavage, enzymatic mechanism, catalysis
INTRODUCTION
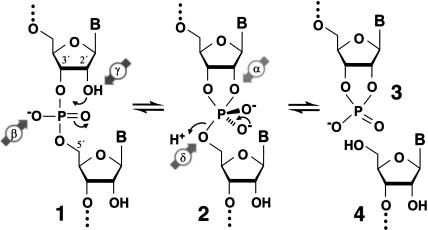
Of the eight known classes of natural ribozymes (Carola and Eckstein 1999; Nissen et al. 2000), four promote sequence-specific RNA cleavage by catalyzing an internal phosphoester transfer reaction (Butcher 2001). This reaction proceeds via a nucleophilic attack by the 2′-oxygen of a ribose moiety on the phosphorus center of the adjacent internucleotide linkage (Fig. 1 ▶). Several distinct catalytic strategies can be used by enzymes to accelerate the otherwise slow spontaneous rate of RNA transesterification. We use abbreviations to designate the four catalytic strategies discussed in this report as follows: α catalysis, in-line nucleophilic attack (Soukup and Breaker 1999); β catalysis, neutralization of negative charge on a nonbridging phosphate oxygen (Emilsson et al. 2003); γ catalysis, deprotonation of the 2′-hydroxyl group (Li and Breaker 1999); and δ catalysis, neutralization of negative charge on the 5′-oxygen atom (Emilsson et al. 2003). We have assumed that other proposed catalytic strategies are not likely to be important for interpreting the function of the ribozymes examined in this study (Emilsson et al. 2003).
FIGURE 1.
Mechanism for RNA cleavage by internal phosphoester transfer involving the 2′-hydroxyl group. The phosphorus center of the RNA linkage (1) is attacked by the 2′-oxygen nucleophile, which generates the dianionic phosphorane species (2). This pentacoordinate structure degrades to yield 2′,3′-cyclic phosphate-terminated (3) and 5′-hydroxyl-terminated (4) RNA fragments. The four main strategies (see text) for catalytic activation of this reaction are denoted (Greek lettering and arrows) in this depiction of an otherwise uncatalyzed phosphoester transfer reaction.
This diversity of catalytic strategies permits different classes of RNA-cleaving ribozymes to exhibit significant complexity in their kinetic characteristics, and numerous biochemical studies have revealed that no two classes of natural self-cleaving ribozymes are kinetically identical (for recent reviews, see Lilley 1999; Warashina et al. 2000; Takagi et al. 2001). Unfortunately, this catalytic diversity has complicated the study of the mechanisms of individual ribozymes. As a result, it is not clear whether self-cleaving ribozymes can engender the same level of catalytic power that is exhibited by most protein enzymes (Radzicka and Wolfenden 1995).
One means by which to measure the catalytic power of an RNA-cleaving enzyme is to compare its maximum observed rate constant (kobs) for phosphoester transfer to that of the corresponding uncatalyzed rate constant. The maximum kobs values under single-turnover conditions for the four classes of natural self-cleaving ribozymes typically do not exceed 10 min−1, even when the most favorable reaction conditions are used (e.g., see Clouet-d’Orval and Uhlenbeck 1997; Fedor 2000; Lafontaine et al. 2001; Nakano et al. 2001). The uncatalyzed rate constant for RNA transesterification is ~10−8 min−1 (Li and Breaker 1999) under near physiological conditions (pH 7.0, 250 mM K+, 5 mM Mg2+, 23°C). Therefore, most natural self-cleaving ribozymes achieve rate enhancements of ~109-fold or less. In contrast, ribonuclease A promotes the same phosphoester transfer reaction with a maximum rate constant of ~80,000 min−1 (DelCardayré and Raines 1994), which corresponds to a minimum rate enhancement of nearly 1013. This difference in catalytic performance means that, whereas ribozymes such as the self-cleaving hammerhead cleave with a half-life measured on a timescale of seconds, substrates for ribonuclease A are cleaved with a half-life measured in milliseconds. Although rate enhancements generated by natural self-cleaving ribozymes and RNase A are impressive, the moderate difference in reaction efficiency has significant biological implications.
RESULTS
Searching engineered enzymes for speed limits
A variety of engineered RNA-cleaving ribozymes and deoxyribozymes were examined to determine whether catalytic nucleic acids are fundamentally prohibited from attaining rate enhancements that are similar to those generated by protein enzymes. We began by conducting a succession of three assays using a histidine-dependent deoxyribozyme, termed HD3 (Roth and Breaker 1998), to define conditions under which to measure the maximum kobs. This is expected to be possible when the substrate is saturated with enzyme, the ionic strength requirements are satisfied, and the cofactor is present in sufficient quantity to saturate the enzyme–substrate complex (Fig. 2A ▶). In addition, all assays in this study were executed using single-turnover conditions because the rate constant under multiple turnover conditions could reflect other potential rate-limiting aspects of catalysis. For example, the rate constant cited for RNase A is likely limited by the speed at which a substrate productively associates with the enzyme (DelCardayré and Raines 1994). For the bimolecular HD3 complex, optimal single-turnover reaction conditions include an enzyme concentration >1 nM, a potassium concentration >300 mM, and a cofactor concentration >100 mM.
FIGURE 2.
Representative examinations of the kinetic characteristics of RNA-cleaving ribozymes and deoxyribozymes. Plots 1 through 3 depict the assay data used to determine the optimal reaction conditions at pH 7.5 and 23°C with regard to the concentration (c) of enzyme, monovalent ion, and cofactor, respectively. Dashed lines identify the concentration of enzyme or cofactor needed to attain half-maximal kobs. Solid lines reflect theoretical activity curves generated using the maximum kobs and the KD values derived from plots 1 and 3. Optimization reactions for HD3 progressed as follows: plot 1 (500 mM KCl, 20 mM L-histidine); plot 2 (100 nM enzyme, 20 mM L-histidine); plot 3 (100 nM enzyme, 1 M KCl). Note: the methyl ester of histidine (HME) was used in place of histidine in plot 3 to increase cofactor solubility (Roth and Breaker 1998). Optimization reactions for MR11 progressed as follows: plot 1 (250 mM KCl, 20 mM MgCl2); plot 2 (100 nM enzyme, 20 mM MgCl2); plot 3 (100 nM enzyme, 500 mM KCl).
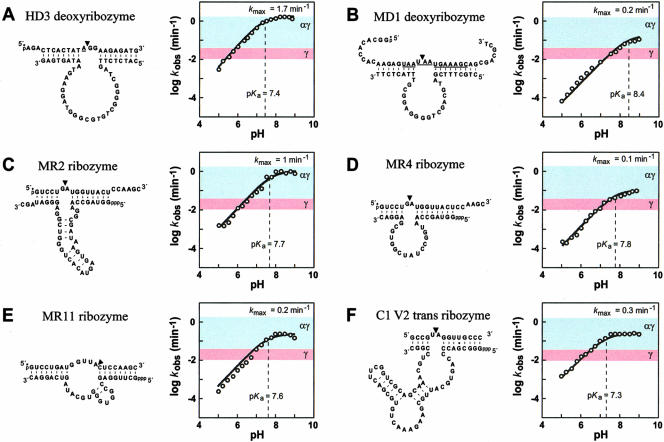
Most likely, the use of these reaction parameters in combination will permit the determination of the rate constant for the chemical step of HD3 function at any given pH, assuming that the use of optimized reaction parameters eliminates all other possible rate-limiting steps such as structural folding or inhibitory effects of reaction mixture constituents. Similarly, the optimal reaction conditions needed for a magnesium-dependent ribozyme, termed MR11 (Fig. 2B ▶; Tang and Breaker 2000), and for the 12 remaining enzymes used in this study have been determined (data not shown). The pH dependency of catalytic function spanning the range from pH 5 to pH 9 was established for each ribozyme and deoxyribozyme whose kinetic profiles met the three requirements described above for determination of the rate constant of the chemical step (Fig. 3 ▶).
FIGURE 3.
The influence of pH on the kobs for six RNA-cleaving ribozymes and deoxyribozymes. In each panel, the sequence and secondary structure model for bimolecular constructs of the ribozyme or deoxyribozyme (bottom strand) and substrate (top strand) is depicted. An arrowhead designates the site of cleavage. The substrates for HD3 and MD1 are DNA oligonucleotides that carry RNA linkages (underlined). The plot in each panel shows the logarithm of the kobs for enzyme catalysis as a function of pH. The dashed line identifies the pH required to attain half-maximal kobs. Colored zones indicate the range of rate constants that are expected from enzymes that fully use the γ catalytic strategy (pink) or a combination of α and γ catalytic strategies (light blue) to the exclusion of other possible strategies. The deoxyribozyme HD3 was analyzed with HME as a cofactor; the remaining enzymes used MgCl2. Reagent concentrations were as follows (enzyme, KCl, and cofactor, respectively): (A) HD3: 100 nM, 1 M, 400 mM; (B) MD1: 1 μM, 200 mM, 10 mM; (C) MR2: 300 nM, 150 mM, 350 mM; (D) MR4: 35 nM, 250 mM, 35 mM; (E) MR11: 100 nM, 500 mM, 750 mM; (F) C1 V2 Trans: 300 nM, 250 mM, 1 M. Below pH 7, assays of MD1 were buffered with succinate, and MR2 and MR4 with Bis-Tris.
The characteristics of enzyme function under varying pH conditions are expected to reveal the presence of protonation or deprotonation events in the chemical transformation that influence kobs. In a previous study, it was determined that a variant of the HD3 deoxyribozyme exhibits an activity profile wherein catalytic activity increases with increasing pH up to the point at which the pH matches the pKa for the imidazole group of the cofactor, either histidine or its methyl ester (Roth and Breaker 1998). The simplest explanation for this pH profile is that the deoxyribozyme uses the cofactor for general base catalysis (Fig. 1 ▶, γ) and has no effect on acid catalysis (Fig. 1 ▶, β and δ). Specific base catalysis produces a 2′-oxyanion that is ~1010-fold more nucleophilic than is oxygen when in the context of the 2′-hydroxyl group (Emilsson et al. 2003). The rate enhancement generated by this catalytic strategy should be dependent on the extent of 2′-hydroxyl deprotonation. Thus, the phosphoester transfer reaction should proceed with increasing speed as pH is increased, up to the point at which this group is completely deprotonated. We observe a similar pH-dependent activity profile for HD3 when examined under otherwise optimal reaction conditions (Fig. 3A ▶), which again suggests that the catalytic strategy used by this deoxyribozyme is likely to be deprotonation of the 2′-hydroxyl group with the assistance of its cofactor.
The kobs of the HD3 deoxyribozyme reaches a plateau at pH values above 7.4 (Fig. 3A ▶), which might correspond to the pKa of the target site 2′-hydroxyl group when it is positioned in the active site of the HD3 deoxyribozyme. Although this kinetic profile is consistent with such a pKa shift at the critical 2′-OH group, we cannot be certain that the pH profile does not reflect some other important proton transfer event. Thus, our discussions regarding the protonation state of the cleavage-site 2′-hydroxyl of this enzyme and of subsequent enzymes must be interpreted cautiously. However, if the pH profile does reflect 2′-hydroxyl deprotonation at the cleavage site, then HD3 could be using histidine (or its methyl ester derivative) to shift the pKa of this group from its normal value of ~13.7 (Li and Breaker 1999) to a new pKa value of 7.4. Using pH values higher than 7.4 would not be expected to lead to significant increases in kobs for this deoxyribozyme, because no additional rate enhancement can be generated once the 2′-hydroxyl group has been completely deprotonated. In this scenario, the enzyme is making use of the γ catalytic strategy to its greatest possible extent. Interestingly, the maximum kobs for HD3 of ~1.7 min−1 is nearly 100-fold greater than the maximum rate constant of ~0.02 min−1, which was measured for RNA transesterification that is accelerated upon total deprotonation of the 2′-hydroxyl group at pH 14.5 (Li and Breaker 1999). Thus, we conclude that HD3 must be using one or more additional strategies to exceed the γ speed limit (Fig. 3A ▶, γ zone) that should be encountered by all enzymes that use only deprotonation of the 2′-hydroxyl group to the exclusion of all other catalytic strategies.
We speculated that HD3 might be augmenting its use of the γ catalytic strategy by positioning the 2′-oxyanion nucleophile for “in-line” attack (Fig. 1 ▶, α). It has been determined (Usher and McHale 1976; Soukup and Breaker 1999) that an RNA linkage that is held in a conformation wherein the 2′ oxygen is positioned in-line (with respect to both the 5′-oxyanion leaving group and the intervening phosphorus center) is at least 10-fold more likely to undergo spontaneous cleavage compared with an unconstrained linkage. However, we speculate that the rate enhancement that can be derived by near perfect use of this α catalytic strategy is unlikely to be greater than 100-fold because of the inherent structural and chemical limitations of RNA phosphodiester linkages (Soukup and Breaker 1999; Emilsson et al. 2003). This rate enhancement by α catalysis reflects the increase in likelihood that the 2′-oxygen atom will be in an appropriate conformation for productive nucleophilic attack on the adjoining phosphorus center. Thus, enzymes that use the combination of α and γ catalytic strategies to promote internal RNA transesterification should achieve a multiple of the two independent rate enhancements. This corresponds to a theoretical maximum kobs value of ~2 min−1 at 23°C.
HD3 has kinetic characteristics that coincide with those expected for an RNA-cleaving enzyme that uses α and γ catalytic strategies to their full potential. Furthermore, the rate constant exhibited by HD3 of ~1.7 min−1 under optimized reaction conditions is congruent with our hypothesis that this deoxyribozyme has attained a form of catalytic perfection for an enzyme that uses an αγ combination of catalytic strategies to the exclusion of all others. Consistent with this view is the fact that we have not been able to isolate (via in vitro selection) variants of this class of deoxyribozymes that achieve higher rate enhancements than the HD3 construct. However, it is important to reiterate that the kinetic profiles of this enzyme, or of the enzymes described below, may not reflect simple αγ catalysis. It is possible (but seemingly far less likely) that a more complex combination of strategies, in combination with structural imperfections, by chance gives rise to the simple kinetic profiles observed.
Six enzymes that share the αγ speed limit
Any catalytic agent or polymer that uses only α and γ catalytic strategies to cleave RNA by phosphoester transfer should be prevented from exceeding this speed limit of ~2 min−1 under similar temperature conditions. If true, then RNA-cleaving ribozymes, deoxyribozymes, or protein enzymes that use α and γ catalytic strategies should not be able to exceed the rate enhancement of the HD3 deoxyribozyme. Because we are not aware of protein ribonucleases that can be used to test this hypothesis, we continued to examine additional ribozymes and deoxyribozymes that have been created by in vitro selection (Breaker 1997) with the expectation that other enzymes might also be limited by their exclusive use of α and γ catalytic strategies. For example, the MR11 ribozyme exhibits a pH profile (Fig. 3E ▶) that is similar to that of HD3, and also attains a maximum kobs (~0.2 min−1) that exceeds the speed limit projected for γ catalysis but falls within the range of rate constants that is projected for combined αγ catalysis. The kinetic characteristics of the MR11 ribozyme are consistent with the hypothesis that this ribozyme approaches the same speed limit as that reached by the HD3 deoxyribozyme, despite the fact that the ribozyme is formed from a different polymer (RNA), has a different catalytic structure (distinctive core sequence), and uses a different cofactor (Mg2+).
Upon examining 12 additional RNA-cleaving enzymes, we identified only four that did not meet the requirements of having saturable enzyme–substrate complexes, rate constants that were invariant at variable ionic strength conditions, and affinities for cofactors to allow saturation without indications of inhibition. These were (1) the 8–17 deoxyribozyme (Santoro and Joyce 1997), which exhibits Mg2+ inhibition at high concentration; (2) the X-motif ribozyme (Tang and Breaker 2000; Lazarev et al. 2003), which cannot be saturated with Mg2+; (3) the MD4 deoxyribozyme (G.M. Emilsson and R.R. Breaker, unpubl.), which is inhibited by increasing ionic strength; and (4) the MR12 ribozyme (class XII; Tang and Breaker 2000), which exhibits significant folding problems (data not shown).
Interestingly, we identified four additional constructs that have kinetic characteristics that precisely coincide with those expected for an enzyme that makes optimal use of α and γ catalytic strategies (Fig. 3B ▶–D,F). Specifically, each enzyme exhibits a linear relationship between rate constant and pH, which yields a plateau in excess of the γ speed limit but does not exceed the combined αγ speed limit maximum. Of the four remaining constructs examined, two, the MD5 deoxyribozyme (G.M. Emilsson and R.R. Breaker, unpubl.) and the C2 V1 trans ribozyme (class II variant 1; Tang and Breaker 1997a), exhibit pH-dependent inhibition (data not shown). Finally, the 10–23 deoxyribozyme (Santoro and Joyce 1997) exhibits a pH profile that is not consistent with αγ catalysis because of a high maximum kobs, and the MR5 ribozyme (class V; Tang and Breaker 2000) exhibits a multiphase pH profile (discussed below).
No thio effect for candidate αγ enzymes
The simplest means by which an enzyme could produce a pH profile and a maximum rate constant as depicted in Figure 3 ▶ would be to use a combination of α and γ catalytic strategies to near perfection. However, it must be emphasized that other catalytic strategies, particularly when combined with cofactor binding and structural folding complexities, could be used by individual ribozymes or deoxyribozymes to produce a pH profile that fortuitously matches that expected for an αγ enzyme. If an enzyme is using α and γ catalytic strategies to the exclusion of others, then such enzymes should not exhibit any indications that other catalytic strategies are being used.
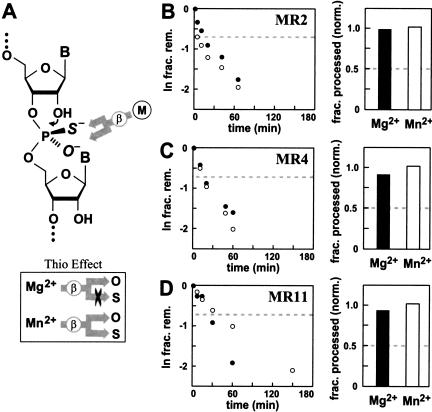
To explore this issue further, we used a chemical analog of the cleavage-site RNA linkage wherein sulfur is used in place of one of the nonbridging phosphate oxygen atoms (Fig. 4A ▶). Certain metal ions are known to have different binding affinities for sulfur compared with oxygen, and this can be exploited to determine whether metal ions are coordinating to specific internucleotide linkages in RNA (Dahm and Uhlenbeck 1991). Specifically, if an enzyme is positioning Mg2+ in contact with a nonbridging phosphate oxygen to achieve β catalysis (Fig. 1 ▶), then the enzyme will exhibit a “thio effect” (loss of activity when a sulfur replaces the oxygen). This thio effect can be substantially reduced or entirely eliminated when a thiophilic metal ion such as Mn2+ is used in place of Mg2+. Because the enzymes depicted in Figure 3 ▶ presumably do not use a β catalytic strategy, we expect that they cannot exhibit a thio effect when a thiophosphate group is used at the cleavage site.
FIGURE 4.
Putative αγ ribozymes do not exhibit metal-mediated β catalysis. (A) Schematic representation of an Sp thiophosphate RNA linkage and the possible contacts made by a metal-ion cofactor during β catalysis. The sulfur atom will be located at either of the two nonbridging positions (Rp or Sp) with near equal distribution. If the enzyme is an obligate metalloenzyme for β catalysis, then the metal ion (M) is expected to coordinate selectively with only one nonbridging position, as established by the active site. The inset reflects the fact that Mn2+ can interact with oxygen or sulfur with similar affinity, whereas Mg2+ has high affinity only for oxygen. (B–D, left) Kinetic characteristics of MR2, MR4, and MR11 ribozymes with thiophosphate substrates in the presence of Mg2+ (filled circles) and Mn2+ (open circles). The dashed line designates the plateau in RNA cleavage that would be expected if a thio effect precluded the processing of one of the two isomers. (Right) The bar graph represents the fraction of total RNA substrate processed in the presence of Mg2+ or Mn2+. The fraction cleaved was corrected and normalized as detailed in Materials and Methods. Again, the dashed line designates the maximum value for RNA cleavage that is expected if Mg2+ could only process one of the two thiophosphate isomers.
To test this prediction, we created substrates for the putative αγ ribozymes MR2, MR4, and MR11 in which the cleavage-site phosphate contains a sulfur atom at one of the two nonbridging atomic positions (Fig. 4A ▶). Chemical synthesis of the thiophosphate linkage (Materials and Methods) should yield a near equal mixture of the diastereomeric substrates wherein the sulfur is located at either the Rp or the Sp position (Burgers and Eckstein 1979). An enzyme that uses a metal ion to achieve β catalysis is likely to be stereoselective with regard to the point of contact between the metal ion and the nonbridging phosphate oxygen. Therefore, such an enzyme would be able to process only about half of the mixed thiophosphate substrate if using Mg2+ as the cofactor, but process all the substrate when the cofactor is the more thiophilic Mn2+ ion. As predicted, we do not observe any significant difference in the yield or the speed of thiophosphate cleavage when the MR2, MR4, and MR11 ribozymes are incubated with either Mg2+ or Mn2+ (Fig. 4B ▶–D). In contrast, the hammerhead ribozyme, which previously had been shown to exhibit a thio effect (e.g., see Scott and Uhlenbeck 1999), generates only 50% yield with Mg2+ compared with Mn2+ (data not shown). Therefore, the MR2, MR4, and MR11 ribozymes do not make use of metal-mediated β catalysis, which is consistent with our hypothesis that these enzymes might be maximally using only α and γ catalytic strategies.
Enzymes that break the αγ speed limit
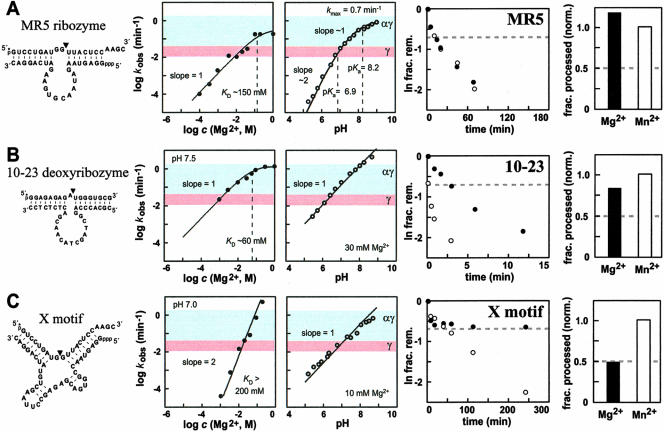
We also examined the function of several enzymes that exhibit more complex kinetic characteristics than those of the six putative αγ ribozymes described above. The MR5 ribozyme has kinetic characteristics that are similar to those of the other αγ candidates (Fig. 5A ▶; data not shown), including a maximum kobs that approaches the αγ speed limit at pH 9. However, the MR5 ribozyme exhibits a multiphase pH profile. Interestingly, this pH profile precisely matches that predicted for a ribozyme that requires two functional groups to be deprotonated to exhibit maximal activity. It is possible that one deprotonation event occurs with the 2′-hydroxyl group at the target linkage whereas the other reflects a structurally important functional group that also has a shifted pKa. If true, then the MR5 ribozyme could represent a seventh class of enzymes that maximally use αγ catalytic strategies. Consistent with this hypothesis is our observation that MR5 also does not show a thio effect (Fig. 5A ▶).
FIGURE 5.
Enzymes with more complicated kinetic profiles can meet or break the αγ speed limit. (A–C) Sequences, secondary-structure models, and characteristics for the MR5, 10–23, and X-motif enzymes, respectively. The cofactor and pH dependencies for each enzyme are depicted in the plots to the left, and the data resulting from thio-effect examination are given in the plots to the right. Other notations are as described in the legends to Figures 2 ▶–4 ▶. Reagent concentrations (enzyme, KCl, and cofactor, respectively) were as follows for each construct: MR5: 24 nM, 250 mM, 200 mM; 10–23: 100 nM, 0 mM, 30 mM; X-motif: 50 nM, 0 mM, 20 mM. Below pH 7, assays of MR5 were buffered with Bis-Tris. Assays of X-motif were buffered with HEPES in the range pH 6.7 to pH 8. Portions of the data presented in Figure 5C ▶ were obtained from Lazarev et al. (2003) for comparison.
In contrast, the 10–23 deoxyribozyme and the X-motif ribozyme exhibit kinetic characteristics that are not consistent with exclusive use of αγ catalysis. The 10–23 deoxyribozyme appears to reach saturation with high concentrations of Mg2+, but the rate constant for this enzyme shows no indication of plateau even at pH 9 (Fig. 5B ▶). Under suboptimal reaction conditions (30 mM Mg2+), the deoxyribozyme achieves a rate constant (~5 min−1) that is in excess of the αγ speed limit (see also Santoro and Joyce 1998). It is possible that the rate constant for the chemical step could be far greater if optimal reaction conditions were used. Similarly, the largest kobs (~7 min−1) of the X-motif ribozyme measured under suboptimal conditions (Fig. 5C ▶) also exceeds the αγ speed limit, which indicates that this enzyme is using additional or alternative catalytic strategies to accelerate RNA cleavage. Indeed, one of the catalytic strategies used by the X-motif ribozyme is β catalysis, as this ribozyme exhibits a thio effect with Mg2+ that can be eliminated by using Mn2+. In contrast, the 10–23 deoxyribozyme does not exhibit a thio effect, indicating that this enzyme achieves its sizeable rate enhancement most likely without using metal coordination to a nonbridging phosphate oxygen.
Prospects for fast RNA and DNA enzymes
Of the 14 classes of engineered ribozymes and deoxyribozymes examined, seven exhibit kinetic characteristics that almost perfectly mimic those expected for an enzyme that is maximally using α and γ catalytic strategies. This is rather striking, because there are at least 11 possible ways that the four major catalytic strategies for RNA cleavage could be used either alone or in combination (Emilsson et al. 2003). Therefore, it is likely that the set of nucleic acid folds sampled by small oligonucleotides at neutral pH has a higher proportion of structures that can use αγ catalysis compared with any other assembly of catalytic strategies. Although enzymes using the αγ combination might be the most common, attempts to engineer these enzymes to achieve rate enhancements that exceed the αγ speed limit are predicted to fail unless additional catalytic strategies can be conferred on the enzyme.
The 10–23 deoxyribozyme has resisted attempts at enhancement by in vitro selection (Santoro and Joyce 1997), but it is unlikely to be conducive to improvement owing to its small size and highly conserved catalytic core. In contrast, ribozymes such as the X-motif hold more promise for high-speed function. The X-motif uses β catalysis, but it must use at least one additional catalytic strategy to achieve such large rate enhancements under suboptimal reaction conditions. The X-motif requires at least two metal ions for activity (the logarithm of the kobs versus Mg2+ profile has a slope of 2; Fig. 5C ▶), and this would be expected if the X-motif makes use of a form of double-metal-ion mechanism (Steitz and Steitz 1993) to achieve β and δ catalysis. This ribozyme class appears to be receptive to mutation and to the addition of appended domains that together might yield variants that produce rate enhancements as large as those of protein enzymes like ribonuclease A. As a result, we expect that ribozymes such as the X-motif and the related MR8 ribozyme (Tang and Breaker 2000; Lazarev et al. 2003) will make excellent candidates for further improvement by in vitro selection.
DISCUSSION
Studies of natural and engineered ribozymes and deoxyribozymes have revealed complex kinetic behavior, which indicates that these enzymes can use multiple catalytic mechanisms. However, this complexity is most likely to be an outgrowth of the combinatorial use of four simple catalytic strategies that in some instances are overlaid with structural folding events or inhibitory interactions with reaction constituents. In this study, we have provided evidence that as many as seven of 14 engineered ribozymes and deoxyribozymes might use only two of these catalytic strategies, positioning for in-line attack and 2′-hydroxyl deprotonation, to near perfection. If correct, then the maximum speed of these RNA and DNA enzymes is limited ultimately by the combination of catalytic strategies in use.
Our results also reveal that some nucleic acid enzymes can produce rate enhancements that exceed the αγ speed limit. The fact that such ribozymes and deoxyribozymes exist is an indication that RNA and DNA enzymes can access combinations of catalytic strategies that, if fully exploited, will produce speeds equivalent to protein enzymes. Therefore, we conclude that RNA and DNA enzymes can be as powerful as their protein counterparts, as long as they are using identical catalytic strategies. In vitro selection under different reaction conditions could be used to favor the isolation of RNA-cleaving enzymes that use other combinations of catalytic strategies, and these constructs would serve as excellent model compounds to further explore mechanisms of biocatalysis. It is interesting to note that all the natural self-cleaving ribozymes are capable of breaking the αγ speed limit of ~2 min−1, and thus biological systems might have a need for ribozymes that use combinations of catalytic strategies that are in parity with protein enzymes.
MATERIALS AND METHODS
Oligonucleotides and chemicals
Synthetic DNAs were purchased from the HHMI Biopolymer–Keck Foundation Biotechnology Resource Laboratory and purified as described (Tang and Breaker 1997b). Synthetic DNAs were used for the deoxyribozymes and as templates to prepare ribozymes by in vitro transcription. The radiolabeled nucleotide [γ-32P]ATP was purchased from Amersham Pharmacia. Synthetic RNAs for substrates were prepared by Dharmacon Research, Inc. Thiophosphate-containing RNAs were prepared by substituting a sulfurylation step in place of the oxidation step of a standard solid-phase RNA synthesis protocol. Sulfur addition typically occurs with the production of near equal amounts of the Rp and Sp isomers of the thiophosphate linkage (S.A. Scaringe, Dharmacon, pers. comm.).
In vitro transcription
Synthetic, negative-strand DNA templates were used for preparative in vitro transcription of ribozyme RNAs by providing a complementary oligonucleotide to create a double-stranded T7 promoter region. RNAs were prepared and purified as described (Soukup and Breaker 1999).
Kinetic assays
Reactions contained trace amounts of 5′ 32P-labeled substrate and excess enzyme. Product yields were determined by quantitating (Molecular Dynamics PhosphorImager) product bands that were generated by PAGE. Each kobs value was determined by plotting the natural logarithm of the fraction of substrate that remained uncleaved versus time (after correction for the amount of substrate that remains uncleaved upon exhaustive incubation), and establishing the negative slope of the resulting line. Individual experimental conditions are detailed in each figure legend. Unless otherwise noted, reactions were buffered with Tris-HCl in the range of pH 7 to pH 9 and MES below pH 7.
Determination of reaction conditions for pH-dependent kinetic analysis
The enzymes in Figures 3 ▶, 4 ▶, and 5 ▶ were each subjected to the analysis outlined in Figure 2 ▶. Additionally, the analysis in Figure 2 ▶ was progressive. Specifically, the concentration of enzyme that was saturating for enzyme–substrate complex, determined in plot 1, was used in plots 2 and 3. The saturating concentration of monovalent ion, determined in plot 2, was used in plot 3. The optimum concentrations of enzyme, monovalent, and cofactor, determined in Figure 2 ▶, were used in Figures 3 ▶, 4 ▶, and 5A ▶. Similarly, the optimum reaction conditions were determined for the 10–23 and X-motif enzymes, but subsaturating amounts of cofactor were used in the pH profile of Figure 5, B and C ▶, to allow for accurate rate constant measurement with manual pipetting.
Thiophosphate cleavage analysis
The mixed thiophosphate isomer RNA substrates (trace; 5′ 32P-labeled) were incubated with enzyme in the presence of either Mg2+ or Mn2+. The fraction of substrate processed was corrected in each case for the amount of substrate that remained uncleaved after exhaustive incubation with Mn2+. The corrected fraction of total RNA substrate processed in the presence of Mg2+ was normalized to the corrected fraction of total RNA substrate processed in the presence of Mn2+. The normalized fraction processed values reflect the ratio of thio substrate cleaved in the presence of Mg2+ versus in the presence of Mn2+ after exhaustive cleavage.
Acknowledgments
We thank members of the Breaker laboratory for helpful discussions. This work was supported by NIH grant GM57500 for deoxyribozyme research and by NIH grant GM559343 for ribozyme research. In vitro selection of new ribozyme and deoxyribozyme motifs was supported by Ribozyme Pharmaceuticals, Inc. RNA science in the Breaker laboratory also is supported through a fellowship to R.R.B. from the David and Lucile Packard Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5670703.
REFERENCES
- Breaker, R.R. 1997. In vitro selection of catalytic polynucleotides. Chem. Rev. 97: 371–390. [DOI] [PubMed] [Google Scholar]
- Burgers, P.M.J. and Eckstein, F. 1979. Diastereomers of 5′-O-adenosyl 3′-O-uridyl phosphorothioate: Chemical synthesis and enzymatic properties. Biochemistry 18: 592–596. [DOI] [PubMed] [Google Scholar]
- Butcher, S.E. 2001. Structure and function of the small ribozymes. Curr. Opin. Struct. Biol. 11: 315–320. [DOI] [PubMed] [Google Scholar]
- Carola, C. and Eckstein, F. 1999. Nucleic acid enzymes. Curr. Opin. Chem. Biol. 3: 274–283. [DOI] [PubMed] [Google Scholar]
- Clouet-d’Orval, B. and Uhlenbeck, O.C. 1997. Hammerhead ribozymes with a faster cleavage rate. Biochemistry 36: 9087–9092. [DOI] [PubMed] [Google Scholar]
- Dahm, S.C. and Uhlenbeck, O.C. 1991. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry 30: 9464–9469. [DOI] [PubMed] [Google Scholar]
- DelCardayré, S.B. and Raines, R.T. 1994. Structural determinants of enzymatic processivity. Biochemistry 33: 6031–6037. [DOI] [PubMed] [Google Scholar]
- Emilsson, G.M., Nakamura, S., Roth, A., and Breaker, R.R. 2003. Ribozyme speed limits. RNA (this issue). [DOI] [PMC free article] [PubMed]
- Fedor, M.J. 2000. Structure and function of the hairpin ribozyme. J. Mol. Biol. 297: 269–291. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D.A., Wilson, T.J., Norman, D.G., and Lilley, D.M.J. 2001. The A730 loop is an important component of the active site of the VS ribozyme. J. Mol. Biol. 312: 663–674. [DOI] [PubMed] [Google Scholar]
- Lazarev, D., Puskarz, I., and Breaker, R.R. 2003. Substrate specificity and reaction kinetics of an X-motif ribozyme. RNA 9: 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. and Breaker, R.R. 1999. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 121: 5364–5372. [Google Scholar]
- Lilley, D.M.J. 1999. Structure, folding and catalysis of the small nucleolytic ribozymes. Curr. Opin. Struct. Biol. 9: 330–338. [DOI] [PubMed] [Google Scholar]
- Nakano, S., Proctor, D.J., and Bevilacqua, P.C. 2001. Mechanistic characterization of the HDV genomic ribozyme: Assessing the catalytic and structural contributions of divalent metal ions within a multichannel reaction mechanism. Biochemistry 40: 12022–12038. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Radzicka, A. and Wolfenden, R. 1995. A proficient enzyme. Science 267: 90–93. [DOI] [PubMed] [Google Scholar]
- Roth, A. and Breaker, R.R. 1998. An amino acid as a cofactor for a catalytic polynucleotide. Proc. Natl. Acad. Sci. 95: 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro, S.W. and Joyce, G.F. 1997. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. 94: 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 37: 13330–13342. [DOI] [PubMed] [Google Scholar]
- Scott, E.C. and Uhlenbeck, O.C. 1999. A re-investigation of the thio effect at the hammerhead cleavage site. Nucleic Acids Res. 27: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup, G.A. and Breaker, R.R. 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5: 1308–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, T.A. and Steitz, J.A. 1993. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. 90: 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, Y., Warashina, M., Stec, W.J., Yoshinari, K., and Taira, K. 2001. Survey and Summary: Recent advances in the elucidation of the mechanisms of action of ribozymes. Nucleic Acids Res. 29: 1815–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, J. and Breaker, R.R. 1997a. Examination of the catalytic fitness of the hammerhead ribozyme by in vitro selection. RNA 3: 914–925. [PMC free article] [PubMed] [Google Scholar]
- ———. 1997b. Rational design of allosteric ribozymes. Chem. Biol. 4: 453–459. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Structural diversity of self-cleaving ribozymes. Proc. Natl. Acad. Sci. 97: 5784–5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher, D.A. and McHale, A.H. 1976. Hydrolytic stability of helical RNA: A selective advantage for the natural 3′,5′-bond. Proc. Natl. Acad. Sci. 73: 1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warashina, M., Takagi, Y., Stec, W.J., and Taira, K. 2000. Differences among mechanisms of ribozyme-catalyzed reactions. Curr. Opin. Biotechnol. 11: 354–362. [DOI] [PubMed] [Google Scholar]