Abstract
The 5′ untranslated leader region of retroviral RNAs contains noncoding information that is essential for viral replication, including signals for transcriptional transactivation, splicing, primer binding for reverse transcription, dimerization of the genomic RNA, and encapsidation of the viral RNA into virions. These RNA motifs have considerable structural and functional overlap. In this study, we investigate the conformational dynamics associated with the use and silencing of a sequence in HIV-2 RNA that is involved in genomic RNA dimerization called stem–loop 1 (SL1) and its relationship with a flanking sequence that is known to be important for encapsidation of viral RNAs. We demonstrate that a long-distance intramolecular interaction between nucleotides located upstream of the primer-binding site domain and nucleotides encompassing the Gag translation start codon functionally silences SL1 as a dimerization element. This silencing can be relieved by mutation or by hybridization of an oligonucleotide that disrupts the long-distance interaction. Furthermore, we identify a palindrome within the packaging/encapsidation signal Ψ (just 5′ of SL1) that can either serve as an efficient dimerization signal itself, or can mediate SL1 silencing through base pairing with SL1. These results provide a tangible link between the functions of genomic RNA dimerization and encapsidation, which are known to be related, but whose physical relationship has been unclear. A model is proposed that accounts for observations of dimerization, packaging, and translation of viral RNAs during different phases of the viral replication cycle.
Keywords: HIV-2, dimerization, RNA structure, antisense oligonucleotides
INTRODUCTION
One unusual characteristic of retroviruses is that two copies of the genome are found in the viral particle. The diploid genome consists of identical positive-strand RNA molecules joined by noncovalent bonds. The dimeric nature of the genomic RNA has been documented by sedimentation analysis (Cheung et al. 1972) and gel electrophoresis (Fu and Rein 1993). Furthermore, electron microscopy studies revealed that a strong intermolecular interaction occurs near the 5′ end in a domain called the Dimer Linkage Structure (DLS; Bender and Davidson 1976; Kung et al. 1976). Besides the DLS, the 5′ end of retroviral genomic RNA is replete with structural and functional elements (Berkhout 1996). It also contains most of the RNA packaging signals that allow the genomic RNA to be selectively recognized and encapsidated in a budding viral particle (for review, see Jewell and Mansky 2000). Upon budding, retroviral particles undergo a maturation process that leads to, among other morphological changes, an increase in the stability of the genomic RNA dimer (Fu et al. 1994; Laughrea et al. 2001).
Since the observation that RNA fragments encompassing the 5′ end of the retroviral genome were able to dimerize in vitro in the absence of any viral or cellular protein (Darlix et al. 1990), precise analyses of the relationships between RNA structural elements involved in dimerization and maturation of the DLS have been attempted. In HIV-1, a short sequence that promotes dimerization was identified and named the dimerization initiation site (DIS; Laughrea and Jette 1994; Skripkin et al. 1994) or stem–loop 1 (SL1; McBride and Panganiban 1996). In vitro, this element maintains two RNA molecules in a dimeric state, either through a kissing-loop interaction or an extended duplex base pairing arrangement (for review, see Greatorex and Lever 1998).
We and others previously demonstrated that two dimerization elements exist within the leader region of HIV-2 genomic RNA (Dirac et al. 2001; Jossinet et al. 2001, 2002; Lanchy and Lodmell 2002). One element is homologous to SL1 in HIV-1 leader RNA. The second element involves the 5′ end of the tRNA-primer binding site (PBS), which was shown to mediate dimerization in the presence of potassium and magnesium at physiological (37°C) or high (55°C) temperatures (Jossinet et al. 2001; Lanchy and Lodmell 2002), but the resulting dimers have properties characteristic of loose dimers (Laughrea and Jette 1996; Muriaux et al. 1996). Loose dimers are functionally defined as those that can be observed on a native gel in Tris-Borate magnesium (TBM) buffer at 4°C but are absent when assayed on a semidenaturing gel in Tris-Borate EDTA (TBE) buffer at 24°C, presumably because the dimers dissociate under these electrophoresis conditions (Shubsda et al. 1999).
PBS-mediated dimerization occurs with a variety of different RNA fragments using several dimerization protocols (Lanchy and Lodmell 2002; Lanchy et al. 2003). However, the use of SL1 as an in vitro dimerization element was observed only for HIV-2 RNA fragments between 444 and 546 nt in length that were incubated at high temperature (Dirac et al. 2002; Lanchy et al. 2003). Contrary to similar HIV-1 RNA fragments, HIV-2 RNAs encompassing the entire untranslated leader region cannot be made to form SL1-dependent tight dimers by incubation at high temperature (Dirac et al. 2002; Lanchy and Lodmell 2002). Tight dimers are defined as stable dimers that withstand semidenaturing electrophoresis in TBE buffer at 24°C. The stable nature of the tight dimers is thought to be caused by an extended base pairing arrangement of SL1 (Laughrea and Jette 1994, Laughrea and Jette 1996; Paillart et al. 1994, 1996; Muriaux et al. 1995, 1996). It has been proposed that the reason HIV-2 RNA does not form tight dimers is that SL1 can be sequestered in a more stable intramolecular interaction (Dirac et al. 2002; Lanchy et al. 2003). This intramolecular interaction is positively regulated by two dimer-interfering elements located upstream and downstream of the major splice donor site (Dirac et al. 2002; Lanchy et al. 2003). Notably, the downstream element contains the start codon from the gag gene, the first major open reading frame in the genome (Fig. 1 ▶). Using site-directed mutagenesis and compensatory mutation analysis, we demonstrated that base pairing between the two elements inhibited the capacity of HIV-2 leader RNA to form SL1-dependent tight dimers (Lanchy et al. 2003).
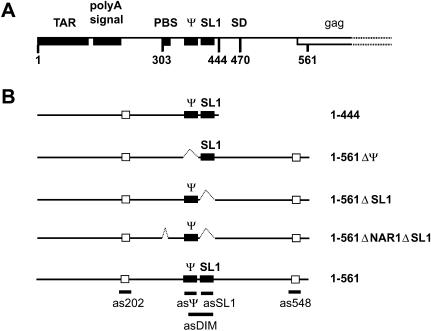
FIGURE 1.
5′ leader region of HIV-2 ROD genomic RNA. (A) The landmark sequences with known functions are indicated by boxes with the name indicated above. TAR, polyA signal, PBS, Ψ, SL1, SD, and gag represent the trans-activation region, the poly(A) signal domain, the primer binding site, the encapsidation signal, the stem–loop 1, the major splice donor site, and the 5′ end of the Gag protein coding region, respectively. (B) RNAs used in this study. The closed boxes represent the in vivo characterized encapsidation signal Ψ (Griffin et al. 2001) and the in vitro characterized dimerization element SL1 (in HIV-2; Dirac et al. 2001; Lanchy and Lodmell 2002). The open boxes represent the cores of the two dimer-interfering elements previously characterized 189–196 and 543–550 (Lanchy et al. 2003). The name of each RNA construct is indicated at the right. The short thick lines below some elements represent the binding sites of antisense DNA oligonucleotides used in this study.
Based on the above observations that dimerization of HIV-2 RNA and the use of dimerization motifs are prone to modulation, in this study we have further analyzed the role of SL1 and flanking regions of HIV-2 leader RNAs using complementary oligonucleotides, deletion mutagenesis, kinetic analysis, and solution structure probing (Fig. 1 ▶). We found that hybridization of oligonucleotides complementary to the dimer-interfering elements were capable of triggering a conformational change that restored the ability of wild-type RNAs to form tight dimers. Surprisingly, an RNA lacking part of SL1 could be induced to dimerize upon hybridization of oligonucleotides complementary to the dimer-interfering elements. Oligonucleotides complementary to the Ψ packaging signal region inhibited this dimerization. Overall, the kinetic and structural data presented here suggest that a physiologically relevant conformational change can be readily induced by oligonucleotide binding, that a conserved autocomplementary sequence within the packaging signal is, in fact, a newly described dimerization motif, and that the use or silencing of tight dimerization is modulated at least in part by a direct interaction of the Ψ and SL1 sequences. These observations suggest a mechanism for coordinating the essential events of dimerization, encapsidation, and translation in vivo.
RESULTS
HIV-2 leader RNA does not form SL1-dependent tight dimers in vitro
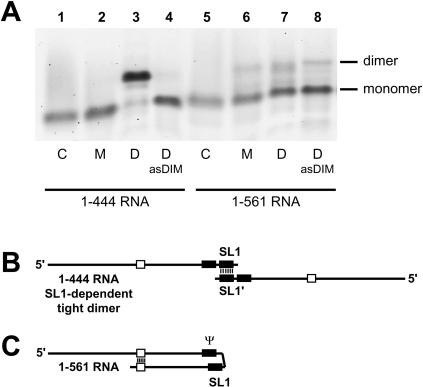
The ability of the 1–444 and 1–561 HIV-2 RNAs to form tight dimers when incubated at 55°C in high potassium (300 mM), high magnesium (5 mM) buffer (dimer buffer) was compared. The 1–444 RNA was shown previously to be especially prone to forming SL1-mediated tight dimers upon incubation at high temperatures (Dirac et al. 2001; Lanchy and Lodmell 2002). The yield of 1–444 RNA tight dimerization was about 90% whereas the level of 1–561 RNA dimers was about 30% (Fig. 2 ▶, cf. lanes 3 and 7). The 1–444 RNA dimerization was inhibited when coincubated with the antisense oligonucleotide asDIM directed against nt 397–426 (Fig. 1 ▶; Table 1 ▶). This oligonucleotide hybridizes to the 5′ stem and loop of SL1 and nucleotides upstream (Table 1 ▶). The low level of 1–561 tight dimer was not affected by the presence of asDIM (Fig. 2 ▶, cf. lanes 7 and 8). These results indicated that the truncated 1–444 HIV-2 leader RNA was able to form very high levels of SL1-dependent tight dimers (Fig. 2B ▶), in contrast to the complete leader 1–561 RNA, which formed mostly monomers (Fig. 2C ▶). The low level of TBE-resistant dimers in 1–561 RNA were SL1-independent.
FIGURE 2.
TBE-resistant dimerization of 1–444 and 1–561 RNAs. (A) 1–444 and 1–561 RNAs were assayed for dimerization at 55°C with monomer (M) or dimer (D) buffer. Dimerization was also assayed in the presence of a 20-fold excess of antisense DNA oligonucleotide asDIM, which is complementary to nucleotides 397–426. After incubation for 30 min, samples were subjected to electrophoresis on a TBE agarose gel at 28°C. Only tight dimers withstand the warm TBE electrophoresis. (Control lanes C) Monomeric RNA that was denatured at 90°C, then quenched on ice immediately prior to loading. (B) Schematic representation of the tight dimer of 1–444 RNA from A, lane 3. The two molecules interact through an extended base pairing of the SL1 elements. (C) Schematic representation of the proposed monomeric form of 1–561 RNA from A, lane 7. Upon incubation of 1–561 RNA at high temperature, there is an intramolecular folding of SL1 that competes with the intermolecular interaction (i.e., dimerization). The SL1 structure is shown partially base paired to the upstream encapsidation signal Ψ, as described in Lanchy et al. (2003).
TABLE 1.
Oligonucleotides used in this study
| as202 | CTAGGAGAGATGGGAGTACACAC |
| asΨ | CTAGGAGCACTCCGTCGTGGTTTG |
| asSL1 | TGGTACCTCGGCCCGCGCCT |
| asDIM | TGGTACCTCGGCCCGCGCCTTTCTAGGAGC |
| as548 | CATCTCCCACAATCTTCTACC |
| sBAMT7R | TAGGATCCTAATACGACTCACTATAGGTCGCTCTGCGGAGAG |
| asECO444 | AAGAATTCGCTCCACACGCTGCCTTTG |
| asECO526 | AAGAATTCGTCTAAAGGTAGGATAG |
| asECO561 | AAGAATTCAGTTTCTCGCGCCCATCTCCC |
The 5′ to 3′ sequence is indicated from left to right.
An antisense DNA oligonucleotide directed against the 3′ end of 1–561 RNA induces tight dimerization
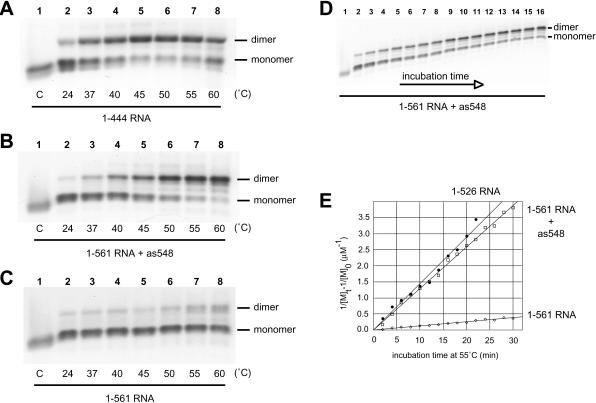
Because two elements located upstream and downstream of SL1 were shown previously to influence the formation of 1–561 RNA tight dimers in vitro (Fig. 2C ▶; Lanchy et al. 2003), we hypothesized that antisense DNA oligonucleotides directed against these interfering elements (as202 and as548; Fig. 1 ▶; Table 1 ▶) should promote 1–561 RNA tight dimerization. We first tested the effect of a 20-fold excess of as548 oligonucleotide on the level of 1–561 RNA tight dimer. After denaturation and quench cooling, the oligonucleotide and the 1–561 RNA were coincubated in dimer buffer at temperatures ranging from 24°C to 60°C and loaded after a 30-min incubation on a TBE gel run at 28°C (see Materials and Methods). As positive and negative controls, the 1–444 (Fig. 3A ▶) and 1–561 (Fig. 3C ▶) RNAs, respectively, were incubated without oligonucleotide as described above. The level of 1–561 RNA tight dimers was low at any temperature (Fig. 3C ▶). However, incubation of the 1–561 RNA together with as548 promoted temperature-dependent dimerization comparable to that of 1–444 RNA (Fig. 3 ▶, cf. A and B). To further characterize the as548-mediated dimerization, we performed kinetic analysis of tight dimerization at 55°C for 1–526 RNA and 1–561 RNA coincubated with or without the as548 oligonucleotide, assayed on a TBE gel at 28°C (Fig. 3D,E ▶). We used 1–526 RNA in this assay because it represents the truncation construct that most nearly resembles the 1–561 RNA with as548 bound; oligonucleotide as548 hybridizes to nt 528–548 to inhibit the long-range interaction, whereas 1–526 RNA lacks these same nucleotides. To determine the dimerization rate constant, the data were fitted using a second-order reaction model (see Materials and Methods). The dimerization rates of 1–526 RNA and 1–561 RNA were 0.061± 0.016 and 0.005± 0.001 μM−1min−1, respectively. The dimerization rate of 1–561 RNA in the presence of as548 was 0.055± 0.013 μM−1min−1 (results from duplicate experiments). Thus, the binding of the as548 oligonucleotide was able to suppress the interference of tight dimerization of 1–561 leader RNA.
FIGURE 3.
Influence of incubation temperature and time on the as548 oligonucleotide-induced dimerization of 1–561 RNA. The 1–444 RNA (A), 1–561 RNA with the as548 oligonucleotide (B), or 1–561 RNA without oligonucleotide (C) were incubated in dimer buffer for 30 min at the indicated temperatures and subjected to electrophoresis on TBE agarose gel run at 28°C. In B, the 1–561 RNA was incubated with a 20-fold excess of the antisense DNA oligonucleotide as548, which is complementary to nt 528–548. The band below the indicated monomer band in A and the two faint bands between monomer and dimer bands in B probably represent low-abundance conformers of monomer (in A) or dimer (in B) species. Control lanes C are described in the previous figure. (D) The time-dependent dimerization of 1–561 RNA at 55°C in the presence of the as548 oligonucleotide was monitored on a TBE agarose gel at 28°C. (Lanes 1–16) Incubation times of 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, 24, 26, 28, 30 min, respectively. (E) Plots of the kinetic data for 1–526 RNA and 1–561 RNA with or without as548 (represented by closed circles, open squares, and open diamonds, respectively). [M]t is the concentration of monomer at time t, and [M]0 is the initial concentration of dimerization-competent monomer.
Induction of 1–561 tight dimerization by antisense oligonucleotides as202 and as548 involves the SL1 region
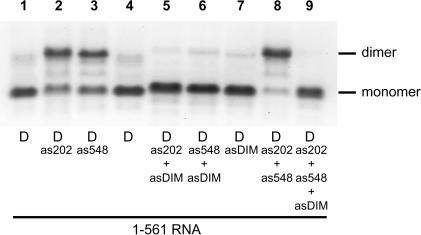
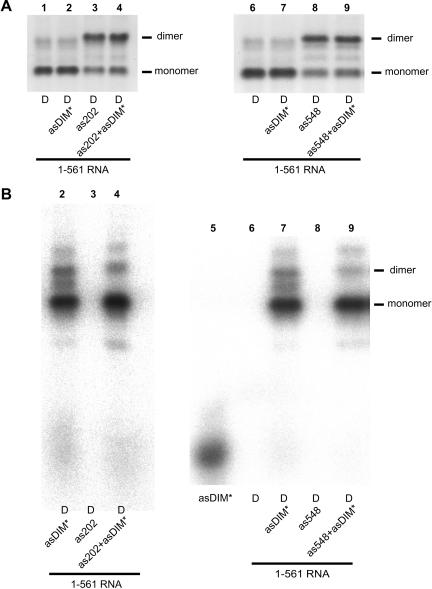
As was observed with oligonucleotide as548 above, an identical dimer-inducing effect was obtained with oligonucleotide as202, which is complementary to the upstream interfering element. To determine whether SL1 was involved in as548 or as202-induced tight dimers, we tested the effect of adding the oligonucleotide asDIM, which is complementary to the 5′ stem of the extended SL1 structure, as previously modeled (McCann and Lever 1997). The 1–561 RNA was incubated for 30 min at 55°C in dimer buffer with or without a 20-fold excess of as202, as548, or both (Fig. 4 ▶). Each of these oligonucleotides promoted tight dimerization of the RNA (Fig. 4 ▶, cf. lanes 2 and 3 and lanes 1 or 4). Coincubation of as202, as548 with asDIM led to an almost complete absence of dimer (Fig. 4 ▶, lanes 5,6,9). Thus, the 1–561 RNA tight dimers induced by targeting both interfering elements used the region complementary to asDIM for dimerization.
FIGURE 4.
Antisense oligonucleotide-mediated activation and suppression of 1–561 RNA tight dimerization. 1–561 RNA was incubated for 30 min at 55°C in dimer buffer with a 20-fold excess of as202, as548, asDIM, or a combination of these oligonucleotides and subjected to electrophoresis on TBE agarose gel at 28°C. Antisense DNA oligonucleotides as202 and as548 target the upstream and downstream dimer-interfering elements, respectively, previously characterized in our laboratory (Lanchy et al. 2003). asDIM is complementary to nt 397–426 and inhibits SL1-dependent tight dimerization of the 1–444 model RNA (Fig. 2 ▶).
Analysis of dimerization domain usage using radiolabeled antisense oligonucleotides
To further analyze whether SL1 and flanking sequences were used in oligonucleotide-induced dimerization, we monitored the partitioning of a limiting amount of radiolabeled asDIM oligonucleotide into the monomer and dimer bands upon induction of 1–561 RNA tight dimerization. Oligonucleotides were added to the 1–561 RNA prior to the initial heat denaturation and quench cooling step. The oligonucleotide–RNA mixtures were then incubated in dimer buffer at 55°C and samples were analyzed on a TBE gel. As before, as202 and as548 oligonucleotides were able to promote dimerization (Fig. 5A ▶, lanes 3,8). There was no detectable inhibition of dimerization by asDIM because the amount of oligonucleotide was low compared to the amount of RNA (Fig. 5A ▶, lanes 4,9). When the gel was visualized for the asDIM-associated radioactivity, the radioactivity was found mostly in the monomer band, even when most of the RNA was shifted to the dimer band by as202 or as548 induction (Fig. 5B ▶, lanes 2,4,7,9). These results confirmed that the dimerization site used in as202- or as548-induced tight dimers involves all or part of the sequence complementary to the asDIM oligonucleotide.
FIGURE 5.
Partitioning of asDIM oligonucleotide with monomer and dimer species of 1–561 RNA upon dimer induction by as202 or as548 oligonucleotides. 1–561 RNA and oligonucleotides were mixed, heated to 90°C, quench cooled, then incubated at 55°C. Reactions shown in lanes 2,4,7,9 included a small amount of radioactively labeled asDIM oligonucleotide. Reactions shown in lanes 4 and 9 additionally contained a 20-fold excess of as202 or as548 oligonucleotides, respectively. The monomer and dimer species were separated on TBE/28°C gel as described in the previous figures. (A) The gel was stained with ethidium bromide and visualized by fluorescence scanning. (B) The same gel was then fixed and dried and the radioactivity associated with the free and bound oligonucleotide was visualized by phosphorimager analysis. Lanes 2,7 suggest that a different interacting region than SL1or Ψ mediated the low level of residual dimers. The exclusion of asDIM* from the dimer bands in lanes 4,9 suggests that nt 397–426 are required for dimerization. Lane 5 corresponds to free radioactively labeled asDIM oligonucleotide loaded without RNA.
An HIV-2 leader RNA lacking SL1 forms tight dimers in the presence of as548 oligonucleotide and uses the region upstream of SL1 as dimerization element
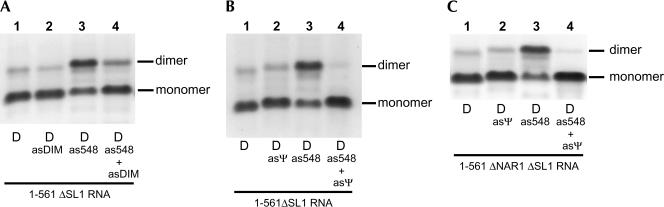
We tested the effect of the as548 oligonucleotide on a construct derived from the 1–561 RNA that lacks nt 409–436 in the SL1 structure. Based on the previous experiments, as548 was not expected to restore dimerization on this construct. The 1–561ΔSL1 RNA formed a very low level of tight dimer upon incubation at 55°C in dimer buffer (Fig. 6A ▶, lane 1). Surprisingly, incubation in the presence of an excess of as548 induced a high yield of tight dimers (Fig. 6A ▶, lane 3). Incubation of the 1–561ΔSL1 RNA with as548 and asDIM virtually eliminated tight dimerization (Fig. 6A ▶, lane 4). Despite the fact that the asDIM-binding site on the 1–561ΔSL1 RNA was reduced to nt 397–408, this oligonucleotide was clearly still able to bind the RNA. Furthermore, this result showed that the region upstream of SL1 can serve as a dimerization element.
FIGURE 6.
as548-Mediated tight dimerization of 1–561RNA lacking SL1and PBS dimerization sites. (A) 1–561 ΔSL1RNA was incubated at 55°C in dimer buffer with a 20-fold excess of as548, asDIM oligonucleotides, or both and subjected to electrophoresis on TBE gel. The 1–561ΔSL1RNA lacks the SL1structure (deletion of nt 409–436). (B) The experiment described in A was reproduced using asΨ instead of asDIM. The asΨ oligonucleotide is complementary to nt 380–404 (Table 1 ▶) and binds to the encapsidation signal Ψ. (C) The experiment described in B was reproduced with the 1–561ΔNAR1ΔSL1RNA. This construct lacks both SL1and the 5′-304-GGCGCC-309-3′ sequence located at the 5′ end of the primer-binding site shown previously to mediate loose dimerization of 1–561 RNA (Jossinet et al. 2001; Lanchy and Lodmell 2002). Loose dimers are not expected to withstand the TBE electrophoresis.
We then tested an antisense oligonucleotide complementary to the in vivo-defined encapsidation signal, Ψ, just upstream of SL1 (Fig. 1B ▶; Griffin et al. 2001) and found that it was also able to inhibit the as548-induced 1–561ΔSL1 RNA tight dimer formation (Fig. 6B ▶, cf. lanes 4 and 3). As a supplementary control, we tested an RNA fragment lacking SL1 and the second dimerization site characterized in vitro, the 5′ end of the primer binding site (1–561ΔNAR1ΔSL1; Jossinet et al. 2001; Lanchy and Lodmell 2002). The result was identical to the one observed with the 1–561ΔSL1 RNA (Fig. 6 ▶, cf. C and B). Thus, this experiment showed that the Ψ sequence was responsible for promoting dimerization under these conditions.
Dimerization through the Ψ region is induced by the binding of an antisense oligonucleotide directed against SL1 and vice versa
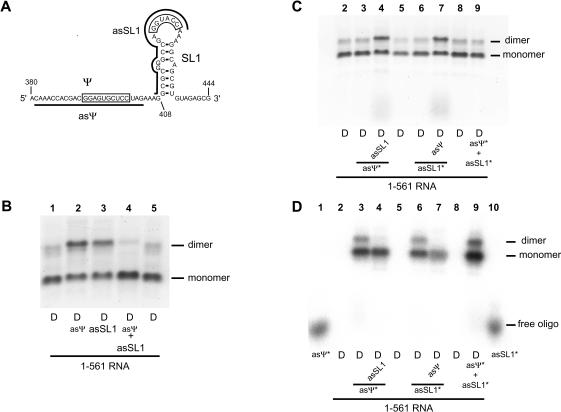
Analysis of the primary sequence of HIV-2 (ROD isolate) leader RNA revealed the presence of a palindromic sequence 5′-GGAGUGCUCC-3′ located upstream of SL1 (nt 392–401; Fig. 7A ▶). In previous work, it was proposed that the lack of use of SL1 as a dimerization element in 1–561 HIV-2 RNA was due to a more favorable intramolecular interaction of SL1 with an upstream sequence that includes the nucleotides 392–401 (Lanchy et al. 2003). Therefore, the next experiment was designed to test whether the Ψ and SL1 sequences possess similar dimer-promoting characteristics. The 1–561 RNA was coincubated with asΨ or asSL1 oligonucleotides in dimer buffer at 55°C, and the resulting dimers were analyzed on a TBE agarose gel (Fig. 7B ▶). Very surprisingly, the presence of either asSL1 or asΨ induced a small but reproducible increase in the level of 1–561 RNA tight dimers (Fig. 7B ▶, cf. lanes 2 or 3 and lanes 1 or 5). However, the presence of both oligonucleotides strongly inhibited dimer formation (Fig. 7B ▶, cf. lane 4 and lanes 1 or 5).
FIGURE 7.
Antisense oligonucleotide directed against SL1induces Ψ-dependent tight dimerization of 1–561 RNA and vice versa. (A) Representation of the Ψ –SL1region and antisense oligonucleotides used in this experiment. The two palindromic sequences involved in dimerization are boxed. asΨ and asSL1antisense oligonucleotides binding sites are represented by thick lines. (B) 1–561 RNA was incubated in dimer buffer at 55°C without or with a 20-fold molar excess of asΨ, asSL1, or both oligonucleotides, and subjected to TBE electrophoresis. (C, D) Partitioning of asΨ or asSL1oligonucleotides in monomer or dimer species of 1–561RNA. The experiment described in B was repeated with one of the oligonucleotides radioactively labeled and present in a small amount and the other present in a 20-fold molar excess. In C, the gel is visualized with ethidium bromide and in D, the same dried gel is visualized by phosphorimager analysis to visualize the radioactivity associated with the labeled oligonucleotide.
To confirm that induction by asΨ was mediated by SL1 and that induction by asSL1 was mediated by the Ψ region, we assayed dimerization in the presence of a small amount of radioactively labeled oligonucleotide with or without an excess of the other oligonucleotide. Because the radioactively labeled oligonucleotide was present in a small substoichiometric amount, there was no detectable induction or inhibition of dimerization by the labeled oligonucleotide alone when the gel was visualized with ethidium bromide (Fig. 7C ▶, lanes 3,6,9). The partitioning of the radioactively labeled oligonucleotide into monomer and dimer species was similar to the proportion of monomer and dimer RNA species (Fig. 7 ▶, cf. lanes 3 in C and D and lanes 6 in C and D). Incubation with a 20-fold molar excess of oligonucleotide (relative to the 1–561 RNA) shifted the radioactivity to the monomer species, indicating that the dimer induction by the binding of one oligonucleotide involved the region targeted by the other oligonucleotide (Fig. 7D ▶, cf. lanes 3 and 4 and lanes 6 and 7). These results support the model of an intramolecular interaction between the Ψ and SL1 region (Fig. 2C ▶), and that the Ψ region could be involved in dimerization in a way similar to SL1. The in vitro interactions between encapsidation and dimerization elements may reflect a functional overlap between encapsidation and dimerization processes observed in vivo (for review, see Greatorex and Lever 1998).
RNase T1 structure probing of the Ψ–SL1 domain
To further demonstrate the differential usage of dimerization elements, we analyzed the structure of the Ψ–SL1 domain of the 1–561 RNA incubated with or without as548 using the RNase T1 (Fig. 8 ▶). The RNase T1 enzyme cuts the phosphodiester backbone of guanines in single-stranded regions. The reactivities of guanines in SL1 suggest it does not form an extended duplex in the presence of as548 for several reasons. First, the G420 and G421 nucleotides in the loop of SL1 remained accessible when the RNA was dimeric (i.e., in the presence of as548; Fig. 8A ▶, lane 2). Second, reactivities of guanines 413–415 and 435 were not compatible with an extended duplex structure of SL1 (Fig. 8B ▶). The lack of reactivity of guanines in the Ψ palindromic sequence and the persistent reactivity of guanines in SL1 in the presence of as548, together with the oligonucleotide binding data (Figs. 6 ▶, 7 ▶) suggested that the Ψ palindrome was the main dimerization element in as548-induced tight dimers of 1–561 RNA.
FIGURE 8.
Structural analysis of the Ψ–SL1region. (A) 1–561 RNA was probed with RNase T1 in the presence (lane 2) or absence (lane 3) of as548. The lanes labeled G, U, A, and C are sequencing lanes. (Lane 1) Control (1–561RNA primer extended without RNase T1 modification); (lane 2) 1–561RNA activated with oligonucleotide as548 during RNase T1 probing; (and lane 3) RNase T1 probing of 1–561RNA without as548 present during modification. Theregion labeled Ψ represents the palindromic sequence GGAGUGCUCC present in this region.The region labeled SL1represents the palindromic sequence GGUACC present in this region.(B) A model representing HIV-2 RNA SL1in an extended duplex conformation. (Open triangles) Cleavages observed in 1–561RNA by RNase T1 in the absence of as548. (Solid circles) RNase T1 cleavages observed in 1–561RNA with as548 present. An increase in nucleotide reactivity is represented by an increase in the symbol present at that location.
DISCUSSION
In retroviral replication, the essential processes of genomic RNA dimerization and encapsidation are thought to be linked. In this study, we demonstrate that in HIV-2 RNA, sequences known separately for their involvement in dimerization and encapsidation are functionally linked. In particular, we show that conformational changes in the leader region RNA can promote or silence the use of dimerization motifs. The conformational changes involve alternative base pairing arrangements affecting the dimerization site SL1, the packaging signal Ψ, and a long-range base-pairing interaction that includes the gag start codon with a sequence just downstream of the polyadenylation signal stem loop, thus potentially providing a mechanism to coordinate the diverse functions of dimerization, encapsidation, and translation.
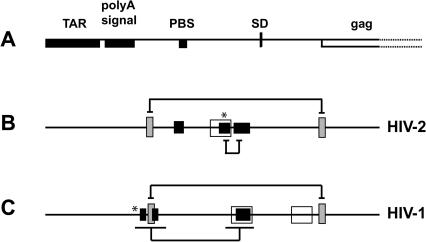
An impetus for this study derived from the apparent functional silence of SL1 in dimerization of HIV-2 RNA fragments under most conditions, whereas its homolog in HIV-1 is clearly a principal player in dimerization (Darlix et al. 1990; Laughrea and Jette 1994; Paillart et al. 1994; Skripkin et al. 1994; Muriaux et al. 1995; Clever et al. 1996; Haddrick et al. 1996). This led to the discovery that in HIV-2 another sequence overlapping the PBS can mediate loose dimerization (Jossinet et al. 2001; Lanchy and Lodmell 2002) and that a specific long-range base-pairing interaction is responsible for silencing SL1 as a dimerization element (Lanchy et al. 2003; a somewhat different interaction was also proposed in Dirac et al. 2002). Significantly, the same long-range interaction that influences HIV-2 SL1-mediated dimerization has been found in HIV-1 (Fig. 9 ▶; Huthoff and Berkhout 2001, 2002; Abbink and Berkhout 2002; Berkhout et al. 2002), and a long-range interaction between nucleotides in the Matrix region of gag with a sequence just downstream of the poly(A) signal in HIV-1 was recently described (Paillart et al. 2002), supporting the idea that the 5′ leader region could serve multiple regulatory purposes.
FIGURE 9.
Structural and functional elements involved in encapsidation and dimerization of HIV-1and HIV-2 genomic RNAs. (A) The landmark sequences with known functions are indicated by boxes (see Fig. 1A ▶). (B, C) The encapsidation, dimerization, and regulatory elements in HIV-2 (Griffin et al. 2001; Dirac et al. 2002; Lanchy et al. 2003) and HIV-1(McBride and Panganiban 1996; Abbink and Berkhout 2002; Berkhout et al. 2002; Clever et al. 2002; Russell et al. 2002) are indicated by open, closed, and hatched boxes, respectively. The described short- or long-range interactions between structural elements are indicated with brackets. The asterisk indicates palindrome (GAGUGCUU/C) locations in HIV-1and HIV-2.
In this work, we used antisense DNA oligonucleotides both to activate and to mask viral RNA sequences involved in dimerization. This strategy allowed us to assess the role of the long-range interaction in the silencing of SL1, to monitor which sequences were required for dimerization depending on the conformation of the RNA, and to easily modulate the conformation of the RNA. Identical kinetic behavior of oligonucleotide as548-induced dimerization, compared to dimerization of wild-type RNAs that did not require an oligonucleotide (e.g. 1–526 RNA), suggested that the oligonucleotide-induced conformation was similar to the wild-type dimerization-competent form. The prospect that other physiological cues could trigger the same or similar conformational changes in vivo is quite intriguing. A ribosome bound to the gag initiation codon would disrupt the long-range interaction described here, and nucleocapsid protein or the nucleocapsid domain of the Gag polyprotein is most certainly capable of catalyzing structural rearrangements in this region. The structural and dynamic intricacies of the dimerization domain are also underscored by the differential magnesium dependence of spontaneous loose and oligonucleotide-induced and TBE-resistant tight dimers. Magnesium dependence is suggestive of tertiary RNA interactions, whereas lack of Mg2+ dependence is more characteristic of simpler secondary structures.
One of the more unexpected findings in this study was the observation that a sequence just upstream of SL1 mediated dimerization. This new dimerization site overlaps a region shown by Lever’s group to be essential for encapsidation in vivo (Griffin et al. 2001). A 10-nt-long palindromic sequence in Ψ, 5′-GGAGUGCUCC-3′, appears to be responsible for the as548-mediated tight dimerization of RNAs lacking functional SL1 for two reasons. First, the antisense oligonucleotides asDIM and asΨ that bind all or part of the palindrome were able to prevent as548-mediated tight dimerization of 1–561ΔSL1 or 1–561ΔNAR1ΔSL1 RNAs. Second, the nucleotide sequence within the palindrome, 5′-GAGUGCUC-3′ is known to form an unusually stable duplex in vitro, and is statistically overrepresented in ribosomal RNA (Gautheret et al. 1995; McDowell et al. 1997; Chen et al. 2000; Deng and Sundaralingam 2000). Furthermore, the unique conformational features of UG base pairs are important recognition motifs in RNA–RNA and RNA–protein interactions (for review, see Varani and McClain 2000). Thus, the palindromic sequence found in the Ψ domain has properties very suggestive of a dimerization element.
The experiments presented here indicate that the Ψ dimerization signal is functionally activated or masked in vitro through its interactions with SL1. When this local interaction occurs, neither sequence is available for dimerization. When the Ψ–SL1 interaction is disrupted, either sequence can mediate tight dimerization. In turn, Ψ–SL1 base pairing is favored by the long-range interaction between nucleotides downstream from the polyadenylation signal (190 region) and at the start codon of gag (550 region). Although the specific secondary structures corresponding to the silenced or dimerization-competent forms of the RNA are not yet specified in this model, the RNase probing data presented here have several important implications. First, the accessibilites of guanines in SL1 in both the dimeric and monomeric RNAs argues against an extended duplex involving SL1 in the dimeric form. On the other hand, the protection of the Ψ palindrome in the as548-induced dimer is consistent with its use as a dimerization element, although it is also not reactive in the monomeric form. Finally, significant changes in guanine reactivity in nucleotides flanking the Ψ–SL1 sequences between the monomeric and dimeric states suggest that a conformational change occurs in this region upon dimerization. It will be interesting to see if and how the dimerization properties of the Ψ domain in vitro are related to its use as a dimerization domain in addition to a packaging signal in vivo.
These findings suggest a model for the role of the HIV-2 Ψ palindromic sequence during encapsidation of the genomic RNA. In this model, the initiation of gag translation disrupts the long-range interaction and promotes conformational changes in the Ψ–SL1 domain (and probably others sites as well). The Ψ domain could thus become a recognition site for other factors, the most obvious candidate being the nucleocapsid domain of the Gag polyprotein. Similarly, the SL1 or Ψ element could become a dimerization site that interacts with another genomic RNA molecule. Experiments by the Lever group showed that HIV-2 selects its genomic RNA for encapsidation cotranslationally (Griffin et al. 2001). It was determined that two main elements mediate this activity: the Ψ domain on the RNA and the nucleocapsid domain of Gag. A similar relationship between genomic RNA translation and encapsidation has been suggested for HIV-1 as well (Liang et al. 2002; Poon et al. 2002). It is still not clear what structural features in the encapsidation signal are specifically recognized by nucleocapsid protein. The palindromic sequence may form a duplex or could form an intramolecular structure with nucleotides located upstream or downstream (Griffin et al. 2001; Lanchy et al. 2003). Nucleocapsid protein is known to have a preference for G/U-containing sequences (Berglund et al. 1997; Fisher et al. 1998; Kim et al. 2002). In addition, in vitro selection studies have shown that RNA ligands selected for nucleocapsid binding end with a self-complementary sequence located in a stem–loop structure (Lochrie et al. 1997). These features are similar to presumed features of the Ψ–SL1 region.
As mentioned above, the long-range interaction involving the start codon of gag apparently also exists in HIV-1. It appears that a palindrome similar to the one we describe here in Ψ exists in HIV-1 (Kuiken et al. 2001). It is located in the 3′ stem of the poly(A) signal domain (nt 98–106; HIV-1 Lai isolate; Fig. 9 ▶). Interestingly, this palindromic sequence is thought to be involved in the regulation of HIV-1 RNA dimerization in seemingly opposite ways. On one hand, it may be responsible for HIV-1 SL1 silencing in vitro through a direct base pairing (Huthoff and Berkhout 2001, 2002; Abbink and Berkhout 2002; Berkhout et al. 2002). On the other hand, it may be directly involved in intermolecular interactions between two genomic RNAs as part of the Dimer Linkage Structure (Hoglund et al. 1997). Furthermore, the importance of the poly(A) signal domain in HIV-1 genomic RNA dimerization is underscored by the characterization of two GU-rich sequences that have recently been shown to be involved in genomic RNA dimerization in vivo (Russell et al. 2002). Similarly to the palindromic sequence, these two GU-rich sequences may be involved in the regulation of HIV-1 RNA dimerization, as they overlap with the upstream element involved in a long-range interaction with the gag start codon, and they could be part of the structure that mediates SL1 silencing in vitro (Fig. 9 ▶). Thus, RNA dimerization and its regulation may be mediated by homologous sequences in HIV-1 and HIV-2.
The newly described features of HIV-2 leader region RNA have experimental and phylogenetic support. HIV-1 and HIV-2 leader RNAs apparently share several functional features, although the relative positions of the structural/functional elements and their roles in dimerization may differ. The next endeavor will be to test the relationships and coordinated regulation of genomic RNA dimerization, encapsidation, and translation in vivo during viral replication.
MATERIALS AND METHODS
Template construction for in vitro transcription
A sense primer containing a BamHI site and the promoter for the T7 RNA polymerase and an antisense primer containing an EcoRI site (Table 1 ▶) were used to amplify the first 444 or 561 nt of HIV-2 genomic RNA sequence, ROD isolate (Guyader et al. 1987). The HIV-2 ROD DNA template (modified plasmid pROD10) was provided by the EU Programme EVA/MRC Centralised Facility for AIDS Reagents, NIBSC, UK (Grant Number QLK2-CT-1999-00609 and GP828102). The numbering is based on the genomic RNA sequence. The digested polymerase chain reaction (PCR) products were cloned in the BamHI and EcoRI sites of the pUC18 plasmid. Mutations in the Ψ or SL1 were introduced by megapriming mutagenesis (Ke and Madison 1997). Nucleotides 380–408 and 409–436 are deleted in the ΔΨ and ΔSL1 fragments, respectively. A PstI fragment of the megaprimed PCR product bearing the mutation was then subcloned into a plasmid containing the AatII to XhoI fragment of the modified pROD10 plasmid (corresponding to the LTR-gag fragment of the proviral DNA; Kaye and Lever 1998). Mutated clones containing the first 444 or 561 nt were then subcloned into pUC18 with a promoter for T7 RNA polymerase as described above.
RNA synthesis and purification
The various plasmids were linearized with EcoRI prior to in vitro transcription. RNAs were synthesized by in vitro transcription of the EcoRI-digested plasmids with the AmpliScribe T7 transcription kit (Epicentre). After transcription, the DNA was digested with the supplied RNase-free DNase, and the RNA was purified by ammonium acetate precipitation followed by size exclusion chromatography (Bio-Gel P-4, Bio-Rad).
In vitro dimerization of HIV-2 RNA
Five to 7 pmole of RNA were denatured in 8 μL water for 2 min at 90°C and quench cooled on ice for 2 min. After the addition of 2 μL monomer buffer (final concentrations: 50 mM Tris-HCl at pH 7.5 at 37°C, 40 mM KCl, 0.1 mM MgCl2) or dimer buffer (final concentrations: 50 mM Tris-HCl at pH 7.5 at 37°C, 300 mM KCl, 5 mM MgCl2), dimerization was allowed to proceed for 30 min at 24°C, 37°C, 40°C, 45°C, 50°C, 55°C, or 60°C. For most of the experiments described here, we used 55°C because the incubation of the 1–444 RNA at 55°C allows a maximal yield of TBE-resistant SL1-dependent dimers. The samples were then cooled on ice to stabilize dimers formed during incubation and loaded on a 0.8% agarose gel with 2 μL of glycerol loading dye 6× (40% glycerol, Tris-borate 44 mM at pH 8.3, 0.25% Bromophenol blue). Electrophoresis was carried out for 90 min at a monitored temperature of 28°C in Tris-borate 44 mM (pH 8.3), EDTA 1 mM (TBE). After electrophoresis, the ethidium bromide-stained gel was scanned on a Fluorescent Image Analyzer FLA-3000 (Fujifilm).
Kinetics of tight dimer formation
One hundred picomoles of RNA with or without a 20-fold excess of as548 oligonucleotide were denatured in 160 μL water for 2 min at 90°C and quench cooled on ice for 2 min. After the addition of 2 μL dimer buffer under the lid of 20 tubes, 8 μL of denatured RNA was aliquoted to each tube. The dimerization was then initiated by a 5-sec spin in a benchtop centrifuge and immediate loading of the tubes in a heating block at 55°C. Dimerization was allowed to proceed for 2–30 min. At each time point, a tube containing 10 μL reaction mixture was removed from the heating block, mixed with 2 μL of glycerol loading dye 6× and loaded on a 0.8% agarose TBE gel. Electrophoresis was carried out at 28°C and 3 V/cm. After electrophoresis, the ethidium bromide-stained gel was scanned on a Fluorescent Image Analyzer FLA-3000 (Fujifilm). Quantification of the extent of dimerization was done using Fujifilm Image Gauge V3.3 software. The data were fitted using a second-order conformation model (Marquet et al. 1994):
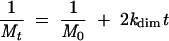 |
where Mt is the concentration of monomer at time t, M0 is the initial concentration of dimerization-competent monomer, and kdim is the second-order rate constant of dimerization (μM−1•min−1).
Antisense oligonucleotides
Five picomoles of RNA with or without 100 pmole of oligonucleotide were denatured in 8 μL water for 2 min at 90°C and quench cooled on ice for 2 min. After the addition of 2 μL fivefold concentrated dimer buffer, dimerization was allowed to proceed for 30 min at 55°C. The samples were then cooled on ice to stabilize dimers formed during incubation and loaded on a 0.8% TBE agarose gel. Electrophoresis was carried out for 90 min at 28°C. To test the monomer/dimer partitioning of the oligonucleotide, 5 pmole of RNA were incubated under dimerization conditions with a small amount of 32P-labeled oligonucleotide (total of 0.02–0.075 pmole) with or without 100 pmole of another unlabeled oligonucleotide. After electrophoresis, the ethidium bromide-stained gel was scanned on a Fluorescent Image Analyzer FLA-3000 (Fujifilm). When radioactive oligonucleotides were used, the gel was fixed after electrophoresis with trichloroacetic acid, then dried. Radioactivity was visualized and quantified with the FLA-3000 instrument using a phosphorimager plate.
Enzymatic RNA probing with RNase T1
In a standard experiment, 5 pmole of 1–561 RNA were dissolved in 4 μL of water with or without 10 pmole of the antisense oligonucleotide as548 (Table 1 ▶), heated for 2 min at 90°C and quench cooled on ice. The samples were incubated in dimer buffer (final concentrations: 50 mM sodium cacodylate at pH 7.5, 5 mM MgCl2, 300 mM KCl) for 15 min or 30 min at 37°C or 55°C, respectively. The samples were quench cooled on ice, 2 μg of Escherichia coli tRNAs (Sigma) and 0.11 units of RNase T1 (Invitrogen) were added, and the samples incubated for 5 min, at 37°C, phenol:chloroform (pH 6.7) extracted, and ethanol precipitated. The samples were pelleted by centrifugation at 15,000 rpm for 30 min, ethanol washed, vacuum dried, and resuspended in RQ1 DNase buffer with 5 units of RQ1 RNase-free DNase (Promega) and incubated for 60 min at 37°C. The DNase was removed using EZ Micropure enzyme removers (Millipore) as per the manufacturer’s directions. The samples were ethanol precipitated, pelleted, ethanol washed, vacuum dried, resuspended in water, and the positions of RNA cleavage were detected by primer extension (Moazed et al. 1986) with avian myeloblastosis reverse transcriptase (Seikagaku America).
Acknowledgments
We acknowledge Hector Valtierra for critical reading of the manuscript. The modified pROD10 plasmids from Drs. J.-M. Bechet and A.M.L. Lever were obtained from the Centralised Facility for AIDS Reagents supported by EU Programme EVA (contract QLK2-CT-1999-00609) and the UK Medical Research Council. This research is supported by the National Institutes of Health grant number AI45388 to J.S.L.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5590603.
REFERENCES
- Abbink, T.E. and Berkhout, B. 2002. A novel long distance base pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 278: 11601–11611. [DOI] [PubMed] [Google Scholar]
- Bender, W. and Davidson, N. 1976. Mapping of poly(A) sequences in the electron microscope reveals unusual structure of type C oncornavirus RNA molecules. Cell 7: 595–607. [DOI] [PubMed] [Google Scholar]
- Berglund, J.A., Charpentier, B., and Rosbash, M. 1997. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 25: 1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54: 1–34. [DOI] [PubMed] [Google Scholar]
- Berkhout, B., Ooms, M., Beerens, N., Huthoff, H., Southern, E., and Verhoef, K. 2002. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem. 277: 19967–19975. [DOI] [PubMed] [Google Scholar]
- Chen, X., McDowell, J.A., Kierzek, R., Krugh, T.R., and Turner, D.H. 2000. Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: One hydrogen bond for each G.U pair in r(GG CGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2). Biochemistry 39: 8970–8982. [PubMed] [Google Scholar]
- Cheung, K.S., Smith, R.E., Stone, M.P., and Joklik, W.K. 1972. Comparison of immature (rapid harvest) and mature Rous sarcoma virus particles. Virology 50: 851–864. [DOI] [PubMed] [Google Scholar]
- Clever, J.L., Wong, M.L., and Parslow, T.G. 1996. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J. Virol. 70: 5902–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever, J.L., Mirandar Jr., D., and Parslow, T.G. 2002. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 76: 12381–12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix, J.L., Gabus, C., Nugeyre, M.T., Clavel, F., and Barre-Sinoussi, F. 1990. Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1. J. Mol. Biol. 216: 689–699. [DOI] [PubMed] [Google Scholar]
- Deng, J. and Sundaralingam, M. 2000. Synthesis and crystal structure of an octamer RNA r(guguuuac)/r(guaggcac) with G.G/U.U tandem wobble base pairs: Comparison with other tandem G.U pairs. Nucleic Acids Res. 28: 4376–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirac, A.M., Huthoff, H., Kjems, J., and Berkhout, B. 2001. The dimer initiation site hairpin mediates dimerization of the human immunodeficiency virus, type 2 RNA genome. J. Biol. Chem. 276: 32345–32352. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 30: 2647–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R.J., Rein, A., Fivash, M., Urbaneja, M.A., Casas-Finet, J.R., Medaglia, M., and Henderson, L.E. 1998. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 72: 1902–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W. and Rein, A. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67: 5443–5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, W., Gorelick, R.J., and Rein, A. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68: 5013–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautheret, D., Konings, D., and Gutell, R.R. 1995. G.U base pairing motifs in ribosomal RNA. RNA 1: 807–814. [PMC free article] [PubMed] [Google Scholar]
- Greatorex, J. and Lever, A. 1998. Retroviral RNA dimer linkage. J. Gen. Virol. 79: 2877–2882. [DOI] [PubMed] [Google Scholar]
- Griffin, S.D., Allen, J.F., and Lever, A.M. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: Cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75: 12058–12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyader, M., Emerman, M., Sonigo, P., Clavel, F., Montagnier, L., and Alizon, M. 1987. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature 326: 662–669. [DOI] [PubMed] [Google Scholar]
- Haddrick, M., Lear, A.L., Cann, A.J., and Heaphy, S. 1996. Evidence that a kissing loop structure facilitates genomic RNA dimerisation in HIV-1. J. Mol. Biol. 259: 58–68. [DOI] [PubMed] [Google Scholar]
- Hoglund, S., Ohagen, A., Goncalves, J., Panganiban, A.T., and Gabuzda, D. 1997. Ultrastructure of HIV-1 genomic RNA. Virology 233: 271–279. [DOI] [PubMed] [Google Scholar]
- Huthoff, H. and Berkhout, B. 2001. Two alternating structures of the HIV-1 leader RNA. RNA 7: 143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry 41: 10439–10445. [DOI] [PubMed] [Google Scholar]
- Jewell, N.A. and Mansky, L.M. 2000. In the beginning: Genome recognition, RNA encapsidation and the initiation of complex retrovirus assembly. J. Gen. Virol. 81: 1889–1899. [DOI] [PubMed] [Google Scholar]
- Jossinet, F., Lodmell, J.S., Ehresmann, C., Ehresmann, B., and Marquet, R. 2001. Identification of the in vitro HIV-2/SIV RNA dimerization site reveals striking differences with HIV-1. J. Biol. Chem. 276: 5598–5604. [DOI] [PubMed] [Google Scholar]
- Kaye, J.F. and Lever, A.M. 1998. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: A possible role for the p2 domain of Gag in RNA encapsidation. J. Virol. 72: 5877–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, S.H. and Madison, E.L. 1997. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 25: 3371–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.J., Kim, M.Y., Lee, J.H., You, J.C., and Jeong, S. 2002. Selection and stabilization of the RNA aptamers against the human immunodeficiency virus type-1 nucleocapsid protein. Biochem. Biophys. Res. Commun. 291: 925–931. [DOI] [PubMed] [Google Scholar]
- Kuiken, C., Foley, B., Hahn, B., Marx, P., McCutchan, F., Mellors, J., Wolinsky, S., and Korber, B. 2001. HIV Sequence Compendium 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory.
- Kung, H.J., Hu, S., Bender, W., Bailey, J.M., Davidson, N., Nicolson, M.O., and McAllister, R.M. 1976. RD-114, baboon, and woolly monkey viral RNA’s compared in size and structure. Cell 7: 609–620. [DOI] [PubMed] [Google Scholar]
- Lanchy, J.M. and Lodmell, J.S. 2002. Alternate usage of two dimerization initiation sites in HIV-2 viral RNA in vitro. J. Mol. Biol. 319: 637–648. [DOI] [PubMed] [Google Scholar]
- Lanchy, J.M., Rentz, C.A., Ivanovitch, J.D., and Lodmell, J.S. 2003. Elements located upstream and downstream of the major splice donor site influence the ability of HIV-2 leader RNA to dimerize in vitro. Biochemistry 42: 2634–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughrea, M. and Jette, L. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33: 13464–13474. [DOI] [PubMed] [Google Scholar]
- ———. 1996. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensable for dimer formation. Biochemistry 35: 1589–1598. [DOI] [PubMed] [Google Scholar]
- Laughrea, M., Shen, N., Jette, L., Darlix, J.L., Kleiman, L., and Wainberg, M.A. 2001. Role of distal zinc finger of nucleocapsid protein in genomic RNA dimerization of human immunodeficiency virus type 1; no role for the palindrome crowning the R-U5 hairpin. Virology 281: 109–116. [DOI] [PubMed] [Google Scholar]
- Liang, C., Hu, J., Russell, R.S., and Wainberg, M.A. 2002. Translation of Pr55(gag) augments packaging of human immunodeficiency virus type 1 RNA in a cis-acting manner. AIDS Res. Hum. Retroviruses 18: 1117–1126. [DOI] [PubMed] [Google Scholar]
- Lochrie, M.A., Waugh, S., Pratt Jr., D.G., Clever, J., Parslow, T.G., and Polisky, B. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res. 25: 2902–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet, R., Paillart, J.C., Skripkin, E., Ehresmann, C., and Ehresmann, B. 1994. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 22: 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, M.S. and Panganiban, A.T. 1996. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 70: 2963–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann, E.M. and Lever, A.M. 1997. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 71: 4133–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.A., He, L., Chen, X., and Turner, D.H. 1997. Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGA UGUCC)2. Biochemistry 36: 8030–8038. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Stern, S., and Noller, H.F. 1986. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J. Mol. Biol. 187: 399–416. [DOI] [PubMed] [Google Scholar]
- Muriaux, D., Girard, P.M., Bonnet-Mathoniere, B., and Paoletti, J. 1995. Dimerization of HIV-1Lai RNA at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J. Biol. Chem. 270: 8209–8216. [DOI] [PubMed] [Google Scholar]
- Muriaux, D., Fosse, P., and Paoletti, J. 1996. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry 35: 5075–5082. [DOI] [PubMed] [Google Scholar]
- Paillart, J.C., Marquet, R., Skripkin, E., Ehresmann, B., and Ehresmann, C. 1994. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 269: 27486–27493. [PubMed] [Google Scholar]
- Paillart, J.C., Skripkin, E., Ehresmann, B., Ehresmann, C., and Marquet, R. 1996. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. 93: 5572–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002. In vitro evidence for a long range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J. Biol. Chem. 277: 5995–6004. [DOI] [PubMed] [Google Scholar]
- Poon, D.T., Chertova, E.N., and Ott, D.E. 2002. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): Functional linkage between translation and RNA packaging. Virology 293: 368–378. [DOI] [PubMed] [Google Scholar]
- Russell, R.S., Hu, J., Laughrea, M., Wainberg, M.A., and Liang, C. 2002. Deficient dimerization of human immunodeficiency virus type 1 RNA caused by mutations of the u5 RNA sequences. Virology 303: 152–163. [DOI] [PubMed] [Google Scholar]
- Shubsda, M.F., McPike, M.P., Goodisman, J., and Dabrowiak, J.C. 1999. Monomer-dimer equilibrium constants of RNA in the dimer initiation site of human immunodeficiency virus type 1. Biochemistry 38: 10147–10157. [DOI] [PubMed] [Google Scholar]
- Skripkin, E., Paillart, J.C., Marquet, R., Ehresmann, B., and Ehresmann, C. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. 91: 4945–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani, G. and McClain, W.H. 2000. The G × U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 1: 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]