Abstract
Ambisense Sendai virus (SeV) was prepared in order to study the control of viral RNA synthesis. In these studies, we found that the relative ratios of genomes/antigenomes formed during infection are largely determined by the relative strengths of the replication promoters, independent of the presence of a functional mRNA start site. We also found that the ability of the viral polymerase (vRdRP) to respond to an mRNA editing site requires prior (re)initiation at an mRNA start site, similar to the acquisition of vRdRP processivity in the absence of nascent chain coassembly. During these studies, the inherent instability of ambisense SeV upon passage in embryonated chicken eggs was noted and was found to be associated with a point mutation in the ambisense mRNA (ambi-mRNA) start site that severely limited its expression. Since the interferon (IFN)-induced antiviral state is mediated in part via double-stranded RNA (dsRNA), the efficiency of the ambi-mRNA poly(A)/stop site was examined. This site was found to operate in a manner similar to that of other SeV mRNA poly(A)/stop sites, i.e., at ∼95% efficiency. This modest level of vRdRP read-through is apparently tolerable for natural SeV because the potential to form dsRNA during infection remains limited. However, when mRNAs are expressed from ambisense SeV antigenomes, vRdRP read-through of the ambi-mRNA poly(A)/stop site creates a capped transcript that can potentially extend the entire length of the antigenome, since there are no further poly(A)/stop sites here. In support of this hypothesis, loss of ambi-mRNA expression during passage of ambisense SeV stocks in eggs is also characterized by conversion of virus that grows poorly in IFN-sensitive cultures and is relatively IFN sensitive to virus that grows well even in IFN-pretreated cells that restrict vesicular stomatitis virus replication, i.e., the wild-type SeV phenotype. The selection of mutants unable to express ambi-mRNA on passage in chicken eggs is presumably due to increased levels of dsRNA during infection. How natural ambisense viruses may deal with this dilemma is discussed.
The order Mononegavirales includes three virus families that replicate in the cytoplasm: Rhabdoviridae, Paramyxoviridae, and Filoviridae. Their nonsegmented negative-strand ([−]) RNA virus (NNV) genomes are 10 to 18 kb in length and contain 5 to 10 tandemly linked genes (or mRNA units) transcribed in turn (1, 3). The genes are separated by conserved junctional sequences that act as mRNA start and poly(A)/stop sites (27). The [−] genomes of all NNV are organized similarly and are thought to express their genes in a similar manner, typified by Sendai virus (SeV), a member of the Respirovirus genus of the subfamily Paramyxovirinae. The first (N) mRNA of all viruses in this subfamily starts precisely 56 nucleotides (nt) from the 3′ end of the genome (nt 56), and the 55 nt upstream are called the leader region (le) (Fig. 1). The last (L) mRNA ends a similar (but variable) distance from the 5′ end of the genome, and the ∼50 nt between the end of the L gene and the 5′ end of the genome are called the trailer region (tr) (Fig. 1 and 2) (31).
FIG. 1.
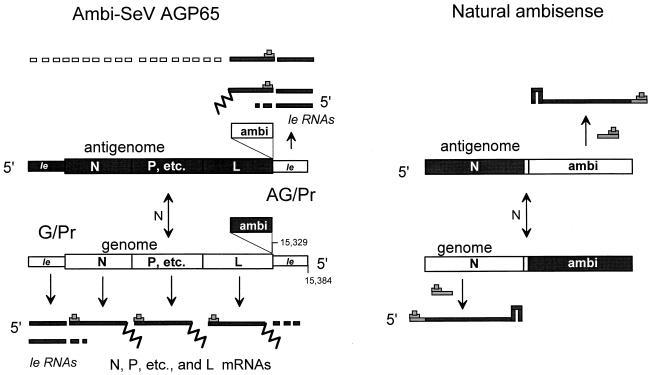
mRNA expression from ambisense SeV and natural ambisense genomes. The genome and antigenome nucleocapsid segments of nonsegmented SeV AGP65 and a typical ambisense segment of a natural (segmented) ambisense virus are shown in the middle as boxes (definitely not drawn to scale); their le regions that are not transcribed into mRNA are slightly thinner. Sequences that can be directly translated into protein ([+]) are shown in dark shading; their complements ([−]) are shown in light shading (le sequences are shaded arbitrarily). Le RNA and mRNA transcripts are shown as very thin boxes or thick lines; the 5′ cap and 3′ poly(A) tail are represented by a hat and squiggly line, respectively. Short boxes extending the transcripts represent RdRP read-through of poly(A)/stop sites. Strong secondary structure at the mRNA 3′ end is shown as a hairpin. The arrows indicate the flow of viral RNA synthesis.
FIG. 2.
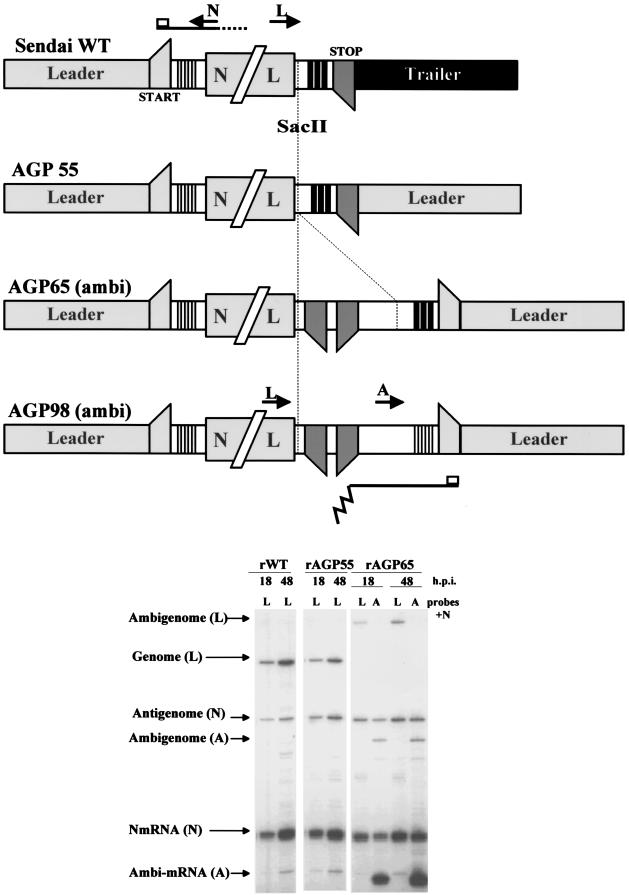
Viral RNA synthesis in wild-type and ambisense SeV infections. The wild-type, AGP55, AGP65, and AGP98 genomes are shown schematically above, not drawn to scale. The central coding region is condensed to a single N/L block, and mRNA start and stop sites are indicated with upward- and downward-pointing trapezoids. The triple bars represent the downstream element of the genomic (light shading) and antigenomic (dark shading) replication promoters (see the text). The oligonucleotide added to AGP65 that contains a poly(A)/stop site for the ambi-mRNA is indicated with a dotted line. The primers N, L, and A, used to detect the various viral RNAs, are shown as short arrows. A primer extension analysis of the RNAs present in wild-type (rWT)-, rAGP55-, and rAGP65-infected cells at 18 and 48 h p.i. is shown below. The various viral RNAs are indicated on the left, along with the relevant primer (in parentheses).
The genomic RNA of [−] RNA viruses functions first as a template for synthesis of mRNAs and then as a template for a full-length complementary copy (the [+] antigenome), which like the genome is found only as assembled nucleocapsids. Viral RNA synthesis is thought to begin at the 3′ end of the [−] genome template (nt 1) with le RNA synthesis (33). During primary transcription or when there is insufficient newly made N protein to assemble the nascent [+] le RNA, viral RNA-dependent RNA polymerase (vRdRP) stops near the end of the leader region and then starts the N mRNA (6). This vRdRP responds to the junctional signals and synthesizes each mRNA in turn. When sufficient newly made N protein is available to assemble the nascent le RNA before it has terminated, antigenome synthesis and assembly with N become coupled. This vRdRP now ignores the junctional signals and synthesizes an exact, complementary copy as a fully assembled nucleocapsid (5). Using in vitro reactions in which genome replication is initiated synchronously, it has finally been possible to provide direct evidence that antigenome synthesis and assembly with N protein take place concurrently (19). Genome synthesis from antigenome templates is thought to take place in a similar fashion, in that the antigenomic promoter (AG/Pr) (at the 3′ end of the antigenome) is also always “on,” and trailer RNAs (tr RNAs) are made from this region independent of N subunit availability (Fig. 1). However, as there are no mRNA initiation sites on the antigenome template, termination of tr RNAs serves only to recycle vRdRP. The leader and trailer sequences, which contain the end element of the bipartite paramyxovirus replication promoters (see below), are thought to promote antigenome and genome synthesis, respectively, in two ways. When present at the 3′ ends of the templates, they direct vRdRP initiation, and the complements of these sequences within the nascent [+] le and [−] tr RNAs promote the initiation of nucleocapsid assembly on these promoter-proximal chains, a prerequisite for genome replication (for a review, see reference 31).
In contrast to bisegmented arenavirus or trisegmented bunyavirus genomes that are organized in an ambisense manner (i.e., mRNAs are expressed from both genome and antigenome templates), there are no natural examples of ambisense NNV. Nevertheless, ambisense rabies viruses (RV) have been prepared by replacing the RV tr region (or AG/Pr) with a second copy of the le region (G/Pr, plus the N mRNA start and stop sites flanking the CAT open reading frame) (13). This RV (SAD Ambi-CAT) antigenome contains an active mRNA promoter whose ambisense mRNA (ambi-mRNA) terminates just before the opposing L mRNA. RV SAD Ambi-CAT infections efficiently express CAT mRNA, and the properties of this ambisense virus are instructive. First, SAD Ambi-CAT replicates similar to wild-type RV in cell culture, suggesting that it is not inherently debilitated and that ambisense NNV might be useful reagents. Also, NNV genomes are always more abundant than antigenomes intracellularly (from 4- to 50-fold among different NNV), presumably because AG/Pr is a stronger replication promoter than G/Pr. RV is an extreme example of this, where antigenomes represent but 2% of the total. However, SAD Ambi-CAT infections (and mature virions) contained equal amounts of the two. Thus, the genome/antigenome ratio for this rhabdovirus appears to be strongly determined by competition between G/Pr and AG/Pr for vRdRP. Moreover, as G/Pr is weak only when AG/Pr is also present, the initiation of genome replication is apparently not rate limiting in RV infections. In contrast, when SeV minigenome replication is reconstituted in a transfected cell system (see below), the initiation of genome replication is clearly rate limiting.
Ambisense SeV can also be used to study viral RNA synthesis in a more natural setting. SeV RNA synthesis has been studied to date either (i) in cell-free reactions (by combining N-RNA templates with cytoplasmic extracts of vaccinia virus [vTF7]-infected, plasmid-transfected cells expressing the viral N, P, and L proteins [11, 25, 29]) or (ii) by reconstituting viral RNA synthesis within vTF7-infected, plasmid-transfected cells (containing DNA copies of minigenomes as model templates) (8). Neither experimental system is ideal, for two reasons. (i) SeV RNA synthesis takes place in a cytoplasmic background strongly conditioned by the vaccinia virus infection, and the possible contribution of the 200 vaccinia virus genes to this process cannot be ruled out. (ii) Unlike natural infection, where the level of genome replication and synthesis of replicase components are interdependent, the replicase components are provided via T7 DdRP in the reconstituted systems, and their availability no longer depends on genome replication. This paper describes the preparation and characterization of ambisense SeV and their use in studying SeV RNA synthesis. During these studies, the inherent instability of ambisense SeV on passage in embryonated chicken eggs was noted.
MATERIALS AND METHODS
Construction of infectious plasmids. (i) AGP55.
To generate the plasmid pAGP55 (AGP55 indicates that the first 55 nt of AG/Pr have been replaced by the equivalent sequences of G/Pr), a DNA fragment was created by fusion PCR for placement between the SacII (end of the L gene) and BamHI (after the hepatitis delta ribozyme) restriction sites. This fusion PCR replaced the first 55 nt of the trailer sequence with 55 nt of the leader sequence (Fig. 2). The PCR fragment was subcloned into pDI E307 (9) using SacII and BamHI restriction sites. To obtain an infectious plasmid, a SphI-SacII fragment comprising the missing N, P, M, F, HN, and L genes was isolated from pFL4 and subcloned into the modified pDI E307.
(ii) AGP65 and AGP98.
The same cloning method was used to create AGP65 and AGP98 except that the fusion PCR was based on a pDI E307 derivative in which the sequence GGTACCCCAGTAAGAAAAACTTACAAAAGTAAGTTTTTCTTAATACGCGTGCGGCCGCAGCGCGCGGGTACC was inserted into the KpnI site between the end of the L open reading frame and the trailer sequence. This fragment comprises a new stop signal for the L gene, followed by a reverse poly(A)/stop signal for the ambisense gene (in bold) (Fig. 2) and then a polylinker with MluI, NotI, and BssHII restriction sites.
(iii) AGP65 ambi-edit.
A 347-bp PCR fragment from the P gene containing the editing site was amplified with MluI AGP (5′ GACGACACGCGTTCTGCATAGTTTGC) and BssHII AGP (5′ GACGCAGCGCGCCCCACCGCTGAATCG) and subcloned into the MluI and BssHII restriction sites of pAGP65. The ambi-mRNA editing site was designed to be in the same hexamer phase as that of the natural P gene editing signal (30).
(iv) AGP65/GFP.
A modified version of our infectious plasmid (pFL5) that contains a unique MluI site in the 5′ untranslated region (UTR) of the F gene was used to introduce additional transcription units into SeV. An oligonucleotide containing the N/P junction was inserted in this site such that only the upstream MluI site was retained, and green fluorescent protein (GFP) was inserted here.
VSV/GFP, which carries the transgene between the M and G genes, was provided by Jacques Perrault, San Diego, Calif. (unpublished data).
Generation of recombinant SeV (rSeV).
Briefly, one 9-cm-diameter dish of A549 cells was infected with 3 PFU of vaccinia virus TF7-3 (14) per cell and transfected 1 h later with 1.5 μg of pGEM-L, 5 μg of pGEM-N, 5 μg of pGEM-PHA (which does not express C proteins), and 15 μg of the various pAGP infectious constructs (16). Twenty-four hours later, 1-β-d-arabinofuranosylcytosine (100 μg/ml) was added to inhibit vaccinia virus replication, and after a further 24 h the cells were scraped into their medium and injected directly into the allantoic cavity of 10-day-old embryonated chicken eggs. Alternatively, BSR-T7 cells that endogenously express T7 RNA polymerase were used to recover GFP-containing viruses without vTF7-3 coinfection or drug treatment (7). Three days later, the allantoic fluids were harvested and reinjected undiluted into eggs. For further passages, the viruses were diluted 1/500 before injection. The presence of viruses was determined by pelleting allantoic fluids through a TNE (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA)-25% glycerol cushion for 20 min at 14,000 rpm in an Eppendorf centrifuge. Virus pellets were then resuspended in sample buffer, and the proteins were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue.
Analysis of viral RNAs by direct primer extension.
HeLa cell monolayers in 9-cm-diameter dishes were infected with 20 PFU of the various rSeV per cell. At 48 h postinfection (p.i.), the cells were solubilized by scraping them into 150 mM NaCl-50 mM Tris (pH 7.4)-10 mM EDTA-0.6% Nonidet P-40. Nuclei were removed by pelleting at 12,000 × g for 5 min. RNA was purified with Trizol (15596-018; Gibco BRL) and then precipitated in propanol. The equivalent RNA of 1/10 of a plate was mixed with 200,000 cpm of 5′ 32P-labeled oligonucleotides N, A, or L and extended using Moloney murine leukemia virus reverse transcriptase for 1 h at 42°C. The labeled cDNA was ethanol precipitated and resuspended in loading solution (95% formamide, 20 mM EDTA, 0.1% bromophenol blue, and xylene cyanol FF). The products were boiled for 1 min and electrophoresed on a 10% sequencing gel.
Analysis of mRNA editing by poisoned primer extension on RT-PCR product.
Extracts of infected A549 cells were prepared as above. To remove encapsidated genomes and antigenomes, cytoplasmic extracts were centrifuged in a step gradient of a 5.7 M CsCl cushion, 40% CsCl, and 20% CsCl at 35,000 rpm overnight in a Beckman centrifuge with an SW55 rotor. Pelleted mRNAs were analyzed by poisoned primer extension after reverse transcription (RT)-PCR amplification. The RT reaction used the oligonucleotide primer Mlu2 (TTTTTCTTAATACGCGTTCTG), and a 1/10 aliquot was used for PCR with 50 pM Mlu2 and PEag primer (5′-CCAGCCAACGGCCGCCC). The PCR products were purified on a 2% agarose gel and annealed to 32P-labeled SeV-edit (5′-GATGTGTTCTCTCCTATG), which hybridizes immediately downstream of the editing site. Primer extension was performed in a 10-μl volume at 37°C for 6 min with 1 U of T7 DNA polymerase (Pharmacia) in the presence of 40 μM dGTP, dTTP, and dCTP and 4 μM ddATP. Then, 300 μM deoxynucleoside triphosphate was added, and the mix was incubated an additional 2 min to chase stalled complexes. The reaction was stopped by adding 4 μl of STOP solution (95% formamide, 20 mM EDTA, 0.1% bromophenol blue, xylene cyanol FF). The products were boiled for 1 min and electrophoresed on a 12.5% sequencing gel.
Real-time PCR.
Infected cell RNAs that were free of genomes and antigenome were prepared as described above. CsCl gradient pelleted RNAs were resuspended in 20 μl of water. One-tenth of this RNA was mixed with 0.5 μg of random hexamer primers and converted to cDNA with Moloney murine leukemia virus reverse transcriptase for 1 h at 37°C. One-tenth of the cDNA was used for real-time PCR, using Perkin-Elmer TaqMan Universal Master mix and the ABI Prism 7700 sequence detector. The program followed was 95°C for 10 min and then 50 cycles of 95°C for 15 s and 60°C for 1 min. The fluorescence-labeled probe was 5′ FAM-CTTCCAGGTACCCGCGCGCTG-3′ TAMRA. The following primers were used: A (CGCGGTACCCCAGTAAGAAA), B (CTTAATACGCGTGCGGCC), C (GTCAAAGACTATTGTCATATGGACAAGTC), and D (GAAAAAACGTGTATGGAATATATAATGAAGTTATA). Several dilutions of nucleocapsid cDNA were tested to construct a standard curve, and the mathematical formula of this curve was used to obtain relative numbers for the samples.
RESULTS
rSeV-AGP55 and ambisense SeV-AGP65 and SeV-AGP98.
The cis-acting sequences that act as genomic and antigenomic replication promoters (G/Pr and AG/Pr) of the Paramyxoviridae are found in the terminal 96 nt of each segment and are bipartite in nature (23, 36, 37, 40, 44). There is both an end element of around the first 20 to 30 nt of the leader or trailer regions (in which the first 12 nt are also conserved) and a downstream element within the 5′ UTR of the N gene or the 3′ UTR of the L gene (Fig. 2). For SeV, the downstream element is a simple but phased sequence repeat (3′ [C1N2N3N4N5N6]3 as [−] RNA) imbedded in what appear to be nonconserved sequences. This simple repeat is found in the 14th, 15th, and 16th N protein subunits that are adjacent to the first two subunits in the helical nucleocapsid with 13 subunits per turn, and this common surface may serve as a recognition site for the initiation of RNA synthesis at nt 1 (36). Three ambisense viruses were initially prepared in which the first 55, 65, and 98 nt of AG/Pr were replaced with the equivalent sequences of G/Pr (AGP55, AGP65, and AGP98) (Fig. 2). Relative to SeV-wt, in SeV-AGP55 only the tr region of AG/Pr has been replaced with that of the le region, which now abuts the L gene poly(A)/stop site. SeV-AGP55 does not contain an ambi-mRNA start site. SeV-AGP65 contains the additional 10 nt of the highly conserved N mRNA start site (followed by a multicloning site and a poly(A)/stop site to terminate the ambi-mRNA) but still contains the downstream replication promoter element of AG/Pr. SeV-AGP98 has replaced this downstream promoter element as well, and both replication promoters of SeV-AGP98 should now be entirely identical. Both SeV-AGP65 and SeV-AGP98 are expected to transcribe a small ambi-mRNA that terminates just before the opposing L mRNA (Fig. 1).
SeV-AGP55, SeV-AGP65, and SeV-AGP98 (and rSeV-wt as a parallel control) were rescued from DNA in A549 cells, and stocks were grown in embryonated chicken eggs. SeV-AGP55 grew to levels in eggs similar to those for rSeV-wt, whereas SeV-AGP65 and SeV-AGP98 both consistently grew to 5- to 10-times-lower levels. To examine the relative levels of the various viral RNAs produced during infection, total (HeLa cell) RNA was isolated at 18 and 48 h p.i. and analyzed by primer extension. Three primers were used in pairs (N + L and N + A). Primer N detects the 5′ ends of the antigenome and the N mRNA, and two primers (L and A) were used to detect the 5′ end of the genome, one of which (primer A) can also be extended to the 5′ end of the ambi-mRNA (Fig. 2). The relative levels of the N and ambisense mRNAs and their respective templates are given in Table 1; part of the data is shown in Fig. 2). Several points are noteworthy.
TABLE 1.
The relative levels of genomes and antigenomes, and their mRNAs, in ambisense SeV-infected cells
| Ratio | Value ± SD (n) for cells infected witha:
|
|||
|---|---|---|---|---|
| Wt | AGP55 | AGP65/98 (early) | AGP65/98 (late) | |
| Genome/antigenome | 8.9 ± 6.6 (7) | 0.8 ± 0.2 (8) | 0.4 ± 0.2 (23) | 0.4 ± 0.1 (16) |
| N mRNA/genome | 4.2 ± 2.9 (9) | 8.1 ± 3.7 (9) | 10.4 ± 6.5 (23) | 12.0 ± 8.9 (16) |
| Ambi-mRNA/antigenome | NAb | NA | 9.8 ± 6.6 (10) | 1.2 ± 0.5 (8) |
n, number of infections analyzed. AGP65/98 (early), early-passage AGP65 and AGP98. AGP65/98 (late), late-passage AGP65 and AGP98. WT, wild type.\
NA, not applicable.
The genome/antigenome ratios of ambisense SeV.
Relative to SeV-wt infections of HeLa cells, where 8 to 10 times as many genomes as antigenomes are found intracellularly at 48 h p.i., SeV-AGP55 infections contain roughly equal levels of the two, and SeV-AGP65 and SeV-AGP98 infections consistently contain slightly more antigenomes than genomes (ca. 2.5-fold). The same ratios were found in the egg-grown virions in all cases (data not shown), as expected (28). Since SeV genomes are not being selectively exported during virion assembly (similar to RV infections [13]), their relative intracellular levels should simply reflect the frequencies with which vRdRP productively initiates at the 3′ end of each template and begins to encapsidate the nascent le or tr RNA chains (i.e., promoter strength). From the data in Table 1, we can conclude that the replacement of the tr region of AG/Pr with the le sequence alone was sufficient to equalize the two SeV-AGP55 replication promoters, i.e., to diminish AG/Pr to the level of G/Pr. The inclusion of an mRNA start site may have had a modest effect in further weakening AG/Pr relative to G/Pr, since SeV-AGP65 and SeV-AGP98 infections contain slightly more antigenomes than genomes. However, when ambi-mRNA expression is suppressed by point mutation in the ambi-mRNA start site (see below), the genome/antigenome ratios remain unaltered (Table 1). The downstream elements of the bipartite promoters also appear to be entirely equivalent with regard to promoter strength, since no differences were found between SeV-AGP65 and SeV-AGP98 (data not shown).
Since SeV-AGP55 replicates to levels similar to those for SeV-wt in several cell lines and in eggs, it would appear that G/Pr is inherently weak only in the presence of the stronger AG/Pr. Thus, as described for RV infections, competition between G/Pr and AG/Pr for SeV replicase appears to be the primary determinant of the relative amounts of genomes and antigenomes formed. However, when SeV defective interfering genome replication is studied in the vTF7-infected, plasmid-transfected cell system, the presence of the tr sequences at G/Pr increased the overall level of minigenome replication 20-fold when either internal deletion or copyback defective interfering minigenomes were examined (9). The inherent weakness of the le sequences at G/Pr in this experimental system indicates that the initiation of genome replication is rate limiting here, unlike the case with natural SeV infections. This difference may reflect the fact that in one case the replicase components (provided via T7 DdRp) are in unlimited supply independent of the level of genome replication, i.e., the system is driven towards replication by unrestricted expression of N subunits; hence, the initiation of genome replication becomes rate limiting here. The modest or negligible contribution of the N mRNA start site to further weakening G/Pr relative to AG/Pr, in contrast, agrees with results of earlier studies (9).
SeV RdRP response to cis-acting sequences in the absence of nascent chain coassembly.
The precise differences between NNV RdRPs that act as transcriptases or replicases are unknown; these polymerases are defined solely by their RNA products (mRNA and antigenome, respectively). Both types of viral RNA synthesis are thought to begin with le RNA synthesis. In the absence of concurrent assembly of the nascent le chain, SeV RdRP mostly terminates near the end of the leader region and starts the N mRNA. However, vRdRP also reads through the le/N junction at a frequency of 10 to 40% for different strains (48). These read-through vRdRP, however, are poorly processive since their transcripts (initiated at nt 1) fall off sharply with length and rarely exceed 350 nt. SeV RdRP's which have initiated the N mRNA, in contrast, finish this mRNA at >90% frequency (48). RdRP (re)initiation at nt 56 is thus associated with its conversion to a form that is processive (independent of concurrent assembly of the nascent chain). This (re)initiation also ensures that the transcript is capped and polyadenylated.
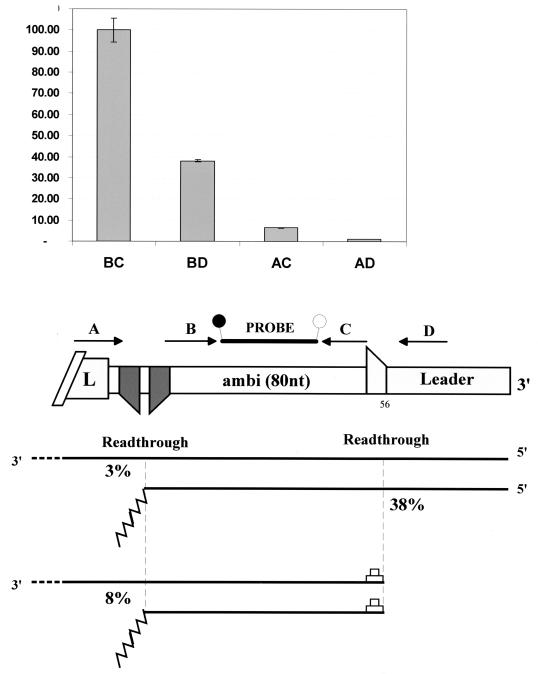
We have used ambisense SeV-AGP65 to further this analysis. We examined whether RdRP responds differently to an mRNA editing or poly(A)/stop site if it has, or has not, reinitiated at the N/ambi-mRNA start site (at the 3′ end of the antigenome). We used quantitative RT-PCR to determine the levels of SeV-AGP65 transcripts initiated at nt 1 or 56 and that had read through the ambi-mRNA poly(A)/stop site, relative to those transcripts that had not extended past the poly(A)/stop site [this ambi-mRNA is 80 nt long exclusive of the poly(A) tail]. Unencapsidated (CsCl pellet) RNAs were isolated from SeV-AGP65 infections and reverse transcribed with random hexamers as primers. This cDNA was then amplified with four primers (two within the ambi-mRNA and one on either side [Fig. 3]). Several dilutions and a standard curve were carried out to ensure that the values were reliable. The results (Fig. 3) showed that (i) viral RNA polymerase read through the antigenome le-N/ambi junction with a frequency of 38%, similar to read-through of the genome le-N junction (48), and (ii) that SeV RdRP responds efficiently to the poly(A)/stop site (>90%) independently of whether it has reinitiated at nt 56.
FIG. 3.
The effect of vRdRP reinitiation at nt 56 on vRdRP response to a poly(A)/stop site. The 3′ end of the AGP65 antigenome is shown schematically in the middle. The four primers (A, B, C, and D) and the derivatized probe (containing a fluorescence emitter and quencher; attached spheres) used in the analysis are shown above the antigenome. The possible products that have initiated either at the antigenome 3′ end or at the ambi-mRNA start site (nt 56) are shown below, along with the calculated levels of RdRP read-through of the two junctions. The relative levels of the various transcripts in three infections were determined by real-time RT-PCR (Materials and Methods) and are shown above (the level of BC was arbitrarily set at 100). The read-through frequency of the le/ambi junction was calculated as [BD]/[BC], and that of the ambi-poly(A)/stop site as [AD]/[BD] and [AC-AD]/[BC-BD] for transcripts initiated at nt 1 and 56, respectively.
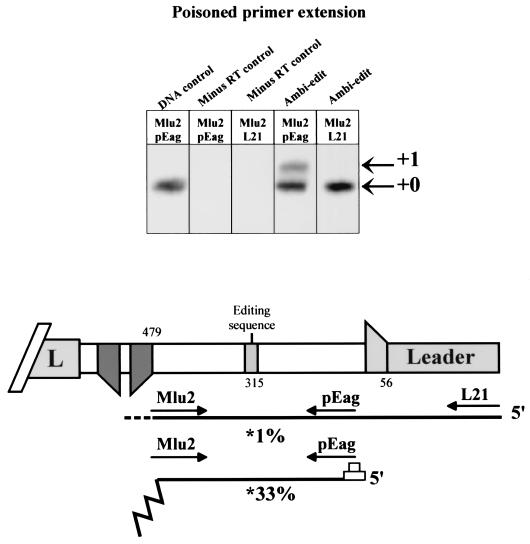
To examine whether SeV RdRP responds differently to an mRNA editing site depending on whether it has reinitiated at nt 56, a fragment of the SeV P gene containing the editing site was inserted into the ambi-mRNA of pSeV-AGP65. The editing site of SeV-AGP65/Ambi-Edit is 250 bp away from the le region in this construction. Nonencapsidated RNAs were isolated as described above and amplified by RT-PCR with two sets of primers (Mlu2 and PEag, the set which amplifies both ambi-mRNAs and le/ambi-mRNA read-through transcripts, and Mlu2 and L21, the set which amplifies only the latter) (Fig. 4). The extent of editing of each transcript was estimated by poisoned primer extension (46). As shown in Fig. 4, RNAs starting at nt 56 were edited in a manner similar to editing of bona fide P mRNAs (at a frequency of ca. 30%, and essentially only the +1G product is seen [47]), whereas the le/ambi-mRNA read-through transcripts were not edited. Thus, the ability of RdRP to respond efficiently to the mRNA editing site requires (re)initiation at nt 56, similar to the acquisition of RdRP processivity in the absence of concurrent assembly of the nascent chain.
FIG. 4.
The effect of vRdRP reinitiation at nt 56 on ambi-mRNA editing. The 3′ end of the AGP65 ambi-edit antigenome is shown schematically below, together with the position of its editing site. The transcripts initiated at either nt 1 or nt 56 and the two primer pairs used to amplify these sequences by RT-PCR are shown below. Note that only the Mlu2/L21 primer pair amplifies transcripts initiated at nt 1. The results of the poisoned primer extension analysis are shown above. Lane “DNA control” shows the results of PCR amplification directly from the infectious plasmid DNA as a negative control. Lanes “minus RT control” are shown for both primer pairs. Lanes Ambi-edit for both primer pairs show the results of RT-PCR amplification of infected CsCl pellet RNA. The results were quantified in a PhosphorImager.
Ambisense SeV instability.
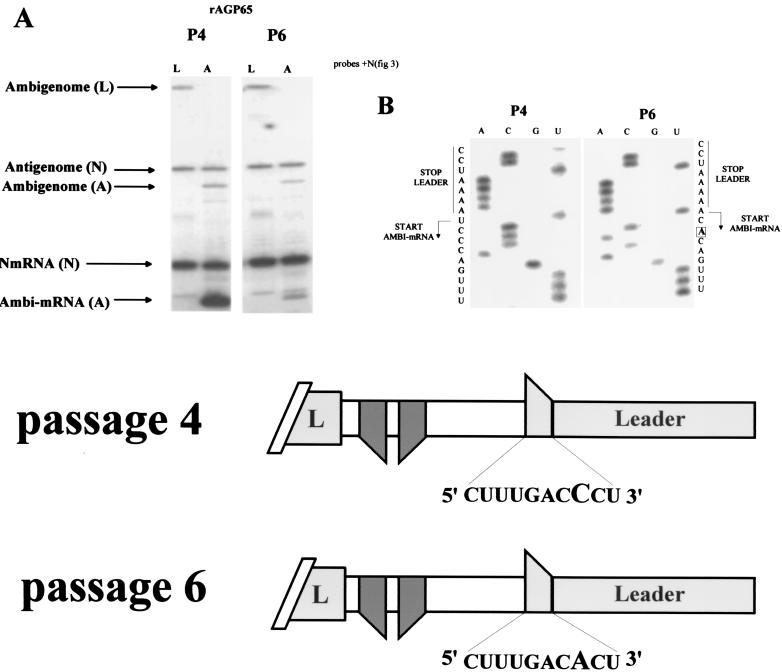
With the notable exception of SeV-AGP55, the other SeV-AGP in this study routinely grew 5- to 10-fold less well in eggs than SeV-wt. This lower-growth phenotype, however, was also routinely lost upon passage in eggs, and later-passage stocks all grew to wild-type levels (data not shown). The increased growth in eggs correlated with a strong reduction in ambi-mRNA levels in cell culture infections, whereas the levels of the genomes, antigenomes, and N mRNAs were all largely unaltered (Fig. 5 and Table 1). The AG/Pr region of six such later-passage SeV-AGP65/98 stocks was amplified by RT-PCR and sequenced. In all cases, C58, the third nucleotide (in bold) of the conserved 3′ 56UCCCANUUUC65 mRNA start signal (written 3′ to 5′ to indicate template status) was mutated to A, and this was the only change found within this region in every case. The reoccurrence of this same mutation suggests that this is the single base substitution within the conserved SeV 3′ UCCCANUUUC mRNA start signal that can most effectively eliminate ambi-mRNA synthesis. When all mRNA start sites of the Paramyxoviridae are aligned, this third nucleotide (C) is also the only one that is strictly conserved, and within the Paramyxoviridae subfamily only 3′ UNCNNNNNNN is strictly conserved (31). The C58A mutation is thus likely responsible for the loss of ambi-mRNA expression.
FIG. 5.
Regaining of “normal-growth-in-eggs” phenotype is associated with the loss of ambi-mRNA expression. (A) Primer extension analysis of the relative levels of the various viral RNAs present in HeLa cells infected with passage four and passage six allantoic fluid stocks (P4 and P6) of AGP65 (see Fig. 2 for details). (B) The ambi-mRNA promoter region of six independently derived passage four and six stocks of AGP65 or AGP98 was amplified by RT-PCR, and their sequences were compared. In all cases, the same mutation (C58A) had occurred during these passages (see the text); this mutation is highlighted below.
Ambi-mRNA expression and IFN sensitivity.
The disadvantage to virus amplification in eggs conferred by ambi-mRNA expression may be due to the increased formation of double-stranded RNA (dsRNA). Since dsRNA is a key mediator of the IFN-induced antiviral state, we examined whether SeV that express ambi-mRNA are more sensitive to IFN action than the later-passage stocks that had lost ambi-mRNA expression. Two IFN-sensitive rodent cell lines were used for this purpose; strongly IFN-competent BF (murine) cells (that both secrete IFN in response to virus infection and respond to IFN treatment) and a more poorly IFN-competent BHK cell line (BHK-LR). Although early-passage SeV-AGP65 stocks grow well in some cell lines (Fig. 2 and 5), obtaining titers by plaque formation is difficult (they form small plaques on MK2 cells [data not shown]). SeV-AGP65/GFP and SeV-wt/GFP, which express a GFP transgene, were therefore also prepared. The presence of the GFP transgene between the M and F genes was found to be neutral with respect to virus phenotype in either virus background. SeV-AGP65/GFP, in particular, also grew poorly in eggs at first and then regained wild-type growth (by P4 in this case, due to the much more efficient BSR-T7 recovery system [see Materials and Methods]). The titers of these stocks were determined by counting infectious centers under a fluorescence microscope. They were then used to infect BF and BHK-LR cells that had, or had not, been pretreated with alpha IFN (IFN-α), with 20 infectious units/cell. The accumulation of GFP intracellularly was followed by fluorescence-activated cell sorting. For reference, the cultures were also infected with SeV-wt/GFP and the related rhabdovirus vesicular stomatitis virus (VSV)/GFP, the latter as an example of a particularly IFN-sensitive virus. As shown in Fig. 6, VSV/GFP growth was strongly suppressed by IFN treatment of both cell lines and was suppressed more strongly in BF than in BHK-LR cells; GFP-positive cells decreased from 77.7 to 13.1% for BF cells and from 88.6 to 27.1% in BHK-LR cells). SeV-wt/GFP replication, in contrast, was relatively IFN insensitive in either cell line; GFP-positive cells decreased from 73.1 to only 54.7% in BF cells, and IFN treatment had an even smaller effect on BHK-LR cells (90.2 to 82.8%). The relative insensitivity of SeV-wt replication to IFN pretreatment is thought to be due to the action of its C gene, whose products dismantle the preexisting IFN-induced antiviral (VSV) state via the inhibition and turnover of Stat1 (15, 17).
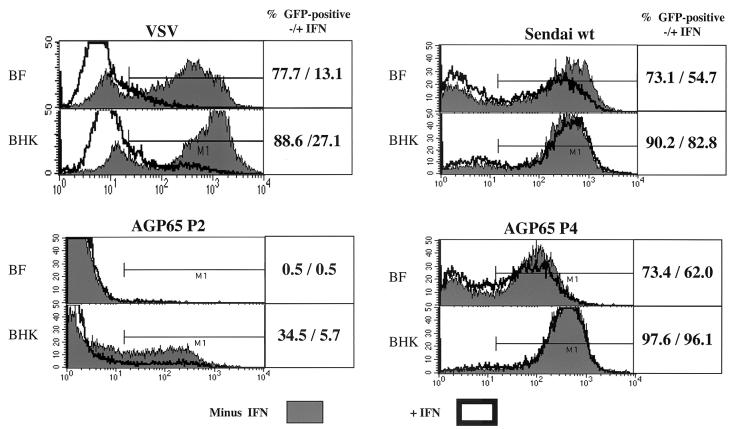
FIG. 6.
Relative IFN sensitivity of early- and later-passage SeV-AGP65/GFP. BF and BHK-LR cell cultures were treated (or not) with 1,000 U of IFN-α2/α1 (49) for 16 h and then infected with 20 infectious units/cell of either VSV/GFP, SeV-wt/GFP, or SeV-AGP65/GFP at passage level 2 (P2) or 4 (P4). The cultures were harvested at 8 p.i. (VSV) or 48 h p.i. (SeV) and analyzed directly for green fluorescence by fluorescence-activated cell sorting. The M1 bar that denotes GFP-positive cells was fixed relative to uninfected cells, and their percentage of the population (−/+ IFN) is shown in each panel.
The growth of early- and later-passage SeV-AGP65/GFP stocks in these cultures was then compared to that of the highly and poorly IFN-sensitive viruses of reference. Early-passage SeV-AGP65/GFP grew very poorly, if at all, in BF cells. This virus did grow to a limited extent in poorly IFN-competent BHK-LR cells, and this growth was relatively IFN sensitive (GFP-positive cells were reduced from 34.5 to 5.7%). The later-passage virus, in contrast, behaved more like SeV-wt/GFP; it grew well in both cell lines, and this growth was relatively IFN resistant. Thus, loss of ambi-mRNA expression during passage of the SeV-AGP65/GFP stocks in eggs is also characterized by conversion of virus that grows poorly in IFN-sensitive cultures and is relatively IFN sensitive to virus that grows well even in IFN-pretreated cells that restrict VSV/GFP replication, i.e., the wild-type SeV phenotype. These results are consistent with the notion that the disadvantage to virus amplification in eggs conferred by ambi-mRNA expression is due to the increased formation of dsRNA.
DISCUSSION
Ambisense SeV were prepared in order to study genome transcription and replication. In these studies, we found that the relative ratios of genomes/antigenomes formed during infection are largely determined by the relative strengths of the replication promoters (similar to RV infections), independent of the presence of an active mRNA start site. We also found that the ability of RdRP to respond to an mRNA editing site requires (re)initiation at nt 56, similar to the acquisition of RdRP processivity in the absence of nascent chain coassembly (48). How RdRP is imprinted for these properties by (re)initiation at nt 56 remains unclear, but work with VSV suggests that the elusive mRNA capping and methylation functions of vRdRP may be involved. The capping of the nascent VSV mRNA 5′ end when it is ca. 50 nt long may well be a checkpoint for continuing mRNA synthesis (43), and poly(A)/stop sites within 50 nt of the start site are ignored (50). We expect that SeV RdRPs that start at nt 56 will respond to the mRNA editing site only if they have already correctly modified the mRNA 5′ end and progressed a minimum distance into the gene, but this remains to be determined.
The requirement for RdRP to reinitiate at nt 56 for mRNA editing competence stands in contrast to RdRP's response to a poly(A)/stop site. This difference was unexpected, since both editing and polyadenylation occur via pseudo-templated transcription, or RdRP stuttering. However, these two events are also different in important aspects. Virtually all mRNAs have long poly(A) tails, i.e., RdRP stuttering is very extensive during polyadenylation. Moreover, mRNA termination, which is essential for initiation of each downstream gene, is apparently coupled to this extensive stuttering. The SeV editing site can be modified so that adenylates rather than guanylates are added, and insertions of up to 20 to 30 adenylates occur during this mRNA synthesis. Even in this case, however, vRdRP continues on at high frequency and does not terminate the mRNA (21). A similar situation (but in reverse) occurs when the upstream tetranucleotide (underlined) of the VSV poly(A)/stop site (3′ AUACU7) is mutated; VSV RdRP reads through the stop site but nevertheless adds a run of 15 to 20 adenylates before continuing strictly templated synthesis (4). RdRP stuttering during paramyxovirus mRNA editing, in contrast, is very limited in extent (1 to 6 Gs are added), only 30 to 70% of the mRNA (determined genetically) is modified, and RdRP then continues mRNA synthesis rather than releasing the chain (10, 39, 45, 46). The SeV poly(A)/stop site thus appears to be a stronger cis-acting element than the mRNA editing site, and this may account for why RdRP efficiently responds to this site independent of whether it has reinitiated at nt 56. Only four mutations (in bold) separate the SeV editing site (3′ GAGUUGUUUUU UCCC) and consensus poly(A)/stop site (3′ UNAUUCUUUUUGRRUCCC; the stutter sites are underlined). These mutations within SeV-AGP65/ambi-edit convert this editing site into an efficient poly(A) site (data not shown). Separation of the U and C runs of the editing site by the GRR insertion is sufficient to destroy editing, but the three upstream substitutions are required as well for efficient polyadenylation. This conversion highlights the very compact nature of these cis-acting sequences and suggests that both sites are organized similarly, with a short downstream “slippery sequence” (U5 or U6C3) and an equally short region upstream that controls the extent of vRdRP stuttering (21, 22).
Early-passage SeV-AGP65 stocks grow poorly in IFN-sensitive cultures, and loss of ambi-mRNA expression during passage in eggs is associated with virus conversion to a wild-type IFN-resistant phenotype (Fig. 6). Thus, although we have not directly shown that dsRNA forms intracellularly due to ambi-mRNA expression, the expected hallmark of its presence (increased IFN sensitivity) is clearly evident. The increased formation of dsRNA may help explain why there are no natural examples of ambisense NNV, but it also raises the question of how the related ambisense RNA viruses have dealt with this dilemma. All natural examples of ambisense genomes are found among segmented [−] RNA viruses, such as bisegmented arenaviruses (where both segments are always ambisense) (41) and trisegmented bunyaviruses (where some of the segments are sometimes ambisense) (42). These segmented RNA viruses differ from NNV in that their single mRNA/template initiates at the very end of each nucleocapsid segment (via a primer-dependent “cap snatching” mechanism); there are no le regions here. For segmented [−] RNA viruses, such as influenza viruses, that never contain ambisense organization, mRNAs are also initiated by cap snatching, but influenza mRNAs are polyadenylated by RdRP stuttering on a short template U run, similar to NNV (32). Arenavirus and bunyavirus mRNAs, in contrast, are not polyadenylated (26, 38). For those bunyavirus segments that are simply [−] sense, mRNAs terminate ca. 100 nt before the 5′ end of the [−] genome, but there are no clues as yet to how this might occur. For ambisense bunyavirus and arenavirus segments, however, the mRNA 3′ ends are predicted to contain strong stem-loop secondary structures (2, 12). These structures may act in part to terminate mRNA synthesis from ambisense segments (Fig. 1) since they play a central role in the one mechanism of mRNA termination that is well established, namely, intrinsic termination in bacteria (20). For Escherichia coli DdRP, the formation of a nascent chain secondary structure by intrinsic termination sites within the RNA exit channel (during chain elongation) is thought to pry open the flaps that tether E. coli DdRp to its template and to simultaneously strip the 3′ end of the nascent RNA from the template (35). A similar termination mechanism appears well adapted to ambisense animal viruses where there is but a single mRNA/nucleocapsid. Termination based on secondary structure of the nascent chain 3′ end could operate at a very high frequency, mRNA termination and release of vRdRP would occur simultaneously, and the potential for dsRNA formation during infection could be reduced to a minimum (Fig. 1).
An intrinsic terminator mechanism, however, is clearly unsuitable for NNV, where there are 5 to 10 mRNAs/genome (Fig. 1). The NNV mRNA expression strategy requires that their RdRPs not detach from the template upon mRNA termination; rather, they scan the template for the next mRNA start site. Another feature of NNV is that mRNA termination does not always operate at maximum efficiency. Dicistronic mRNAs due to RdRP read-through of a poly(A)/stop site are frequent in some paramyxovirus infections, and this RdRP read-through is thought to play a role in modulating the infection by specifically silencing expression of the second cistron (reviewed in reference 31). SeV expresses relatively little dicistronic mRNA (up to 5%), but even this modest level of RdRP read-through has important consequences in the case of the ambi-mRNA. Read-through of a recombinant NNV ambi-mRNA stop site generates a capped transcript that can potentially extend the entire length of the antigenome, as there are no mRNA stop sites on the NNV antigenome (Fig. 1). Moreover, even though only ∼5% of SeV RdRP reads through the ambi-mRNA stop site, the ambi-mRNA is initiated >20 times as frequently as the L mRNA (24), generating ample amounts of stable, complementary mRNAs. The consequence of this read-through is thus a strong potential to form dsRNA. dsRNA is thought to activate cellular kinases (including constitutively expressed PKR) that stimulate several transcription factors that promote IFN expression, such as IFN regulatory factors, NF-κB, and activating transcription factor 2/c-Jun (34). It is also an essential cofactor for some IFN-stimulated genes whose products are important components of the antiviral state (such as PKR and 2-5 oligo-adenylate synthetase), and dsRNA thereby also acts as a general indicator of the level of virus infection for the cellular innate immune system (18). We presume that this is why the adaptation of AGP65 and AGP98 for growth in the developing chicken embryo is associated with the loss of ambi-mRNA synthesis.
Acknowledgments
This work was supported by a grant from the Swiss National Science Fund.
REFERENCES
- 1.Abraham, G., and A. K. Banerjee. 1976. Sequential transcription of the genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 73:1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auperin, D. D., M. Galinski, and D. H. Bishop. 1984. The sequences of the N protein gene and intergenic region of the S RNA of pichinde arenavirus. Virology 134:208-219. [DOI] [PubMed] [Google Scholar]
- 3.Ball, L. A., and C. N. White. 1976. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 73:442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, J. N., and G. W. Wertz. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J. Virol. 75:6901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumberg, B. M., C. Giorgi, and D. Kolakofsky. 1983. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell 32:559-567. [DOI] [PubMed] [Google Scholar]
- 6.Blumberg, B. M., M. Leppert, and D. Kolakofsky. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23:837-845. [DOI] [PubMed] [Google Scholar]
- 7.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calain, P., and L. Roux. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 67:4822-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calain, P., and L. Roux. 1995. Functional characterisation of the genomic and antigenomic promoter of the Sendai virus. Virology 211:163-173. [DOI] [PubMed] [Google Scholar]
- 10.Cattaneo, R., K. Kaelin, K. Baczko, and M. A. Billeter. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759-764. [DOI] [PubMed] [Google Scholar]
- 11.Curran, J., J.-B. Marq, and D. Kolakofsky. 1992. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology 189:647-656. [DOI] [PubMed] [Google Scholar]
- 12.De Haan, P., L. Wagemakers, D. Peters, and R. Goldbach. 1990. The S RNA segment of tomato spotted wilt virus has an ambisense character. J. Gen. Virol. 71:1001-1007. [DOI] [PubMed] [Google Scholar]
- 13.Finke, S., and K.-K. Conzelmann. 1997. Ambisense gene expression from recombinant rabies virus: random packaging of positive- and negative-strand ribonucleoprotein complexes into rabies virions. J. Virol. 71:7281-7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin, D., J. Curran, M. Itoh, and D. Kolakofsky. 2001. Longer and shorter forms of Sendai virus C proteins play different roles in modulating the cellular antiviral response. J. Virol. 75:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcin, D., T. Pelet, P. Calain, L. Roux, J. Curran, and D. Kolakofsky. 1995. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 14:6087-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcin, D., J. B. Marq, L. Strahle, P. Le Mercier, and D. Kolakofsky. All four Sendai virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology, in press. [DOI] [PubMed]
- 18.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 19.Gubbay, O., J. Curran, and D. Kolakofsky. 2001. Sendai virus genome synthesis and assembly are coupled: a possible mechanism to promote viral RNA polymerase processivity. J. Gen. Virol. 82:2895-2903. [DOI] [PubMed] [Google Scholar]
- 20.Gusarov, I., and E. Nudler. 2001. Control of intrinsic transcription termination by N and NusA. The basic mechanisms. Cell 107:437-449. [DOI] [PubMed] [Google Scholar]
- 21.Hausmann, S., D. Garcin, C. Delenda, and D. Kolakofsky. 1999. The versatility of paramyxovirus RNA polymerase stuttering. J. Virol. 73:5568-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hausmann, S., D. Garcin, A. S. Morel, and D. Kolakofsky. 1999. Two nucleotides immediately upstream of the essential A6G3 slippery sequence modulate the pattern of G insertions during Sendai virus mRNA editing. J. Virol. 73:343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman, M. A., and A. K. Banerjee. 2000. Precise mapping of the replication and transcription promoters of human parainfluenza virus type 3. Virology 269:201-211. [DOI] [PubMed] [Google Scholar]
- 24.Homann, H. E., P. H. Hofschneider, and W. J. Neubert. 1990. Sendai virus gene expression in lytically and persistently infected cells. Virology 177:131-140. [DOI] [PubMed] [Google Scholar]
- 25.Horikami, S. M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iapalucci, S., N. Lopez, and M. T. Franze-Fernandez. 1991. The 3′ end termini of the Tacaribe arenavirus subgenomic RNAs. Virology 182:269-278. [DOI] [PubMed] [Google Scholar]
- 27.Iverson, L. E., and J. K. Rose. 1982. Sequential synthesis of 5′-proximal vesicular stomatitis virus mRNA sequences. J. Virol. 44:356-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolakofsky, D., and A. Bruschi. 1975. Antigenome in Sendai virions and Sendai virus-infected cells. Virology 66:185-191. [DOI] [PubMed] [Google Scholar]
- 29.Kolakofsky, D., J. Curran, T. Pelet, and J. P. Jacques. 1993. Paramyxovirus P gene mRNA editing, p. 105-123. In R. Benne (ed.), RNA editing. Ellis Horwood, London, England.
- 30.Kolakofsky, D., T. Pelet, D. Garcin, S. Hausmann, J. Curran, and L. Roux. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J. Virol. 72:891-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 32.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1532. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 33.Leppert, M., L. Rittenhouse, J. Perrault, D. F. Summers, and D. Kolakofsky. 1979. Plus and minus strand leader RNAs in negative strand virus-infected cells. Cell 18:735-747. [DOI] [PubMed] [Google Scholar]
- 34.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 35.Mooney, R. A., and R. Landick. 1999. RNA polymerase unveiled. Cell 98:687-690. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, S. K., Y. Ito, and G. D. Parks. 1998. A functional antigenomic promoter for the paramyxovirus Simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J. Virol. 72:10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy, S. K., and G. D. Parks. 1999. RNA replication for the paramyxovirus simian virus 5 requires an internal repeated (CGNNNN) sequence motif. J. Virol. 73:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattnaik, A. K., and G. Abraham. 1983. Identification of four complementary RNA species in Akabane virus-infected cells. J. Virol. 47:452-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelet, T., J. Curran, and D. Kolakofsky. 1991. The P gene of bovine parainfluenza virus 3 expresses all three reading frames from a single mRNA editing site. EMBO J. 10:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pelet, T., C. Delenda, O. Gubbay, D. Garcin, and D. Kolakofsky. 1996. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology 224:405-414. [DOI] [PubMed] [Google Scholar]
- 41.Salvato, M. S. 1993. Molecular biology of the prototype arenavirus, lymphocytic choriomeningitis virus, p. 133-156. In M. S. Salvato (ed.), The arenaviruses. Plenum Press, New York, N.Y.
- 42.Schmaljohn, C. S., and J. W. Hooper. 2001. Bunyaviridae: the viruses and their replication, p. 1581-1633. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 43.Stillman, E. A., and M. A. Whitt. 1999. Transcript initiation and 5′-end modifications are separable events during vesicular stomatitis virus transcription. J. Virol. 73:7199-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tapparel, C., D. Maurice, and L. Roux. 1998. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif, (GNNNNN)3, is essential for replication. J. Virol. 72:3117-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas, S. M., R. A. Lamb, and R. G. Paterson. 1988. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the paramyxovirus SV5. Cell 54:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vidal, S., J. Curran, and D. Kolakofsky. 1990. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 9:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vidal, S., J. Curran, and D. Kolakofsky. 1990. Editing of the Sendai virus P/C mRNA by G insertion occurs during mRNA synthesis via a virus-encoded activity. J. Virol. 64:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vidal, S., and D. Kolakofsky. 1989. Modified model for the switch from Sendai virus transcription to replication. J. Virol. 63:1951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber, H., D. Valenzuela, G. Lujber, M. Gubler, and C. Weissmann. 1987. Single amino acid changes that render human IFN-alpha 2 biologically active on mouse cells. EMBO J. 6:591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whelan, S. P., J. N. Barr, and G. W. Wertz. 2000. Identification of a minimal size requirement for termination of vesicular stomatitis virus mRNA: implications for the mechanism of transcription. J. Virol. 74:8268-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]