Abstract
Cobalamin in the form of adenosylcobalamin (Ado-CBL) is known to repress expression of genes for vitamin B12 biosynthesis and be transported by a posttranscriptional regulatory mechanism, which involves direct binding of Ado-CBL to 5′untranslated gene regions (5′UTR). Using comparative analysis of genes and regulatory regions, we identified a highly conserved RNA structure, the B12-element, which is widely distributed in 5′UTRs of vitamin B12-related genes in eubacteria. Multiple alignment of approximately 200 B12-elements from 66 bacterial genomes reveals their common secondary structure and several extended regions of sequence conservation, including the previously known B12-box motif. In analogy to the model of regulation of the riboflavin and thiamin biosynthesis, we suggest Ado-CBL-mediated regulation based on formation of alternative RNA structures including the B12-element. In Gram-negative proteobacteria, as well as in cyanobacteria, actinobacteria, and the CFB group, the cobalamin biosynthesis and vitamin B12 transport genes are predicted to be regulated by inhibition of translation initiation, whereas in the Bacillus/Clostridium group of Gram-positive bacteria, these genes seem to be regulated by transcriptional antitermination. Phylogenetic analysis of the B12-elements reveals a large number of likely duplications of B12-elements in several bacterial genomes. These lineage-specific duplications of RNA regulatory elements seem to be a major evolutionary mechanism for expansion of the vitamin B12 regulon.
Keywords: Bacteria, comparative genomics, regulatory RNA, B12-element, cobalamin
INTRODUCTION
Synthesized only by prokaryotic organisms, vitamin B12 or cobalamin (CBL) is an essential cofactor for several important enzymes that catalyze a variety of transmethylation and rearrangement reactions (Martens et al. 2002). Expression of the Salmonella typhimurium cob operon, encoding the CBL biosynthetic pathway, and of the btuB gene of Escherichia coli and S. typhimurium, encoding the vitamin B12 transporter, is repressed by addition of vitamin B12 by a post-transciptional regulatory mechanism (Lundrigan et al. 1991; Richter-Dahlfors and Andersson 1992). As shown by deletion analysis, this regulation requires unusually long 5′-untranslated leader sequences of the corresponding mRNAs, which contain several conserved elements. The leader mRNAs of the cob and btuB genes contain an evolutionarily conserved sequence known as the B12-box (Franklund and Kadner 1997). Translational regulation of these genes requires also a conserved RNA hairpin that would mask the ribosome-binding site (RBS), thus inhibiting translation initiation and gene expression (Richter-Dahlfors et al. 1994; Nou and Kadner 1998). In addition, a cis-acting translational enhancer element in the cob leader mRNA is absolutely required to unfold the inhibitory RBS hairpin in the absence of coenzyme B12 (Ravnum and Andersson 2001). In the same work, a secondary structure model for the cob leader mRNA was obtained from the chemical probing expriments with single-stranded RNA-modifying agents and combined with the output of a RNA folding computer program. No cobalamin-regulatory genes were identified in bacteria, but it was shown that adenosylcobalamin (Ado-CBL) is an effector molecule involved in the regulation of CBL genes (Nou and Kadner 2000). Recently, the structure-dependent spontaneous cleavage of RNA technique was applied to the E. coli btuB leader sequence in the presence and absence of Ado-CBL, and it was shown that this sequence fragment can directly bind Ado-CBL, consequently undergoing conformational changes in the secondary and tertiary structure of the RNA (Nahvi et al. 2002). The investigators suggested that the mechanism of the btuB regulation involves formation of two alternative RNA structures, repressing and antirepressing, in the presence and absence of Ado-CBL, respectively.
The comparative analysis is a powerful approach to identification of regulatory patterns in bacterial genomes (Gelfand et al. 2000). Comparative sequence analysis was used for prediction of conserved RNA secondary structures (Eddy and Durbin 1994; Marck and Grosjean 2002) and detection of novel regulatory RNA elements, for instance, iron-responsive elements in E. coli (Dandekar et al. 1998) or S-boxes in Gram-positive bacteria (Grundy and Henkin 1998). In such studies, analysis of complementary substitutions in aligned sequences is used to construct a single conserved structure. The number of known noncoding RNA families is expanding rapidly. This resulted in development of a number of specialized databases, in particular the RNA family database Rfam (Griffiths-Jones et al. 2003), the database of known RNA structures RNABase (Murthy and Rose 2003), and the noncoding RNAs database (Szymanski et al. 2003). Current availability of multiple complete genomes provides an opportunity to identify consensus RNA elements upstream of co-regulated genes. In particular, highly conserved RFN and THI elements were identified in the regulatory regions of genes involved in the biosynthesis of two different vitamins, riboflavin and thiamin, respectively (Vitreschak et al. 2002; Rodionov et al. 2002). Recently it was confirmed that these RNA elements control expression of the target genes through a post-transciptional regulatory mechanism (Mironov et al. 2002; Winkler et al. 2002a,b).
Here we applied the comparative approach to analysis of 5′-untranslated regions (5′UTRs) of vitamin B12-related genes in ~100 prokaryotic genomes. We report identification of a novel conserved RNA element involved in regulation of B12-related genes in eubacteria. This new 5′UTR regulatory RNA, named the B12-element, is highly conserved on the sequence and structural levels and includes the previously defined B12-box motif. The B12-elements are widely distributed in bacteria: ~200 elements were identified in 5′UTRs of B12-related genes in 67 bacterial genomes. The common structure of the B12-element was inferred and a possible mechanism of the B12-element-mediated regulation involving either transcriptional or translational attenuation was proposed for different groups of bacteria.
RESULTS AND DISCUSSION
Conserved structure of the B12-element
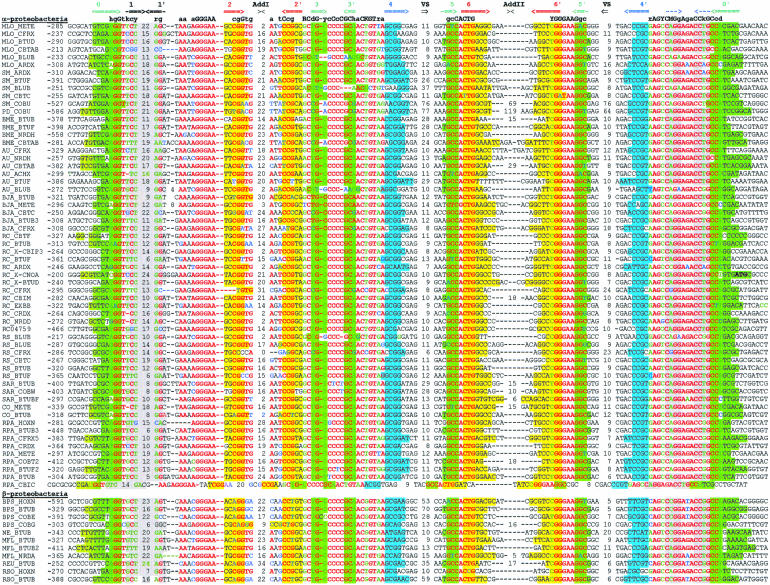
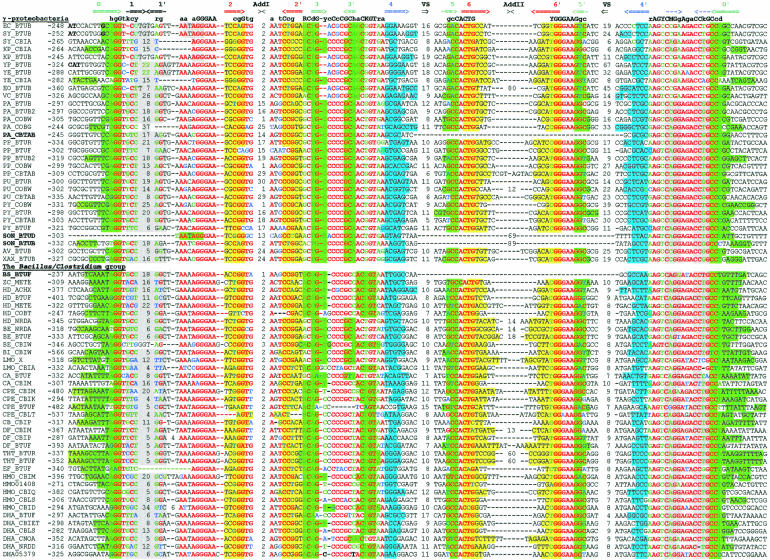
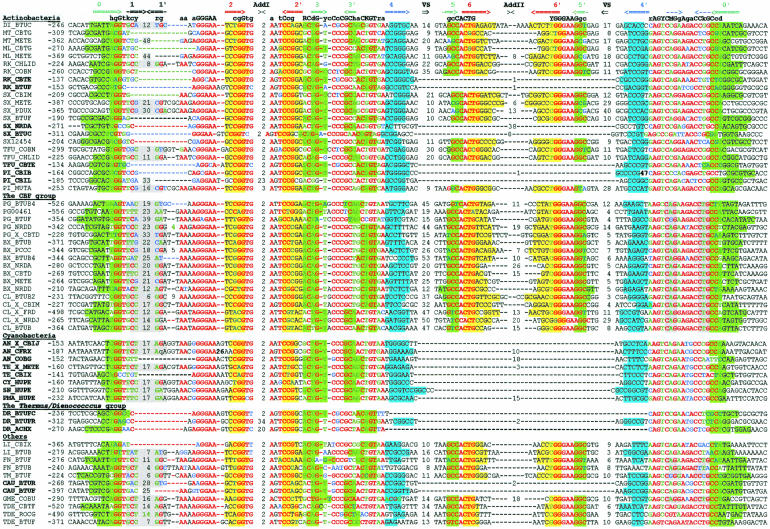
The btuB genes of E. coli and S. typhimurium have extensive regulatory regions including the conserved B12-box sequence (Ravnum and Andersson 1997; Nou and Kadner 2000). We started with identification of orthologs of btuB in related bacteria. The upstream regions of btuB orthologs were aligned by the RNAMultAln program and the conserved RNA secondary structure was identified. This novel RNA structure, named the B12-element, consist of a number of helices and conserved sequence motifs, including the known B12-box. We constructed a pattern, which corresponded to the identified B12-element and scanned 107 genomic sequences using the RNA-PATTERN program. As a result, we found ~200 B12-elements in 67 bacterial genomes. Multiple alignment of these elements is shown in Figure 1 ▶. The B12-element is widely distributed in eubacteria, but it has not been observed in Archaea and Eukaryota. Most eubacterial genomes, containing the CBL biosynthesis or transport genes, have 1 to 13 B12-elements. The distribution of the elements is different in various taxonomic groups. On the average, Gram-negative α-proteobacteria have five B12-elements per genome, γ- and β-proteobacteria have two elements, Gram-positive bacteria have three elements, and cyanobacteria have only one B12-element per genome. All of these elements are located upstream of B12-related genes. The detailed functional, positional, and phylogenetic analysis of the CBL genes is submitted elsewhere (D.A. Rodionov, A.G. Vitreschak, A.A. Mironov, and M.S. Gelfand, in prep.).
FIGURE 1.
Alignment of B12-element sequences. The first column contains the genome abbreviations (listed in Table 3 ▶) and the names of the proximal downstream genes. The second column contains the position of the first base relative to the start codon. The complementary stems of the RNA secondary structure are shown by arrows in the upper line. Base-paired positions are highlighted in matching colors. Conserved positions are set in red; degenerate conserved positions, in green; nonconserved positions, in black; nonconsensus nucleotides in conserved positions, in blue. The lengths of the additional and variable stem–loops are given.
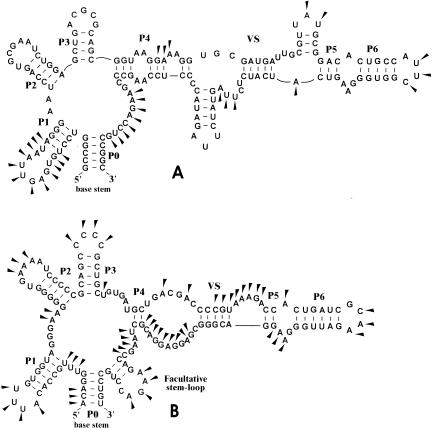
Previously, we described highly conserved 5′UTR structures, namely the RFN and THI elements, involved in regulation of riboflavin- and thiamin-related genes, respectively (Rodionov et al. 2002; Vitreschak et al. 2002). Similarly to these elements, the B12-element has a set of helices closed by a single base stem, and regions of high sequence conservation that are distributed along the entire element (Fig. 2 ▶). The conserved core of B12-element consists of seven helices (P0 to P6) and single-stranded regions with a high degree of sequence conservation. Existence of the conserved helices is confirmed by compensatory substitutions in base-paired positions. In addition to the conserved core, the B12-element has a number of facultative nonconserved stem-loops, designated Add-I and Add-II, and one internal variable structure, named VS. The previously defined B12-box is situated within the long internal loop between P0 and P4, slightly overlapping the base stem P0. In a number of cases a part of the B12 box can fold into a facultative stem-loop structure (Fig. 2 ▶). Other identified conserved regions of the B12-element are located upstream of the B12-box. The first one stretches from P1 to P4 and contains an AG-rich internal loop between P1 and P2 with consensus 5′-AAN AGGGAA-3′. Interestingly, AG-rich conserved regions were also observed in internal loops of the RFN and THI elements. Two other conserved regions, with consensi 5′-GC CACTG-3′ and 5′-YGGGAAGGC-3′, are located between P5 and P6, partially overlapping these helices.
FIGURE 2.
Conserved structure of the B12-element. Capital letters indicate invariant positions. Lowercase letters indicate strongly conserved positions. Degenerate positions: R = A or G; Y = C or U; K = G or U; M = A or C; H = not G; D = not C; N = any nucleotide. Dashes and black dots indicate obligatory and facultative base pairs, respectively. The conserved helices are numbered P0 to P6. Stem–loops of variable and constant lengths are shown by broken and sold lines, respectively. Additional stem–loops are designated Add-I and Add-II. The sequence of the conserved B12-box motif is shaded in gray. The variable structure VS separates two conserved parts of the B12-element, BI and BII. Taxonomic variations of the VS topology are shown in the inset.
Beside the conserved core structure, the B12-element includes additional nonconserved stem-loops, Add-I and Add-II. The former seems to be correlated with phylogeny, as it occurs only in genomes of proteobacteria. In contrast, the presence of the latter stem-loop does not depend on the phylogenetic position of a genome and occurs in <10% of cases. The maximal observed lengths of the Add-I and Add-II stem-loops are 34 and 122 nucleotides, respectively.
The B12-elements can be classified into two major types based on the existence of a highly conserved stem-loop region, named BII. This part of the B12-element includes conserved regions, 5′-GCCACTG-3′ and 5′-YGGGAAGGC-3′, which partially overlap the P5 and P6 helices (Fig. 2 ▶). Although most B12-elements are complete, the BII part is absent in a number of genomes: in all B12-elements of cyanobacteria, bacteria Deinococcus radiodurans, Bacillus subtilis, Shewanella oneidensis, Chloroflexus auranatiacus, as well as in some B12-elements of actinobacteria and Pseudomonas aeruginosa (Table 1 ▶). Short B12-elements without the BII part were found upstream of some B12-related genes and, therefore, are functional. In complete B12-elements, the BII part is separated from other conserved parts of the B12-element by a variable linker, which can fold into a nonconserved variable structure, named VS. This structure has different topologies in various taxonomic groups of bacteria. For example, the VS structures are represented by one helix in bacteria from the Bacillus/Clostridium group, whereas VS of γ-proteobacteria consist of two adjacent helices. In other cases the VS structure is more complex (see the lower frame in Fig. 2 ▶). Thus, the additional highly conserved BII part can play either an auxiliary role in the function of the B12-element or perform some other function.
TABLE 1.
Phylogenetic distribution of B12-elements in bacteria
| Type of B12-elements | |||||
| Taxonomic group | Genomes with B12-elements | Number of B12-elements | complete | without BII | Proposed type of regulation |
| α-proteobacteria | 11 | 58 | all | no | Translational |
| β-proteobacteria | 5 | 11 | all | no | Translational |
| γ-proteobacteria | 14 | 31 | yes | 3 | Translational |
| δ-proteobacteria (GME) | 1 | 1 | all | no | Translational/transcriptional |
| The Bacillus/Clostridium group | 14 | 37 | yes | 1 | Transcriptional |
| Actinobacteria | 7 | 22 | yes | 6 | Translational |
| Cyanobacteria | 5 | 8 | no | all | Translational |
| The Thermus/Deinococcus group | 1 | 3 | no | all | Translational |
| The CFB group (BX, PG, CL) | 3 | 17 | all | no | Translational |
| Spirochetes (LI, TDE) | 2 | 5 | all | no | ? |
| Chloroflexaceae (CAU) | 1 | 2 | no | all | Translational |
| Fusobacteriaceae (FN) | 1 | 2 | all | no | Transcriptional |
| Thermotogales (TM) | 1 | 1 | all | no | Transcriptional |
| Total | 66 | 198 | 174 | 23 | Transcriptional ~20%; translational ~80% |
The previously proposed secondary structures for the cbiA (Ravnum and Andersson 2001) and btuB (Nahvi et al. 2002) leader mRNAs from enterobacteria differ in some details in their topology. Here we propose a conserved secondary structure for 5′UTRs of B12-regulated genes. Similarly to the riboflavin- and thiamin-specific 5′UTR regulatory RNAs, RFN and THI elements, this new B12-specific RNA has a compact secondary structure, consisting of a set of conserved helices closed by a single base stem. The previously proposed RNA secondary structure of the cbiA regulatory region (Ravnum and Andersson 2001) is not compact, although a number of helices in both structures coincide (namely, P1, P2, P3, and P6). Moreover, despite of compactness of the predicted secondary structure of the btuB regulatory region (Nahvi et al. 2002) and similarity in topology of both structures, this structure differs from the B12-element in regions of base pairing of the base stem. However, the suggested conserved structure of the B12-element is mostly consistent with the chemical probing data for the btuB and cbiA leader regions (Fig. 3 ▶).
FIGURE 3.
Predicted RNA secondary structure of the B12-elements upstream of btuB from E. coli and cbiA from S. typhimurium genes. Chemical modifications of the B12-loop region of the btuB (A) and cbiA (B) 5′UTRs are shown by filled triangles (Ravnum and Andersson 2001; Nahvi et al. 2002).
Regulation of transcription/translation mediated by B12-element
Analysis of the leader sequences of B12-regulated genes allowed us to predict additional RNA regulatory hairpins downstream of all B12-elements. In some cases these additional hairpins are followed by runs of thymidines and therefore are candidate ρ-independent terminators of transcription. In other cases the hairpins overlap the ribosome-binding site (RBS) of the first gene in the B12-regulated operon and are candidate translational sequestors that prevent ribosome binding to the RBS. Previously, it has been shown that RBS-sequestering hairpins are involved in the regulation of translation of the btuB gene of E. coli and S. typhimurium, as well as the cbiA gene of S. typhimurium. (Ravnum and Andersson 1997; Nou and Kadner 2000). Most Gram-positive bacteria from the Bacillus/Clostridium group, Chloroflexus aurantiacus, Fusobacterium nucleatum, and Termotoga maritima have a candidate terminator hairpin, whereas most Gram-negative bacteria (proteobacteria and the CFB group), as well as actinomycetes, cyanobacteria, and D. radiodurans have candidate RBS-sequestering hairpins downstream of B12-elements. Therefore, the regulation of B12-related genes is likely to operate mainly at the level of transcription in the former group of bacteria and at the level of translation in the latter group (Table 1 ▶). Thus, the phylogenetic distribution of the proposed terminators and sequestors in Gram-positive and Gram-negative bacteria, respectively, is similar to previously observed distribution of the regulatory hairpins in the riboflavin and thiamin regulons (Vitreschak et al. 2002; Rodionov et al. 2002). Analysis of the 5′UTR of the cobalamin biosynthetic operon from Geobacter metallireducens reveals two possibilities of regulation. In this case the predicted terminator hairpin overlaps the RBS sequence of the first gene in the operon, and therefore, it can function as both a terminator and a sequestor.
The mechanism of the Ado-CBL-dependent regulation involves formation of two alternative RNA conformations. It has been shown in experiments that the RBS sequestor of the S. typhimurium cbi operon forms in the presence of Ado-CBL, leading to inhibition of the translation initiation, whereas the antisequestor conformation formed in the absence of Ado-CBL opens the RBS sequence and allows translation to initialize (Ravnum and Andersson 2001).
Previously we proposed the riboswitch mechanism involving the RFN and THI elements. These elements are stabilized by effector molecules (flavin mononucleotide or thiamin pyrophosphate, respectively), allowing formation of downstream regulatory hairpins (transcriptional terminator or a hairpin sequestering the RBS site) that leads to repression of transcription or translation, respectively. In the absence of the effector, a more energetically favorable RNA structure that is alternative to parts of the vitamin-specific element and the regulatory hairpin can fold, thus releasing gene expression (Rodionov et al. 2002; Vitreschak et al. 2002). This riboswitch mechanism was experimentally confirmed for thiamin and riboflavin regulons (Mironov et al. 2002; Winkler et al. 2002a,b). In analogy with the riboflavin- and thiamin-specific riboswitches, we propose here the cobalamin-specific riboswitch.
It seems to have some specific properties. It has been experimentally shown that an additional regulatory element is located between the B12-element and the sequestor hairpin of the cbiA gene of S. typhimurium (Ravnum and Andersson 2001). This element, a translational enhancer, plays the key role in the regulation of translation initiation of cbiA. In the absence of cobalamin it interacts with the region, corresponding to the left stem of the RBS sequestor, and thus releases the translation initiation of cbiA. Moreover, it has been suggested (Ravnum and Andersson 2001) that in the presence of cobalamin, the enhancer element interacts with the B12-element, thus the RBS sequestor forms and repression the translation initiation is repressed.
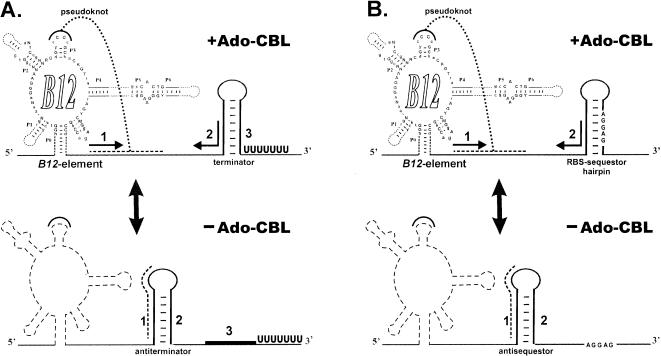
To elucidate the mechanism of regulation, we analyzed multiple alignments of upstream regions of close ortologs of btuB genes from enterobacteria and some α-proteobacteria. It turned out that these alignments contained islands of conservation, corresponding to B12-elements, sequestor hairpins, and some additional elements in between (Fig. 4 ▶). The first additional conserved element is complementary to the left stem of the RBS sequestor. Thus, it can form an antisequestor structure whose possible role is similar to the translational enhancer. Another conserved region can form a pseudoknot by interacting with the loop region of the P3 helix of the B12-element. Similar additional elements in 5′UTRs of B12-regulated genes were found in most Gram-positive and Gram-negative bacteria (Fig. 5 ▶). In some cases the pseudoknot element overlaps the left base stem of the antisequestor/antiterminator, in other cases (including the cases shown in Fig. 4 ▶), it is located within the antisequestor/antiterminator. Thus, the predicted mechanism of the cobalamin riboswitch differs from the thiamin and riboflavin riboswitches, as it involves formation of the pseudoknot (Fig. 6 ▶). The B12-element stabilized by Ado-CBL interacts with the antisequestor/antiterminator region, forms the pseudoknot, and promotes the sequestor formation. In the absence of Ado-CBL, the nonstabilized B12-element cannot form the pseudoknot and the more favorable of antisequestor/antiterminator structure folds, thus allowing for the translation initiation or transcriptional readthrough.
FIGURE 4.
Multiple alignment of the btuB upstream regions from enterobacteria (A) and some α-proteobacteria (B). Gray background denotes the P0 and P3 stems of the B12-element and the proposed RBS sequestors. Bold black denotes the main stem of the antisequestor. Arrows show the complementary stems of RNA secondary structures. Candidate pseudoknots are underlined and designated Pkn. RBSs and start codons are shown in bold letters.
FIGURE 5.
Conserved RNA elements upstream of some B12-regulated genes. The P0 and P3 stems of the B12-element are highlighted in green. Proposed regulatory hairpins (sequestors in proteobacteria and actinobacteria; terminators in the Bacillus/Clostridium group) are highlighted in blue. Red denotes the base stem of the antiterminator/antisequestor. Arrows in the upper line show the complementary stems of RNA secondary structures. Candidate pseudoknots that overlap the P3 loop and the antiterminator/antisequestor are underlined. RBSs and the start codons are set in blue.
FIGURE 6.
Predicted mechanism of the B12-mediated regulation of CBL genes: (A) transcriptional attenuation; (B) translational attenuation (inhibition of translation initiation).
Phylogeny of the B12-elements
Global analysis of the B12-elements in available bacterial genomes demonstrates that this conserved RNA regulatory element is widely distributed in eubacteria and regulates most genes required for the CBL biosynthesis and some other genes (Table 2 ▶; for details see D.A. Rodionov, A.G. Vitreschak, A.A. Mironov, and M.S. Gelfand, in prep.). First, various cbi and cob biosynthetic genes are regulated by B12-elements in most CBL-synthesizing bacteria. These genes either form single CBL gene clusters or are scattered along the chromosome. Moreover, genes for most known and predicted cobalt transporters, as well as cobalt chelatases and reductases, are often preceded by candidate B12-elements. Indeed, cobalt ions are required for the de novo CBL biosynthesis. Second, the vitamin B12 transport systems, btuBFCD in Gram-negative bacteria and btuFCD in Gram-positive bacteria, are mostly B12-regulated. Third, B12-regulons of various bacteria are predicted to include enzymes from known B12-dependent or alternative pathways, as well as several hypothetical enzymes from unknown pathways. In particular, B12-independent izozymes of the methionine synthase and ribonucleotide reductase are B12 regulated in bacteria that have both B12-dependent and B12-independent izozymes.
TABLE 2.
Phylogenetic distribution of gene clusters regulated by B12-elements
| Gene cluster | Function | Taxonomic group |
| 1. CBL biosynthesis | ||
| cbi and cob | cobalamin biosynthesis | proteobacteria, the Bacillus/Clostridium group |
| cbt, hoxN, cbiMNQO, hupE | cobalt transporters | all CBL-synthesizing bacteria |
| orfl-cobW-cobN-chlID | cobalt chelation | α-, β-proteobacteria, Pseudomonadaceae, actinobacteria |
| bluB | cobalt reduction | α-proteobacteria |
| btuR | CBL adenosyltransferase | α-, β-proteobacteria, Pseudomonadaceae |
| 2. Vitamin B12 transport | ||
| btuB | vitamin B12 receptor | proteobacteria |
| btuFCD | vitamin B12 transporter (ABC components) | α-, β-proteobacteria, Pseudomonadaceae, the Bacillus/Clostridium and CFB groups, Deinococcusradiodurans, actinobacteria, spirochetes, Fusobacteriaceae, Thermotogales, Chloroflexaceae |
| 3. B12-dependent or alternative metabolic pathways | ||
| metE | methionin synthase | various groups |
| nrd | ribonucleotide reductase | various groups |
| ardX-frdX | predicted enzymes | α-proteobacteria |
| achX | predicted enzymes | Deinococcus radiodurans and some other species |
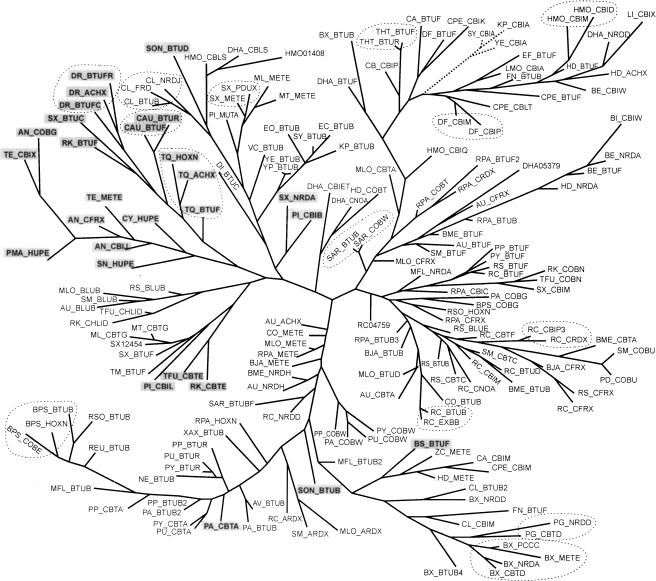
Using multiple alignment of identified B12-element sequences without additional nonconserved interior loops and the BII part, we constructed the maximum likelihood phylogenetic tree for these RNA elements (Fig. 7 ▶). The BII part was not used as it is not obligatory for all B12-elements. The tree of B12-elements has a number of branches that correspond to taxonomic groups, for instance, the Bacillus/Clostridium group or cyanobacteria. Comparison of the B12-element phylogenetic tree with the standard trees for ribosomal proteins (Wolf et al. 2001) reveals both lineage-specific and gene-specific branches, as well as recent genome-specific duplications and horizontal transfer events.
FIGURE 7.
Phylogenetic tree of B12-elements. The names of the proximal genes of the B12-regulated operons are given. The genome abbreviations are listed in Table 3 ▶. B12-elements without the BII part are set in bold and shaded in gray. Groups of genome-specific B12-elements are circled by dotted lines.
The branch of B12-elements found upstream of the cbi operons of enterobacteria is the most obvious example of possible horizontal transfer, as it clusters with various B12-elements from the Bacillus/Clostridium group (Fig. 7 ▶), and the same holds for phylogenetic trees constructed for each gene from the cbi operon (data not shown). These observations allow us to suggest that the complete transcriptional unit, including the regulatory B12-element and the cbi operon, has been likely transferred from the Bacillus/Clostridium group to three enterobacterial genomes.
The phylogenetic tree of B12-elements contains a number of genome-specific branches. In particular, four B12-elements found upstream of the pccC, cbtD, nrdA, and metE genes from Bacteroides fragilis form a separate branch (Fig. 7 ▶). Furthermore, in D. radiodurans, Burkholderia pseudomallei, Rhodopseudomonas palustris, Rhodobacter capsulatus, Heliobacillus mobilis, Sphingomonas aromaticivorans, Streptomyces colelicolor, Clostridium difficile, Porphyromonas gingivalis, Chlorobium tepidum, and Chloroflexus aurantiacus, we observed other organism-specific branches containing two or three B12-elements. In all of these cases, the regulatory elements are located upstream of various nonhomologous genes. Thus, it seems likely that the evolution of the B12-elements often involved independent lineage-specific duplications with subsequent transfer to a position upstream of a new gene. On the other hand, the phylogenetic tree has branches that correspond to B12-elements occurring upstream of orthologous genes, for example, the btuB genes from enterobacteria, the cobW genes from pseudomonads, and the bluB and metE genes from α-proteobacteria. These observations indicate that these B12-elements have co-evolved with the corresponding B12-regulated genes.
Finally, the constructed phylogenetic tree allows us to propose a possible origin of the B12-elements lacking the highly conserved BII part (see bold elements in Fig. 7 ▶). Most B12-elements without the BII part, for example, all B12-elements from early-diverged bacteria (D. radiodurans, Chloroflexaceae, and cyanobacteria), form a separate branch in the phylogenetic tree. The BII part seems to appear after divergence of these taxonomic groups (recall that the BII part was not used in constructing the tree). However, a B12-element from B. subtilis lacking the BII part is clustered with several complete B12-elements from other Bacillus species. Moreover, another BII-deficient B12 element from Pseudomonas aeruginosa clusters with other Pseudomonas B12-elements with both BI and BII parts. The absence of BII parts in these two B12-elements likely is a consequence of late deletions in these species. Thus, although the additional BII part is highly conserved in a large number of B12-elements, it is possibly not obligatory and can play an auxiliary role in the functioning of the B12-element.
CONCLUSIONS
Regulation of most B12-related genes in bacteria appears to operate through a unique RNA structural element. The B12-element is characterized by its compact secondary structure with a number of conserved helices and extended regions of sequence conservation, which could be necessary for specific metabolite binding. Recently, it has been shown that Ado-CBL specifically binds the leader mRNAs of CBL-related genes from enterobacteria (Nahvi et al. 2002). However, the B12-element also contains the highly conserved BII structure, which is not obligatory for all bacterial B12-elements, being absent in deeply branching bacterial groups and in some other cases. The role of this additional part of the B12-element is not clear.
On the whole, the mechanism of regulation of vitamin B12-related genes is similar to the previously proposed mechanisms of regulation of riboflavin- and thiamin-related genes (Rodionov et al. 2002; Vitreschak et al. 2002). At that, a highly conserved RNA element (RFN, THI, or B12) is stabilized by direct binding of the effector (flavin mononucleotide, thiamin pyrophosphate, or adenosylcobalamin, respectively). Thus, an adjacent regulatory hairpin, terminator or sequestor, can fold that leads to transcriptional or translational repression of vitamin-related genes. In the absence of the effector, the unstable vitamin-specific RNA element is replaced by an alternative antiterminator or antisequestor RNA conformation allowing for transcription readthrough or translation initiation. Interestingly, the observed transcriptional and translational types of regulation have similar taxonomic distribution for each of the analyzed vitamin regulons. At that, the transcriptional termination occurs mostly in Gram-positive organisms, whereas the inhibition of translation initiation happens in Gram-negative proteobacteria, D. radiodurans, the CFB group and cyanobacteria.
This study once again demonstrates the power of the simultaneous positional and phylogenetic analysis of regulatory elements and genes. This approach provides an opportunity for identification of new RNA regulatory elements in bacterial genomes and prediction of the regulation. Moreover, it uncovers the traces of evolutionary events such as horizontal transfer and lineage-specific duplication of genes and regulatory elements.
MATERIALS AND METHODS
Complete and partial sequences of bacterial genomes were downloaded from GenBank (Benson et al. 2003). Preliminary sequence data were also obtained from the www sites of the Institute for Genomic Research (http://www.tigr.org), the University of Oklahoma’s Advanced Center for Genome Technology (http://www.genome.ou.edu/), the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/), the DOE Joint Genome Institute (http://jgi.doe.gov), and the ERGO Database (http://ergo.integratedgenomics.com/ERGO) (Overbeek et al. 2003). The genome abbreviations from the ERGO database are used throughout and listed in Table 3 ▶.
TABLE 3.
Taxonomy and abbreviations for bacterial genomes used in this work
| Tax | Genome | Abbreviation |
| α | Agrobacterium tumefaciens | AU |
| Bradyrhizobium japonicum | BJA | |
| Brucella melitensis | BME | |
| Caulobacter crescentus | CO | |
| Mesorhizobium loti | MLO | |
| Rhodobacter capsulatus | RC | |
| Rhodobacter sphaeroides | RS | |
| Rhodopseudomonas palustris | RPA | |
| Sinorhizobium meliloti | SM | |
| Sphingomonas aromaticivorans | SAR | |
| β | Burkholderia pseudomallei | BPS |
| Methylobacillus flagellatus | MFL | |
| Nitrosomonas europaea | NE | |
| Ralstonia eutropha | REU | |
| Ralstonia solanacearum | RSO | |
| γ | Escherichia coli | EC |
| Salmonella typhimurium | SY | |
| Klebsiella pneumoniae | KP | |
| Yersinia enterocolitica | YE | |
| Yersinia pestis | YP | |
| Erwinia carotovora | EO | |
| Vibrio cholerae | VC | |
| Pseudomonas aeruginosa | PA | |
| Pseudomonas putida | PP | |
| Pseudomonas fluorescens | PU | |
| Pseudomonas syringae | PY | |
| Shewanella oneidensis | SON | |
| Azotobacter vinelandii | AV | |
| Xanthomonas axonopodis | XAX | |
| δ | Geobacter metallireducens | GME |
| Chloroflexus aurantiacus # | CAU | |
| Fusobacterium nucleatum | FN | |
| Thermotoga maritima | TM | |
| B/C | Bacillus subtilis | BS |
| Bacillus cereus | ZC | |
| Bacillus megaterium | BI | |
| Bacillus halodurans | HD | |
| Bacillus stearothermophilus | BE | |
| Listeria monocytogenes | LMO | |
| Clostridium acetobutylicum | CA | |
| Clostridium perfringes | CPE | |
| Clostridium botulinum | CB | |
| Clostridium difficile | DF | |
| Thermoanaerobacter tengcongensis | THT | |
| Enterococcus faecalis | EF | |
| Heliobacillus mobilis | HMO | |
| Desulfitobacterium halfniense | DHA | |
| Act | Corynebacterium diphtheriae | DI |
| Mycobacterium tuberculosis | MT | |
| Mycobacterium leprae | ML | |
| Thermobifida fusca | TFU | |
| Rhodococcus str. | RK | |
| Streptomyces coelicolor | SX | |
| Propionicibacterium shermanii | PI | |
| Cya | Anabaena sp. | AN |
| Prochlorococcus marinus. | PMA | |
| Synechocystis sp. | CY | |
| Synechococcus sp. | SN | |
| Thermosynechococcus elongatus | TEL | |
| CFB | Porphyromonas gingivalis | PG |
| Bacteroides fragilis | BX | |
| Chlorobium tepidum | CL | |
| T/D | Deinococcus radiodurans | DR |
| SP | Treponema denticola | TDE |
| Leptospira interrogans | LI |
The names of taxonomic groups in Tax column (α, β, γ, δ, B/C, Act, Cya, CFB, T/D and SP) stand for α-, β-, γ-, and δ-proteobacteria, the Bacillus/Clostridium group, actinobacteria, cyanobacteria, the CFB group, the Thermus/Deinococcus group and spirochetes, respectively.
The conserved secondary structure of the B12-element was derived using the RNAMultAln program (A.A. Mironov, unpubl.). This program simultaneously creates a multiple alignment and a conserved secondary structure for a set of RNA sequences. The RNA-PATTERN program (Vitreschak et al. 2001) was used to search for new B12-elements in bacterial genomes. The input RNA pattern described both the RNA secondary structure and the sequence consensus motifs. The RNA secondary structure was described as a set of the following parameters: the number of helices, the length of each helix, the loop lengths, and the description of the topology of helix pairs. Additional RNA secondary structures, in particular antiterminators and antisequestors, were predicted using Zuker’s algorithm of free energy minimization (Lyngso et al. 1999) implemented in the Mfold program (http://bioinfo.math.rpi.edu/~mfold/rna).
Protein similarity search was done using the Smith-Waterman algorithm implemented in the GenomeExplorer program (Mironov et al. 2000). Orthologous proteins were initially defined by the best bidirectional hit criterion (Tatusov et al. 2000) and if necessary, confirmed by construction of phylogenetic trees. The phylogenetic trees of the B12-elements and B12-related proteins were created by the maximum likelihood method implemented in PHYLIP (Felsenstein 1981). Multiple sequence alignments were constructed using CLUSTALX (Thompson et al. 1997).
Complete FASTA sequences of B12-elements, including the additional and variable regions are available from the authors (L.V. at l_veter@mail.ru or M.G. at gelfand@ig-msk.ru).
Acknowledgments
We are grateful to Andrei Osterman for attention, advice, and encouragement. This study was partially supported by grants from the Ludwig Institute for Cancer Research and the Howard Hughes Medical Institute (55000309).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., and Wheeler, D.I. 2003. GenBank. Nucleic Acids Res. 31: 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar, T., Beyer, K., Bork, P., Kenealy, M.R., Pantopoulos, K., Hentze, M., Sonntag-Buck, V., Flouriot, G., Gannon, F., and Schreiber, S. 1998. Systematic genomic screening and analysis of mRNA in untranslated regions and mRNA precursors: Combining experimental and computational approaches. Bioinformatics 14: 271–278. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. and Durbin, R. 1994. RNA sequence analysis using covariance models. Nucleic Acids Res. 22: 2079–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. 1981. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17: 368–376. [DOI] [PubMed] [Google Scholar]
- Franklund, C.V. and Kadner, R.J. 1997. Multiple transcribed elements control expression of the Escherichia coli btuB gene. J. Bacteriol. 179: 4039–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand, M.S., Novichkov, P.S., Novichkova, E.S., and Mironov, A.A. 2000. Comparative analysis of regulatory patterns in bacterial genomes. Brief Bioinform. 1: 357–371. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones, S., Bateman, A., Marshall, M., Khanna, A., and Eddy, S.R. 2003. Rfam: An RNA family database. Nucleic Acids Res. 31: 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, F.J. and Henkin, T.M. 1998. The S box regulon: A new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30: 737–749. [DOI] [PubMed] [Google Scholar]
- Lundrigan, M.D., Koster, W., and Kadner, R.J. 1991. Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12. Proc. Natl. Acad. Sci. 88: 1479–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngso, R.B., Zuker, M., and Pedersen, C.N. 1999. Fast evaluation of internal loops in RNA secondary structure prediction. Bioinformatics 15: 440–445. [DOI] [PubMed] [Google Scholar]
- Marck, C. and Grosjean, H. 2002. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8: 1189–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens, J.H., Barg, H., Warren, M.J., and Jahn, D. 2002. Microbial production of vitamin B12. Appl. Microbiol. Biotechnol. 58: 275–285. [DOI] [PubMed] [Google Scholar]
- Mironov, A.A., Vinokurova, N.P., and Gelfand, M.S. 2000. GenomeExplorer: Software for analysis of complete bacterial genomes. Mol. Biol. 34: 222–231. [Google Scholar]
- Mironov, A.S., Gusarov, I., Rafikov, R., Lopez, L.E., Shatalin, K., Kreneva, R.A., Perumov, D.A., and Nudler, E. 2002. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell 111: 747–756. [DOI] [PubMed] [Google Scholar]
- Murthy, V.L. and Rose, G.D. 2003. RNABase: An annotated database of RNA structures. Nucleic Acids Res. 31: 502–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi, A., Sudarsan, N., Ebert, M.S., Zou, X., Brown, K.L., and Breaker, R.R. 2002. Genetic control by a metabolite binding mRNA. Chem. Biol. 9: 1043–1049. [DOI] [PubMed] [Google Scholar]
- Nou, X. and Kadner, R.J. 1998. Coupled changes in translation and transcription during cobalamin-dependent regulation of btuB expression in Escherichia coli. J. Bacteriol. 180: 6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Adenosylcobalamin inhibits ribosome binding to btuB RNA. Proc. Natl. Acad. Sci. 97: 7190–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek, R., Larsen, N., Walunas, T., D’Souza, M., Pusch, G., Selkov Jr., E., Liolios, K., Joukov, V., Kaznadzey, D., Anderson, I., et al. 2003. The ERGO(TM) genome analysis and discovery system. Nucleic Acids Res. 31: 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnum, S. and Andersson, D.I. 1997. Vitamin B12 repression of the btuB gene in Salmonella typhimurium is mediated via a translational control which requires leader and coding sequences. Mol. Microbiol. 23: 35–42. [DOI] [PubMed] [Google Scholar]
- ———. 2001. An adenosyl-cobalamin (coenzyme-B12)-repressed translational enhancer in the cob mRNA of Salmonella typhimurium. Mol. Microbiol. 39: 1585–1594. [DOI] [PubMed] [Google Scholar]
- Richter-Dahlfors, A.A. and Andersson, D.I. 1992. Cobalamin (vitamin B12) repression of the cob operon in Salmonella typhimurium requires sequences within the leader and the first translated open reading frame. Mol. Microbiol. 6: 743–749. [DOI] [PubMed] [Google Scholar]
- Richter-Dahlfors, A.A., Ravnum, S., and Andersson D.I. 1994. Vitamin B12 repression of the cob operon in Salmonella typhimurium: Translational control of the cbiA gene. Mol. Microbiol. 13: 541–553. [DOI] [PubMed] [Google Scholar]
- Rodionov, D.A., Vitreschak, A.G., Mironov, A.A., and Gelfand, M.S. 2002. Comparative genomics of thiamin biosynthesis in procaryotes: New genes and regulatory mechanisms. J. Biol. Chem. 277: 48949–48959. [DOI] [PubMed] [Google Scholar]
- Szymanski, M., Erdmann, V.A., and Barciszewski, J. 2003. Noncoding regulatory RNAs database. Nucleic Acids Res. 31: 429–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Galperin, M.Y., Natale, D.A., and Koonin, E.V. 2000. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak, A.G., Mironov, A.A., and Gelfand, M.S. 2001. The RNApattern program: Searching for RNA secondary structure by the pattern rule. In Proceedings of the 3rd International Conference on “Complex Systems: Control and Modeling Problems.”, September 4–9, 2001, pp. 623–625. The Institute of Control of Complex Systems, Samara, Russia.
- Vitreschak, A.G., Rodionov, D.A., Mironov, A.A., and Gelfand, M.S. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30: 3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, Y.I., Rogozin, I.B., Grishin, N.V., Tatusov, R.L., and Koonin, E.V. 2001. Genome trees constructed using five different approaches suggest new major bacterial clades. BMC Evol. Biol. 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, W., Nahvi, A., and Breaker, R.R. 2002a. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956. [DOI] [PubMed] [Google Scholar]
- Winkler, W., Cohen-Chalamish, S., and Breaker, R.R. 2002b. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. 99: 15908–15913. [DOI] [PMC free article] [PubMed] [Google Scholar]