Abstract
The 3′–5′ riboexonuclease Rrp6p, a nuclear component of the exosome, functions with other exosome components to produce the mature 3′ ends of 5.8S rRNA, sno- and snRNAs, and to destroy improperly processed precursor (pre)-rRNAs and pre-mRNAs. Rrp6p is a member of the RNase D family of riboexonucleases and displays a high degree of homology with the active site of the deoxyriboexonuclease domain of Escherichia coli DNA polymerase I, the crystal structure of which indicates a two-metal ion mechanism for phosphodiester bond hydrolysis. Mutation of each of the conserved residues predicted to coordinate metal ions in the active site of Rrp6p abolished activity of the enzyme in vitro and in vivo. Complete loss of Rrp6p activity caused by the Y361F and Y361A mutations supports the critical role proposed for the phenolic hydroxyl of Tyr361 in the reaction mechanism. Rrp6p also contains an helicase RNase D C-terminal (HRDC) domain of unknown function that is similar to domains in the Werner’s and Bloom’s Syndrome proteins. A point mutation in this domain results in Rrp6p that localizes to the nucleus, but fails to efficiently process the 3′ ends of 5.8S pre-rRNA and some pre-snoRNAs. In contrast, this mutant retains the ability to degrade rRNA processing intermediates and 3′-extended, poly(A)+ snoRNAs. These findings indicate the potential for independent control of the processing and degradation functions of Rrp6p.
Keywords: Exosome, Rrp6p, riboexonuclease, RNA processing
INTRODUCTION
Synthesis of RNA molecules in the nuclei of eukaryotic cells produces precursor RNAs (pre-RNAs) that require RNA processing reactions to produce the mature functional RNAs. For transcripts synthesized by RNA polymerase II, such as mRNAs, snRNAs, and snoRNAs, maturation begins with cotranscriptional 5′ capping and, for pre-mRNAs, continues with splicing and finishes in most cases with cleavage and polyadenylation of the pre-mRNA to produce the mature 3′ end. For snRNAs and snoRNAs in yeast, 3′ end processing of the transcripts follows: (1) cleavage of the precursor by the RNase III-type enzyme Rnt1p, or cleavage by components of the mRNA 3′ end processing system; or (2) release of the intron encoded snoRNA by the debranching enzyme (Allmang et al. 1999a; Fatica et al. 2000; van Hoof et al. 2000). Experimental evidence indicates that after these cleavage events, the mature 3′ end of these small RNAs results from 3′–5′ exonucleolytic trimming of the pre-RNAs by the nuclear form of a conserved complex of riboexonucleases called the exosome (Mitchell et al. 1997). RNA polymerase I synthesizes a single pre-rRNA transcript that undergoes a complex series of nucleolytic and base modification reactions that produce the mature 18S, 5.8S, and 25S rRNAs (Venema and Tollervey 1999). The nuclear form of the exosome plays a critical role in these maturation events by trimming the last 152 nt from the 3′ end of 5.8S pre-rRNA in a two-step reaction that first requires the core exosome to trim the molecule to within 30 nt of the mature 3′ end, and then the exosome subunit Rrp6p to trim the pre-rRNA to its mature 3′ end (Briggs et al. 1998; Allmang et al. 1999b).
In these reactions, the exosome plays a productive role in that it processes the 3′ ends of these small stable RNAs to their mature sites. In addition, the exosome plays a destructive role in that it degrades improperly processed intermediates in the rRNA processing pathway and it may function to degrade abnormal poly(A)+ snRNA and snoRNA processing intermediates (Allmang et al. 1999a, 2000; van Hoof et al. 2000). The destructive activity of the exosome also plays a prominent role in the cytoplasm, where it functions in the normal turnover pathway for mRNAs, in a pathway that degrades mRNAs lacking stop codons, and, in the case of yeast, in a pathway that controls the levels of the dsRNA virus LA (Widner and Wickner 1993; Anderson and Parker 1998; Frischmeyer et al. 2002; van Hoof et al. 2002). In the nucleus, the exosome, along with its nuclear components Rrp6p and Dob1p/Mtr4p, plays a role in degrading inefficiently spliced or polyadenylated mRNAs, and this nuclear mRNA surveillance function appears to be intimately linked to the transcription, processing, and transport of mRNAs out of the nucleus (Bousquet-Antonelli et al. 2000; Burkard and Butler 2000; Hilleren 2001).
The exosome functions in the nucleus and the cytoplasm and appears to exist as a core particle comprised of nine 3′–5′ riboexonucleases and a protein of unknown function (Csl4p/Ski4p; Mitchell and Tollervey 2000; Butler 2002). In the cytoplasm, the core exosome interacts with the Ski complex, which appears to mediate the interaction of the exosome with substrate mRNAs (Brown et al. 2000; Araki et al. 2001; van Hoof et al. 2002). In the nucleus, the core exosome interacts with the putative RNA helicase Dob1p/Mtr4p and the 3′–5′ riboexonuclease Rrp6p, both of which play essential roles in the nuclear processing and degradation functions of the exosome (Mitchell et al. 1997; Allmang et al. 1999b). Current models to explain the regulation of the inherently destructive power of the exosome feature the interaction of the complex with other proteins, such as those in the Ski complex, which would serve to activate the exosome allosterically or facilitate access of the exosome to the substrate through the unwinding activity of a helicase, such as Dob1p/Mtr4p or the Ski complex subunit Ski2p (van Hoof and Parker 1999; Mitchell and Tollervey 2000; Butler 2002). Indeed, the binding of specific proteins to the AU-rich instability elements of unstable mRNAs recruits the mammalian exosome to degrade these transcripts (Chen et al. 2001; Mukherjee et al. 2002).
One approach to understanding the function and regulation of the exosome features the development of a clear picture of the contribution of individual riboexonuclease components to the various exosome activities. This is particularly true for the nuclear subunit Rrp6p because this protein appears to function differently from the core exosome components. For example, Rrp6p removes the last 30 nucleotides from the 3′ end of pre-5.8S rRNA after the core exosome produces this substrate by trimming a longer precursor, and Rrp6p removes the last three or so nucleotides from some snoRNAs after the core exosome removes their long 3′ extensions (Allmang et al. 1999a; van Hoof et al. 2000). On the other hand, all of the nuclear exosome components, including Rrp6p, are required for degrading dead-end pre-rRNA processing intermediates such as the 5′ external transcribed spacer (ETS) RNA produced by an early cleavage of the rRNA precursor (Allmang et al. 1999a,b). The separable activities of Rrp6p and the core exosome in RNA processing, but not in RNA degradation, raises the question as to whether the activity of Rrp6p might be regulated differently during the processing and degradation of RNA substrates. As an approach to answer this question, we undertook a structure/function analysis of Rrp6p, in which we altered conserved amino acids in the enzyme and assessed the effects on the localization and function of the mutant proteins. These experiments indicate that Rrp6p contains, at a minimum, a core riboexonuclease domain related to the deoxyriboexonuclease domain of DNA polymerase I, a domain involved in regulating its RNA 3′ end processing function and a nuclear localization domain.
RESULTS
Rrp6p requires a highly conserved exonuclease domain for function
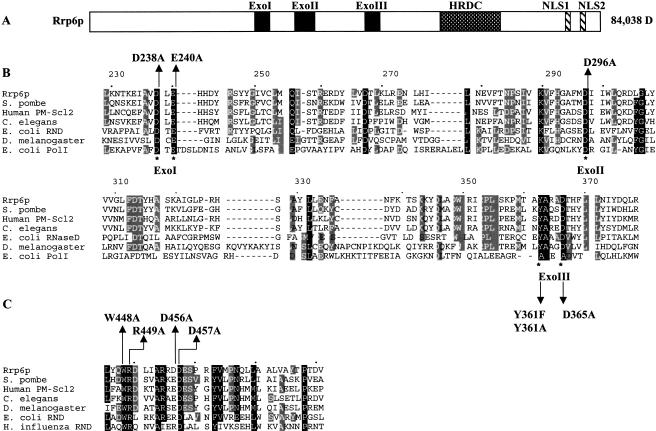
Comparison of the amino acid sequence of Rrp6p with proteins in existing databases reveals a general domain structure, illustrated in Figure 1A ▶. Rrp6p contains three putative exonuclease motifs, a so-called HRDC domain and two potential nuclear localization signals (NLSs). We assessed the functional importance of these regions after site-directed mutagenesis of conserved residues followed by functional analysis of Gfp-Rrp6p fusion proteins in vivo. First, we mutated amino acids in Rrp6p based on its homology with the deoxyriboexonuclease domain of E. coli DNA polymerase I, the crystal structure of which indicates that four basic side-chains coordinate two metal ions in the active site (Fig. 1B ▶; Beese et al. 1993). The model for phosphodiester bond hydrolysis proposes that one of the metal ions acts as a Lewis acid to activate a water molecule, which carries out nucleophilic attack of the phosphodiester backbone, whereas the others, along with the tyrosine corresponding to position 361 of Rrp6p, serve to orient the nucleophile and stabilize the transition state intermediate.
FIGURE 1.
Domain structure of Rrp6p. (A) Diagram of Rrp6p indicating the exonuclease domain containing three exonuclease motifs (ExoI, ExoII, and ExoIII), the helicase-RNase D C-terminal (HRDC) domain, and two putative nuclear localization signals. (B) Amino acid sequence homology between the exonuclease domain of Rrp6p and several other closely related proteins: Saccharomyces pombe, Q10146; Human PM-Scl2, Q01780; Caenorhabditis elegans, P34607; Escherichia coli rnd, P09155; Drosophila melanogaster, AAB49975; and E. coli PolI, 1KFD. (C) Amino acid sequence homology between the HRDC domain of Rrp6p and several other closely related proteins: S. pombe, Q10146; Human PM-Scl2, Q01780; C. elegans, P34607; D. melanogaster, AAF55107; E. coli rnd, P09155; and Haemophilus influenza rnd, P44442.
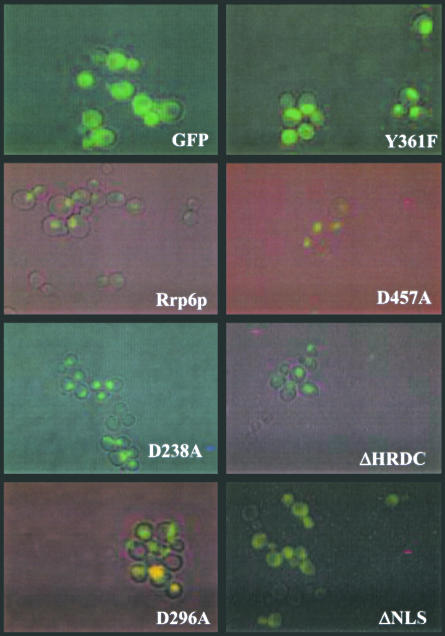
We expressed mutant alleles encoding Rrp6p as Gfp-Rrp6p fusion proteins in an rrp6-Δ strain and analyzed their expression, localization, and activity by a variety of techniques. Fluorescence microscopy of growing cells revealed expression of the mutant proteins as well as their accumulation in the nucleus (Fig. 2 ▶; data not shown). This pattern of accumulation matches that of the normal protein and coincides with nuclear staining of the DNA (Fig. 2 ▶; data not shown). In contrast, these experiments show disperse cytoplasmic and nuclear fluorescence of Gfp and partial cytoplasmic accumulation of Gfp-Rrp6-15p with its NLSs deleted (Fig. 2 ▶; ΔNLS). These findings indicate that Rrp6p proteins with mutations in the exonuclease motifs accumulate within the nucleus at levels similar to the normal protein, whereas ΔNLS results in a defect in nuclear localization.
FIGURE 2.
Subcellular localization of Gfp-Rrp6p mutants in logarithmically growing yeast cells. Strain BPO2-KAN (MATα ade1 ade2 lys2 gal1 ura3-52 rrp6::KAN) carrying yeast CEN plasmids expressing each of the indicated RRP6 alleles as GFP-fusion proteins were grown in synthetic complete glucose media lacking uracil and methionine at 30°C to a density of ~106 cell/mL. Cells were visualized for green fluorescence or by direct interference contrast, and the images were overlayered by using Adobe Photoshop 5.0.
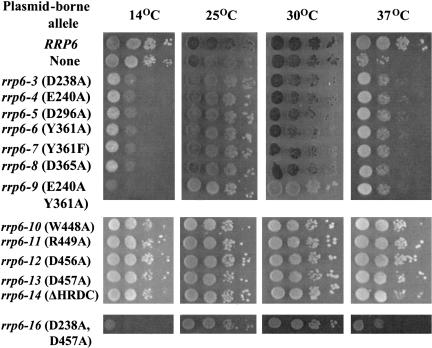
As a second functional test, we analyzed the ability of the Rrp6p exonuclease motif mutants to complement the heat sensitivity of an rrp6-Δ strain. We also analyzed the ability of the strains to grow at 14°C because two previously characterized recessive alleles, rrp6-1 (D238N) and rrp6-2 (D238N), caused cold-sensitive growth (Burkard and Butler 2000). Figure 3 ▶ shows that expression of the RRP6 gene fused to Gfp allows yeast cells to grow at all temperatures tested, whereas the absence of RRP6 causes heat-sensitive growth at 37°C. Expression of each of the exonuclease-motif mutant proteins causes cold-sensitive growth at 14°C. Subsequent analysis demonstrated that the cold-sensitive phenotype of each mutant, such as the rrp6-1 and rrp6-2 mutants, is recessive to RRP6 (Burkard and Butler 2000). Interestingly, each of the new mutations partially complements the heat-sensitive phenotype associated with the rrp6-Δ mutation (Fig. 3 ▶). The ability of the exonuclease mutant proteins to complement the heat sensitivity of rrp6-Δ and to cause cold-sensitive growth indicates that the mutant proteins are expressed as functional, albeit abnormal, proteins in the cell. The previously observed cold sensitivity caused by chromosomal mutations rrp6-1 and rrp6-2 in exonuclease motif I, as well as their ability to complement the heat sensitivity of rrp6-Δ, suggests that the phenotypes observed for the exonuclease motif mutants in Figure 3 ▶ do not result from fusion of the proteins to Gfp or from their expression from a plasmid (Burkard and Butler 2000).
FIGURE 3.
Growth phenotypes of RRP6 mutants. BPO2-KAN carrying yeast CEN plasmids expressing each of the indicated RRP6 alleles as Gfp-fusion proteins were spotted in 10-fold serial dilutions beginning with 104 cells on synthetic complete glucose plates lacking uracil and methionine and incubated at the indicated temperature.
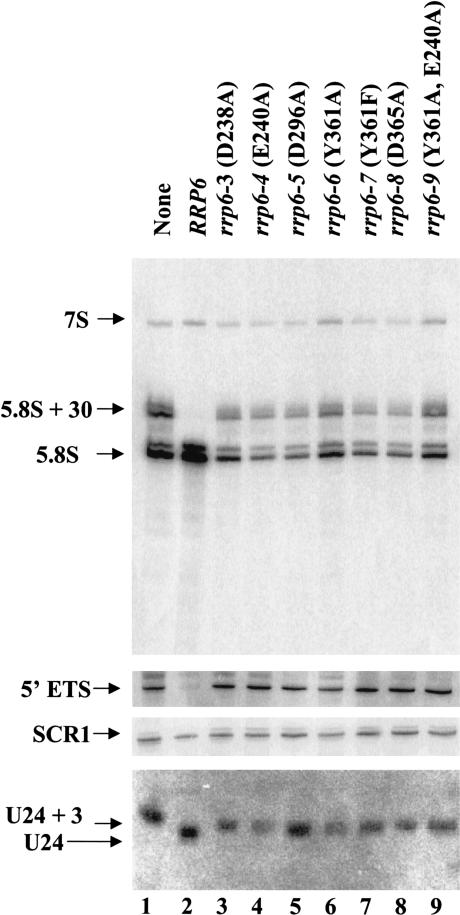
Rrp6p functions in the nucleus to form the mature 3′ ends of 5.8S rRNA and various snRNAs and snoRNAs and to degrade certain aberrant RNAs (Briggs et al. 1998; Allmang et al. 1999a; van Hoof et al. 2000). Previous studies showed that the rrp6-Δ mutation results in 3′ end processing phenotypes distinct from those observed for mutation or depletion of other exosome components. For example, rrp6-Δ causes the accumulation of 5.8S rRNA extended by ~30 nt at its 3′ end and U24 snRNA extended by approximately three nucleotides at its 3′ end, whereas defects in other exosome components result in 5.8S rRNA and U24 RNA with even longer 3′ extensions (Allmang et al. 1999a; van Hoof et al. 2000). Northern blot analysis of the amounts and lengths of these RNAs in strains containing exonuclease motif mutations shows that each mutation abolishes the ability of Rrp6p to carry out 3′ end processing of these pre-RNAs because the extent of each defect equals that observed in the rrp6-Δ strain (Fig. 4 ▶; Table 1 ▶). In contrast, expression of Gfp-Rrp6p complements these defects. These findings indicate that each of the exonuclease motif mutations abolishes Rrp6p-specific RNA processing in vivo.
FIGURE 4.
Northern blot analysis of RNA from exonuclease domain mutants. Strain BPO2-KAN carrying yeast CEN plasmids expressing each of the indicated RRP6 alleles as GFP-fusion proteins were grown in synthetic complete glucose media lacking uracil and methionine at 30°C. Total RNA was isolated, and 10 μg was analyzed by Northern blotting for the RNA species indicated at the left.
TABLE 1.
Summary of RNA processing phenotypes of RRP6 mutants
| Allele | Mutation | 5.8S + 30 | 5′ ETS | U24 + 3 | poly(A)+ U24/snr72 | Exonuclease activity in vitro |
| RRP6 | none | − | − | − | − | + |
| rrp6-Δ | deletion | + | + | + | + | ND |
| rrp6-3 | D238A | + | + | + | + | − |
| rrp6-4 | E240A | + | + | + | ND | − |
| rrp6-5 | D296A | + | + | + | ND | − |
| rrp6-6 | Y361A | + | + | + | ND | − |
| rrp6-7 | Y361F | + | + | + | ND | − |
| rrp6-8 | D365A | + | + | + | ND | − |
| rrp6-9 | Y361A, E240A | + | + | + | ND | − |
| rrp6-10 | W448A | − | − | − | ND | + |
| rrp6-11 | R449A | − | − | − | ND | − |
| rrp6-12 | D456A | − | − | − | ND | + |
| rrp6-13 | D457A | + | − | + | − | − |
| rrp6-14 | ΔHRDC | + | + | + | ND | − |
| rrp6-16 | D457A, D238A | + | + | + | ND | ND |
| rrp6-15 | ΔNLS | − | − | − | ND | ND |
5.8S + 30 and 5′ETS indicate the presence of these 3′ extended precursors in the mutants. U24 + 3 refers to the three nucleotide 3′ extended U24 snoRNA (see Figs. 4 ▶, 5 ▶). Poly(A)+ U24/snr72 indicates the presence of 3′ extended, poly(A)+ snoRNAs. ND indicates not determined. Exonuclease activity in vitro was analyzed as described in Materials and Methods.
Rrp6p, along with other exosome components, plays a role in degrading the 5′ ETS released from the rRNA precursors during the earliest steps of rRNA processing (Allmang et al. 1999a,b). The RRP6 exonuclease motif mutations inhibit the degradation of the 5′ ETS but do not affect the levels of SCR1 RNA, the RNA component of the signal recognition particle (Fig. 4 ▶).
A mutation in the Rrp6p HRDC domain affects 3′ end processing, but not degradation of precursor RNAs
Rrp6p contains a domain homologous to the so-called HRDC regions of a variety of proteins involved in RNA and DNA processing (Fig. 1A,C ▶; Morozov et al. 1997; Briggs et al. 1998). Figure 1C ▶ illustrates the conservation of this domain among members of the RNase D family of proteins (Mian 1997; Moser et al. 1997). We deleted this region and mutated conserved amino acids within it to alanine followed by analysis of the effects of expression of these proteins as Gfp fusions in an rrp6-Δ strain. Each of the mutant proteins localized to a subcellular region coincident with the DNA-stained nucleus, indicating expression and correct localization of the proteins in vivo (Fig. 2 ▶; data not shown). Analysis of the temperature dependence of growth of these strains showed growth at all temperatures, including the ability of the mutant fusion proteins to complement the heat-sensitive growth defect caused by the rrp6-Δ mutation (Fig. 3 ▶).
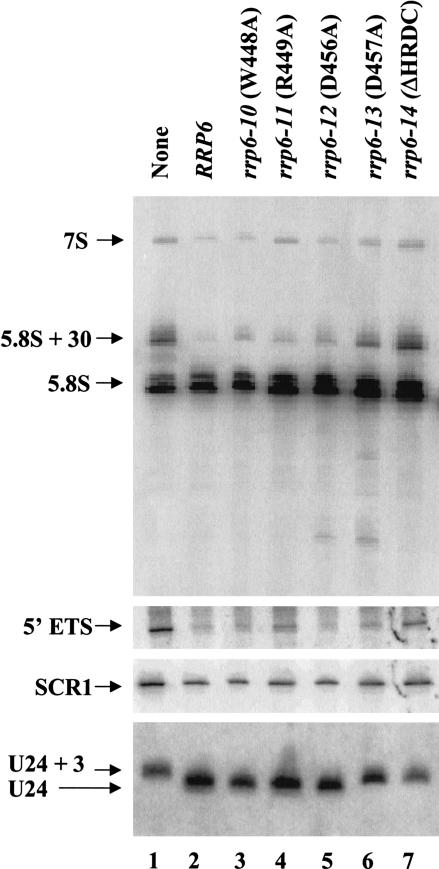
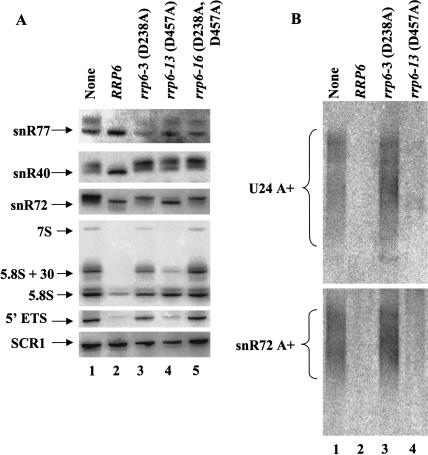
Analysis of 5.8S rRNA and U24 snoRNA processing in strains containing the HRDC-region mutations shows that the rrp6-13 and the HRDC deletion (rrp6-14) mutations have the most significant effects on processing of U24 and 5.8S rRNA, with the deletion mutant displaying the more severe defect in 5.8S rRNA processing (Fig. 5 ▶; Table 1 ▶). Although the deletion in rrp6-14 and the rrp6-Δ mutation alone yield levels of 5.8S + 30, 10-fold greater than that found in normal cells, the rrp6-13 mutation produces about three times the normal level. In contrast, the level of 5.8S+30 in the rrp6-10, rrp6-11, and rrp6-12 appears similar to that found in normal cells. The rrp6-13 and rrp6-14 mutations abolish correct 3′ end formation of U24 snRNA (Fig. 5 ▶) and result in a partial defect in snR72 3′ end processing (Fig. 6A ▶, lane 4). Analysis of snR77 and snR40 reveals processing defects caused by rrp6-13 equal to that caused by the absence of RRP6 (Fig. 6A ▶, cf. lanes 1 and 4). These findings indicate that the HRDC region, and in particular aspartate 457, plays an important role in role in 3′ end processing of pre-5.8S rRNA and pre-snoRNAs in vivo. In contrast, the rrp6-13 mutation does not cause a significant defect in the degradation of the 5′ ETS rRNA, despite its inability to efficiently process the 3′ ends of 5.8S and pre-snoRNAs (Fig. 5 ▶, lane 6; Fig. 6A ▶, lane 4; Table 1 ▶). This finding prompted us to investigate the effects of these mutations on the levels of poly(A)+ snoRNA precursors found in some exosome mutants, including rrp6-Δ (Allmang et al. 1999a; van Hoof et al. 2000). Northern blot analysis of oligo(dT)-selected poly(A)+ RNAs shows the accumulation of adenylated forms of U24 and snR72 RNAs in the deletion and rrp6-3 mutants, but not in the rrp6-13 mutant (Fig. 6B ▶). We have observed the same pattern for snR40 and snR77 RNAs (data no shown). These findings indicate that the properties of Rrp6p responsible for the degradation of 5′ ETS rRNA and poly(A)+ snoRNA precursors are not significantly disrupted by the rrp6-13 mutation.
FIGURE 5.
Northern blot analysis of RNA from HRDC domain mutants. Strain BPO2-KAN carrying yeast CEN plasmids expressing each of the indicated RRP6 alleles as GFP-fusion proteins were grown in synthetic complete glucose media lacking uracil and methionine at 30°C. Total RNA was isolated, and 10 μg was analyzed by Northern blotting for the RNA species indicated at the left.
FIGURE 6.
Northern blot analysis of RNA from RRP6 mutants. (A) Strain BPO2-KAN carrying yeast CEN plasmids expressing each of the indicated RRP6 alleles as GFP-fusion proteins were grown in synthetic complete glucose media lacking uracil and methionine at 30°C. Total RNA was isolated, and 10 μg was analyzed by Northern blotting for the RNA species indicated at the left. (B) Sixty μg of total RNA from strains grown as described in A was fractionated by oligo(dT)-celluose chromatography (Patel and Butler 1992), and the poly(A)+ fractions were analyzed by Northern blotting as described in A.
The contrasting RNA processing and growth phenotypes observed with the rrp6-3 and rrp6-13 mutations led us to ask which of these effects would predominate in a double mutant. Analysis of such a mutant (rrp6-16 [D238A, D457A]) reveals cold-sensitive growth, as well as defective 5.8S rRNA and U24 snRNA 3′ end processing and 5′ ETS degradation (Fig. 6A ▶). Analysis of the levels of poly(A)+ snoRNAs in the rrp6-13 mutant showed amounts equal to that observed in the rrp6-3 mutant (data not shown). These findings indicate intragenic dominance of rrp6-3 over rrp6-13 and imply that the rrp6-13 mutation may affect the 3′ end processing, but not the degradation function of Rrp6p.
Enhancement of Rrp6p nuclear localization by the carboxyl-terminal domain
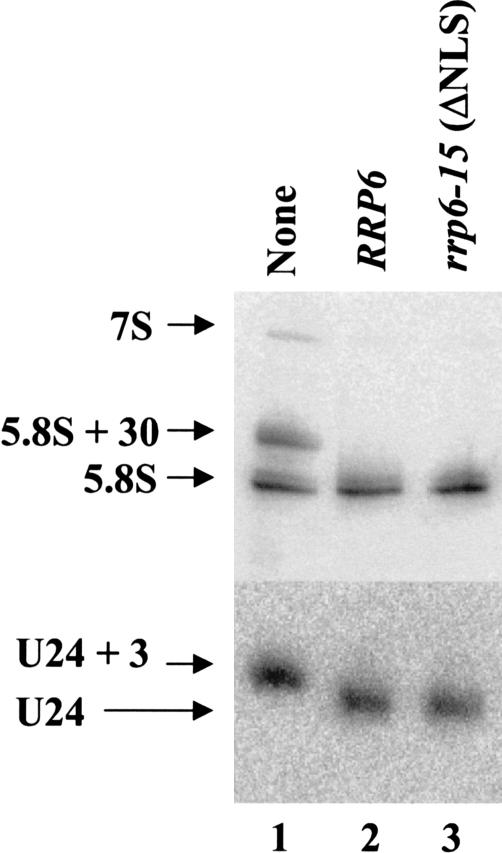
Rrp6p contains two sequences near its carboxyl terminus that resemble consensus NLSs (Fig. 1 ▶; Briggs et al. 1998). Deletion of both of these signals resulted in an Rrp6p mutant (rrp6-15) that localizes to the cytoplasm and the nucleus as judged by GFP fluorescence (Fig. 2 ▶) and confocal microscopy (data not shown). However, the mislocalization of this fraction of Rrp6p does not affect the growth or RNA processing phenotypes in this strain (Figs. 3 ▶, 7 ▶). These findings indicate that the sequences deleted in rrp6-15 play an important role in targeting or retaining the protein in the nucleus, but Rrp6p has other properties that contribute to its ability to enter and function in the nucleoplasm.
FIGURE 7.
Northern blot analysis of RNA from the RRP6 NLS mutant. Strain BPO2-KAN carrying yeast CEN plasmids expressing the rrp6-15 mutant as GFP-fusion proteins was grown in synthetic complete glucose media lacking uracil and methionine at 30°C. Total RNA was isolated, and 10 μg was analyzed by Northern blotting for the RNA species indicated at the left.
Analysis of the riboexonuclease activity of Rrp6p mutants in vitro
We expressed each of the mutant Rrp6p proteins in E. coli, purified them as glutathione-S-transferase (Gst) fusions, and assayed their riboexonuclease activity in vitro (Table 2). None of the proteins with mutations in the exonuclease motifs showed measurable riboexonuclease activity. Among the HRDC mutants, Gst-Rrp6p-10 (W448A) and Gst-Rrp6p-12 (D456A) displayed riboexonuclease activity in vitro. Two of the 12 mutant proteins tested, Rrp6-11p and Rrp6-13p, had no activity in vitro, despite the fact that each mutation produces enzyme with apparent activity in vivo. The RNA processing data in vivo clearly predicts that Gst-Rrp6-11p should function in vitro (Fig. 5 ▶). However, these results do not provide a clear prediction for Gst-Rrp6-13p because strains producing it fail to efficiently process the 3′ ends of 5.8S and U24 pre-RNAs, but do degrade 5′ ETS (Figs. 5 ▶, 6 ▶). Repeated attempts to purify active enzyme from these mutants resulted in inactive preparations, indicating that their purification may result in loss of exonuclease activity.
DISCUSSION
The experiments reported here reveal functional information about the domain structure of Rrp6p, a nuclear 3′–5′ riboexonuclease that functions as a subunit of the exosome in yeast. Functional analysis of specific Rrp6p mutants identified motifs required for riboexonuclease activity, efficient nuclear localization, and discrimination between RNA substrates destined for 3′ end processing or degradation.
Rrp6p functions in the nucleus of yeast cells as a riboexonuclease required for 3′ end processing of 5.8S pre-rRNA and a variety of snRNAs and snoRNAs. In these cases, Rrp6p appears to act downstream of the core exosome because defects in core components cause the accumulation of pre-RNAs with 3′ extensions longer than those found in RRP6 mutants (Allmang et al. 1999a,b; van Hoof et al. 2000). Rrp6p belongs to the ribonuclease D family of 3′–5′ riboexonucleases and bears striking homology with the 3′–5′ deoxyriboexonuclease domain of E. coli DNA polymerase I (Moser et al. 1997). Comparison of the crystal structure of DNA polymerase I with functional analysis of mutants with site-directed amino acid changes within the active site produced a model in which negatively charged side-chains coordinate two metal ions required for phosphodiester bond cleavage (Beese and Steitz 1991). One metal ion acts as a Lewis acid to deprotonate a water molecule that carries out nucleophilic attack on the phosphodiester backbone between the final and penultimate nucleosides. Site-specific mutagenesis and mapping of the metal ion binding sites in the mammalian poly(A)-nuclease PARN recently provided strong evidence that it uses a similar two-ion mechanism of phosphodiester bond cleavage (Ren et al. 2002). Rrp6p, like other members of the RNase D family, including PARN, contains all of the conserved amino acids expected for enzymes by using this mechanism as well as conserved spacing between each side-chain. Analysis of Rrp6p mutants in which each of these side-chains was changed to alanine showed that the altered enzymes were defective in vivo and in vitro. In particular, the conservative change of tyrosine 361 to phenylalanine abolished Rrp6p activity, as does the homologous change (Y497F) in DNA polymerase I (Derbyshire et al. 1991). The phenolic hydroxyl of tyrosine 497 in DNA polymerase I is thought to act as a proton donor that orients and stabilizes the activated hydroxyl nucleophile. Replacement of the hydroxyl with hydrogen in Rrp6p and DNA polymerase I resulted in substantial loss of exonuclease activity. The similarities of the phenotypes of each of the exonuclease motif mutations indicate that each plays an essential role in the catalytic activity of the enzyme and supports the hypothesis that these motifs fold to form the active site domain in the three-dimensional structure of the protein in a manner similar to that demonstrated for the exonuclease domain of DNA polymerase I. Thus, the analysis of the function of conserved amino acid side-chains within each of the three predicted exonuclease motifs of Rrp6p is consistent with the two-metal ion mechanism of phosphodiester bond cleavage and provides evidence that this mechanism operates in riboexonucleolytic and deoxyriboexonucleolytic enzymes (Moser et al. 1997).
Rrp6p also contains a less-conserved domain called HRDC found in other proteins that interact with nucleic acids, in particular the Werner’s and Bloom’s Syndrome proteins, Saccharomyces cerevisiae Sgs1p, and E. coli RNase D (Morozov et al. 1997). The HRDC domain may play a role in nucleic acid binding based on the NMR spectra chemical shifts induced upon addition of DNA to the HRDC domain of Sgs1p (Liu et al. 1999). We have been unable to observe stable binding of Rrp6p or nuclease-deficient mutants to RNA by gel electrophoretic mobility analysis or nitrocellulose filter binding assays. Nevertheless, our results indicate that this domain may play an important role in the activity of Rrp6p. In particular, the rrp6-13 (D457A) and the rrp6-14 mutation (ΔHRDC) reduce the ability of the enzyme to carry out 5.8S rRNA and snoRNA 3′ end formation. Both of these mutations also abolish enzymatic activity in vitro, indicating that the HRDC region may play a critical role in Rrp6p function. On the other hand, the rrp6-13 mutation does not significantly affect the degradation of the 5′ ETS RNA or poly(A)+ snoRNAs intermediates in vivo, indicating that the mutant enzyme may play a direct or indirect role in the degradation of these molecules. The 5.8S rRNA and U24 snoRNA processing defects observed for the rrp6-13 mutation represent the loss of Rrp6p-specific 3′ end processing activities because these defects do not occur in other exosome mutants (Allmang et al. 1999a; van Hoof et al. 2000). In contrast, accumulation of 5′ ETS RNA, occurs in all exosome mutants tested, indicating that degradation of this fragment may be a highly cooperative process. The ability of the rrp6-13 mutants to degrade 5′ ETS RNA, but not to efficiently process the 3′ ends of 5.8S rRNA or snoRNAs, indicates that 5′ ETS degradation may not require the exonuclease activity of Rrp6p. However, each of the exonuclease motif mutants fail to process 5.8S rRNA and U24 snoRNA, as well as fail to degrade 5′ETS RNA. This apparent contradiction may reflect the possibility that the presence of catalytically inactive Rrp6p in the exosome could negatively affect the ability of the complex to degrade RNAs, such as 5′ ETS. These considerations indicate that the rrp6-13 mutation may allow Rrp6p to function as an allosteric activator of the exosome, but not as a riboexonuclease in Rrp6p-specific 3′ end processing reactions.
Alternatively, the ability of the rrp6-13 mutant to degrade 5′ ETS could indicate that the enzyme is active in vivo and that its inactivity in vitro reflects technical problems during protein purification. In this view, the mutant enzyme would fail to efficiently process 5.8S rRNA and snoRNAs in vivo for reasons other than loss of exonuclease activity, such as inability to efficiently bind the substrate. This binding model differs from the allosteric model proposed above in that it predicts that combining the rrp6-13 mutation and an exonuclease motif mutation, such as rrp6-3 would yield a double mutant unable to degrade 5′ ETS, and unable to process the 3′ ends of 5.8S RNA or snoRNAs. In contrast, the allosteric model predicts that the double mutant should have the same phenotype as rrp6-13 alone. Analysis of the double mutant (rrp6-16) revealed defects in both the 3′ end processing and degradation functions of Rrp6p, indicating that both of these functions require the riboexonuclease activity of the enzyme. These findings indicate that the rrp6-13 mutation affects a domain required for the efficient activation of Rrp6p in its role as an RNA 3′ end processing factor, but not in its role in RNA degradation. Thus, this mutation may allow the classification of Rrp6p mutant phenotypes into those resulting from inhibition of RNA 3′ end processing or RNA degradation. In this light, the poly(A)+ snoRNAs observed in the rrp6-3, but not the rrp6-13, mutant would represent molecules destined for degradation rather than precursors destined to become mature snoRNAs. Experiments currently underway in our laboratory focus on the comparison of the mRNA processing and localization phenotypes of the rrp6-3 and rrp6-13 mutations.
Finally, comparison of the growth and molecular phenotypes of these mutants raises questions about the essential function of Rrp6p. The riboexonuclease domain mutations confer cold-sensitive growth and fail to process or degrade RNA substrates. Their ability to complement the heat sensitivity of rrp6-Δ implies that this phenotype does not result from loss of the known enzymatic functions of Rrp6p, or growth at high temperature requires only very low Rrp6p activity. The rrp6-13 mutation in the HRDC domain affects processing, but not degradation, and does not cause either heat or cold sensitivity. This indicates that the cold sensitivity caused by the riboexonuclease domain mutations does not result merely from the inability of the cell to degrade aberrant RNAs in the nucleus because an rrp6-Δ strain fails to degrade RNAs but is not cold sensitive. This view indicates that the cold-sensitive phenotype of the exonuclease domain mutants may require the presence of an enzyme that fails to function in the degradation pathways of the exosome. Indeed, cold sensitivity often reflects an assembly defect and indicates that Rrp6p exonuclease domain mutants may form incomplete or inactive exosomes. Such mutants often result in a dominant phenotype when expressed in the presence of the normal allele; yet, expression of the exonuclease domain mutants in an RRP6 background does not cause obvious cold sensitivity. Despite the lack of an observable growth defect, we observed a slight rRNA processing defect in an RRP6/rrp6-1 (D238N) strain, indicating partial dominance of a cold-sensitive exonuclease domain mutation (Briggs et al. 1998).
In summary, this report presents evidence for three functional domains for the riboexonuclease Rrp6p that are required for efficient nuclear localization of the protein, a two-metal ion exonuclease activity, and the ability to discriminate between RNA substrates destined for 3′ end processing or degradation.
MATERIALS AND METHODS
Plasmids and site-directed mutagenesis
Mutations in RRP6 were made by oligonucleotide-directed site-specific mutagenesis (Kunkel et al. 1991) on ssM13mp18RRP6, which contains the KpnI-EcoRI fragment from pRST66 (Burkard and Butler 2000). First, a silent mutation producing a HindIII site was created by changing A1136 to a G (where 1 is the A in the RRP6 start codon), which produced M13mp18RRP6H. Subsequently, the following changes were made to M13mp18RRP6H: A712C to produce rrp6-3 (D238A); A718C to produce rrp6-4 (E240A); A866C to produce rrp6-5 (D296A); T1080G, A1081C to produce rrp6-7 (Y361A); A1081T to produce rrp6-8 (Y361F); A1093C to produce rrp6-9 (D365A); T1341G, G1342C to produce rrp6-10 (W448A); A1344G, G1345C to produce rrp6-11 (R449A); A1366C to produce rrp6-12 (D456A); and A1369C to produce rrp6-13 (D457A). Mutations were verified by DNA sequence analysis of the entire region between the KpnI (634 bp) and NsiI (1524 bp) sites. Sequence analysis of putative A866C mutants revealed one isolate containing an additional A1081T mutation, which was named rrp6-9 (Y361A, E240A). Plasmids for expression of Gst-Rrp6p in E. coli were constructed by replacing the KpnI-EcoRI fragment of pRST66 (Burkard and Butler 2000) with the corresponding fragment from M13mp18RRP6H to produce pRST66H (HindIII), followed by replacement of the KpnI-NsiI fragment of pRST66H with the HindIII-NsiI fragments of the appropriate M13mp18RRP6H mutants to produce pRST66H1 (D238A), pRST66H2 (E240A), pRST66H3 (D296A), pRST66H4 (Y361A), pRST66H5 (Y361F), pRST66H6 (D365A), pRST66H24 (Y361A, E240A), pRST66H7 (W448A), pRST66H8 (R449A), pRST66H9 (D456A), and pRST66H10 (D457A).
Yeast centromere plasmids expressing each of the RRP6 mutants fused to green fluorescent protein (GFP) were constructed by gap repair (Orr-Weaver et al. 1983). The rrp6-3 to rrp6-10 mutations were transferred to pGFP-FOR11 (CEN6, URA3, MET17::GFP; Burkard and Butler 2000) by cotransformation of BPO2-KAN (rrp6::KAN, ura3-52; Burkard and Butler 2000) with BglII-linearized pGFP-FOR11 and the appropriate BamH1 fragments from pRST66H thru pRST66H24. Clones were selected on SCD–URA plates, the plasmids were screened for the presence of the HindIII marker, and the DNA sequence of the repaired region was verified. The resulting plasmids were named pGFP-RRP6H1 (D238A), pGFP-RRP6H2 (E240A), pGFP-RRP6H3 (D296A), pGFP-RRP6H4 (Y361A), pGFP-RRP6H5 (Y361F), pGFP-RRP6H6 (D365A), and pGFP-RRP6H24 (Y361A, E240A). The rrp6-14 deletion mutation was created by PCR-deletion mutagenesis of pGFP-RRP6H, which replaced nucleotides 1323 to 1456 of RRP6 with a ClaI restriction site to create pGFP-RRP6ΔHRDC. The entire RRP6 open reading frame of pGFP-RRP6ΔHRDC was verified by DNA sequence analysis. Point mutations were inserted into the deleted region by cotransforming ClaI-linearized pGFP-RRP6ΔHRDC with the appropriate BamH1 fragments from pRST66H7 to pRST66H10 to create pGFP-RRP6H7 (W448A), pGFP-RRP6H8 (R449A), pGFP-RRP6H9 (D456A), and pGFP-RRP6H10 (D457A). The entire DNA sequence of each gap-repaired region was verified after recovery of the plasmids.
The rrp6-16 allele was constructed by replacing the BglII fragment of pGFP-RRP6H10 with the BglII fragment containing the rrp6-3 mutation to produce pGFP-RRP6H01.
Northern blot analysis
Total RNA or poly(A)+ RNA was isolated from yeast strains grown to an A600 of 0.5–2.0 as described (Patel and Butler 1992), and Northern blot analysis was carried out as described in Briggs et al. 1998. 5′ 32P-labeled DNA oligonucleotide probes for 5.8S and SCR1 RNA were described in Briggs et al. (1998). U24 was detected with OSB 138 (5′-TCAGAGATCTTGGTGATAAT-3′), snR72 was detected with OSB267 (5′-ATCAGACTGACGTG CAAATCAT-3′), snR40 was detected with OSB275 (5′-ACCT GAGTACTTGTGGCATC-3′), and snR77 was detected with OSB280 (5′-CTCGTTCAGCCAGTAATTCCAGC-3′).
Purification and assay of Gst-Rrp6p
Gst-Rrp6p was purified from E. coli carrying the pRST66H-series of plasmids as previously described (Burkard and Butler 2000). The purified proteins were dialyzed against 100 volumes of dialysis buffer (100 mM Tris-Cl at pH 7.5, 5 mM Mg2acetate, 1 mM DTT, and 50% glycerol) and stored at −20°C. Riboexonuclease assays were carried out at 30°C in 60 μL reactions containing Gst-Rrp6p (4 nM) and 5′ 32P-labeled RNA 11mer (GACGUAUGGUC; 12 nM) in 10 mM Tris-Cl (pH 7.5), 12.5 mM KOAc, 5 mM Mg2OAc, 1 mM DTT, and 5% glycerol. Ten μL samples were removed at the appropriate times and mixed with 5 μL 40 mM Na2EDTA to stop the reaction, and the products were separated by electrophoresis on a 16% polyacrylamide and 8 M urea gel. The reaction products were analyzed by storage PhosphorImager analysis.
Acknowledgments
We are grateful to Mark Dumont, Elizabeth Grayhack, and Eric Phizicky for helpful discussions, and to the members of our laboratory for comments on the manuscript. We thank Nathaniel Hanson for excellent technical assistance. This work was supported by a grant from the Public Health Service (NIH-GM59898). The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Allmang, C., Kufel, J., Chanfreau, G., Mitchell, P., Petfalski, E., and Tollervey, D. 1999a. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18: 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang, C., Petfalski, E., Podtelejnikov, A., Mann, M., Tollervey, D., and Mitchell, P. 1999b. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes & Dev. 13: 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang, C., Mitchell, P., Petfalski, E., and Tollervey, D. 2000. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 28: 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.S.J. and Parker, R.P. 1998. The 3prime to 5prime degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, Y., Takahashi, S., Kobayashi, T., Kajiho, H., Hoshino, S., and Katada, T. 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 20: 4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese, L.S. and Steitz, T.A. 1991. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: A two metal ion mechanism. EMBO J. 10: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese, L.S., Friedman, J.M., and Steitz, T.A. 1993. Crystal structures of the Klenow fragment of DNA polymerase I complexed with deoxynucleoside triphosphate and pyrophosphate. Biochemistry 32: 14095–14101. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli, C., Presutti, C., and Tollervey, D. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775. [DOI] [PubMed] [Google Scholar]
- Briggs, M.W., Burkard, K.T., and Butler, J.S. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273: 13255–13263. [DOI] [PubMed] [Google Scholar]
- Brown, J.T., Bai, X., and Johnson, A.W. 2000. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA 6: 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard, K.T. and Butler, J.S. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 20: 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J.S. 2002. The yin and yang of the exosome. Trends Cell. Biol. 12: 90–96. [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464. [DOI] [PubMed] [Google Scholar]
- Derbyshire, V., Grindley, N.D., and Joyce, C.M. 1991. The 3′-5′ exonuclease of DNA polymerase I of Escherichia coli: Contribution of each amino acid at the active site to the reaction. EMBO J. 10: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A., Morlando, M., and Bozzoni, I. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19: 6218–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischmeyer, P.A., van Hoof, A., O’Donnell, K., Guerrerio, A.L., Parker, R., and Dietz, H.C. 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261. [DOI] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Roshbash, M., Parker, P., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing requires the nuclear exosome. Nature 413: 538–542. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A., Bebenek, K., and McClary, J. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204: 125–139. [DOI] [PubMed] [Google Scholar]
- Liu, Z., Macias, M.J., Bottomley, M.J., Stier, G., Linge, J.P., Nilges, M., Bork, P., and Sattler, M. 1999. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure Fold Des. 7: 1557–1566. [DOI] [PubMed] [Google Scholar]
- Mian, I.S. 1997. Comparative sequence analysis of ribonucleases HII, III, II, PH and D. Nucleic Acids Res. 25: 3187–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2000. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 7: 843–846. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., and Tollervey, D. 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466. [DOI] [PubMed] [Google Scholar]
- Morozov, V., Mushegian, A.R., Koonin, E.V., and Bork, P. 1997. A putative nucleic acid-binding domain in Bloom’s and Werner’s syndrome helicases. Trends Biochem. Sci. 22: 417–418. [DOI] [PubMed] [Google Scholar]
- Moser, M., Holley, W., Chatterjee, A., and Mian, I. 1997. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 25: 5110–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, D., Gao, M., O’Connor, J.P., Raijmakers, R., Pruijn, G., Lutz, C.S., and Wilusz, J. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T.L., Szostak, J.W., and Rothstein, R.J. 1983. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 101: 228–245. [DOI] [PubMed] [Google Scholar]
- Patel, D. and Butler, J.S. 1992. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell. Biol. 12: 3297–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Y.G., Martinez, J., and Virtanen, A. 2002. Identification of the active site of poly(A)-specific ribonuclease by site-directed mutagenesis and Fe2+-mediated cleavage. J. Biol. Chem. 277: 5982–5987. [DOI] [PubMed] [Google Scholar]
- van Hoof, A. and Parker, R. 1999. The exosome: A proteasome for RNA? Cell 99: 347–350. [DOI] [PubMed] [Google Scholar]
- van Hoof, A., Lennertz, P., and Parker, R. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof, A., Frischmeyer, P.A., Dietz, H.C., and Parker, R. 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264. [DOI] [PubMed] [Google Scholar]
- Venema, J. and Tollervey, D. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33: 261–311. [DOI] [PubMed] [Google Scholar]
- Widner, W.R. and Wickner, R.B. 1993. Evidence that the SKI antiviral system of Saccharomyces cerevisiae acts by blocking expression of viral mRNA. Mol. Cell. Biol. 13: 4331–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]