Abstract
Synthetic analogs of the 5′-terminal caps of eukaryotic mRNAs and snRNAs are used in elucidating such physiological processes as mRNA translation, pre-mRNA splicing, intracellular transport of mRNA and snRNAs, and mRNA turnover. Particularly useful are RNAs capped with synthetic analogs, which are produced by in vitro transcription of a DNA template using a bacteriophage RNA polymerase in the presence of ribonucleoside triphosphates and a cap dinucleotide such as m7Gp3G. Unfortunately, because of the presence of a 3′-OH on both the m7Guo and Guo moieties, up to half of the mRNAs contain caps incorporated in the reverse orientation. Previously we designed and synthesized two “anti-reverse” cap analogs (ARCAs), m73′dGp3G and m27,3′-OGp3G, that cannot be incorporated in the reverse orientation because of modifications at the C3′ position of m7Guo. In the present study, we have synthesized seven new cap analogs modified in the C2′ and C3′ positions of m7Guo and in the number of phosphate residues, m27,2′-OGp3G, m72′dGp3G, m72′dGp4G, m27,2′-OGp4G, m27,3′-OGp4G, m7Gp5G, and m27,3′-OGp5G. These were analyzed for conformation in solution, binding affinity to eIF4E, inhibition of in vitro translation, degree of reverse capping during in vitro transcription, capping efficiency, and the ability to stimulate cap-dependent translation in vitro when incorporated into mRNA. The results indicate that modifications at C2′, like those at C3′, prevent reverse incorporation, that tetra- and pentaphosphate cap analogs bind eIF4E and inhibit translation more strongly than their triphosphate counterparts, and that tetraphosphate ARCAs promote cap-dependent translation more effectively than previous cap analogs.
Keywords: Cap-dependent translation, T7 RNA polymerase, in vitro transcription, capping efficiency, dinucleoside tetra- and pentaphosphates, cap analog inhibition, translational efficiency, anti-reverse cap analogs, eIF4E
INTRODUCTION
The 5′-terminus of all eukaryotic cellular (i.e., not organellar or viral) mRNAs is modified with a 5′–5′ m7GTP moiety, and U-type snRNAs are modified with a 5′–5′ m32,2,7GTP moiety (Mattaj 1986; Furuichi and Shatkin 2000). This modification plays a major role in the function of these RNAs in a variety of cellular processes including translation (Gingras et al. 1999; Rhoads 1999), splicing (Izaurralde et al. 1994), intracellular transport (Izaurralde et al. 1992), and turnover (Beelman et al. 1998). Synthetic cap analogs have played an important role in elucidating these physiological processes. Studies on the inhibition of protein synthesis by cap analogs modified in the m7G, ribose, or phosphate moieties delineated the structural requirements for cap interactions with the cap-binding protein eIF4E (Adams et al. 1978; Darzynkiewicz et al. 1985, 1987, 1989). The conclusions drawn from these studies were further validated when the tertiary structure of eIF4E bound to m7GDP was solved (Marcotrigiano et al. 1997; Matsuo et al. 1997). m32,2,7Gp3G and its analogs were instrumental in the discovery that the cap of some U-type snRNPs serve a signaling role for their nuclear import (Hamm et al. 1990; Fischer et al. 1991). Synthetic mRNAs capped with m32,2,7Gp3G were used to show enhanced translational efficiency in extracts derived from Ascaris lumbricoides (Maroney et al. 1995). Dinucleotide cap analogs were used to show that the cap specificity of the CBC factor involved in pre-mRNA splicing differs from that of eIF4E (Izaurralde et al. 1994). Affinity resins and free cap analogs based on m7GTP and m32,2,7GTP were used to purify five eIF4E isoforms from Caenorhabditis elegans and to show they differed in cap-binding specificity (Jankowska-Anyszka et al. 1998; Keiper et al. 2000; Miyoshi et al. 2002; Stachelska et al. 2002).
Interestingly, some synthetic analogs are more active as inhibitors of translation than the corresponding natural compounds, for example, those created by addition of a methyl group to the N2 group of guanine, replacement of the N7 methyl group with an aryl moiety, substitution of this aryl group at the para position with Cl or F, or addition of phosphate residues to the 5′–5′ bridge in addition to the natural three residues (Cai et al. 1999 and references therein). Some of these modified cap analogs have also been tested for direct binding affinity to eIF4E (Niedzwiecka et al. 2002). The most profound effect is produced by extending the native cap analog m7Gp3G with additional phosphate moieties. The tetraphosphate analog m7Gp4G binds both unphosphorylated and phosphorylated eIF4E with 10-fold higher affinity than m7Gp3G (Niedzwiecka et al. 2002; Zuberek et al. 2003).
In addition to their use as inhibitors of physiological processes, cap analogs have been used in synthetic reactions to create capped mRNAs. This is achieved by in vitro transcription of DNA templates with either a bacterial (Contreras et al. 1982) or bacteriophage (Konarska et al. 1984; Yisraeli and Melton 1989) RNA polymerase in the presence of all four ribonucleoside triphosphates and a cap dinucleotide such as m7Gp3G. The polymerase initiates transcription with a nucleophilic attack by the 3′-OH of the Guo moiety in m7Gp3G on the α-phosphate of the next templated nucleoside triphosphate, resulting in the initial product m7G(5′)p3(5′)GpN. Unfortunately, the nucleophilic attack can also occur by the 3′-OH of m7Guo, producing RNAs containing a “reverse cap,” that is, G(5′)p3(5′)m7GpN (Pasquinelli et al. 1995). Reverse-capped pre-U1 RNA transcripts, when injected into Xenopus laevis oocyte nuclei, are exported more slowly than natural transcripts (Pasquinelli et al. 1995). Similarly, cytoplasmic reverse-capped U1 RNAs fail to be imported into the nucleus.
Previously we designed and synthesized two “anti-reverse” cap analogs (ARCAs), m73′dGp3G and m27,3′-OGp3G, that cannot be incorporated in the reverse orientation because of modifications at the 3′-O position of m7Guo (Stepinski et al. 2001). Transcripts produced with SP6 polymerase using ARCAs were of the predicted length and indistinguishable in size and homogeneity from those produced with m7Gp3G or Gp3G. Analysis of the transcripts with RNase T2 and tobacco acid pyrophosphatase (TAP) indicated that reverse caps were formed with m7Gp3G but not with ARCAs. The ARCAs inhibited cell-free translation with a KI similar to that of m7Gp3G. Most significantly, the in vitro translational efficiency of ARCA-capped transcripts was considerably higher than that of m7Gp3G-capped mRNAs. In a subsequent study, Peng et al. (2002) synthesized m27,3′-OGp3G by a different route and demonstrated that it was incorporated into RNA solely in the normal orientation during in vitro transcription by T7 RNA polymerase.
In the present work, we have tested the idea that cap analogs with higher intrinsic affinity for eIF4E may produce mRNAs with higher translational efficiency if further modified to prevent reverse incorporation. Furthermore, we have examined chemical modifications other than at the 3′-O position of m7Guo that may prevent reverse incorporation. Surprisingly, analogs modified on the 2′-O position of m7Guo are also incorporated solely in the natural orientation, despite the fact that this position does not participate in phosphodiester formation. Some of these new cap analogs yield mRNAs with higher translational efficiency than previously observed.
RESULTS
Synthesis of novel ARCAs
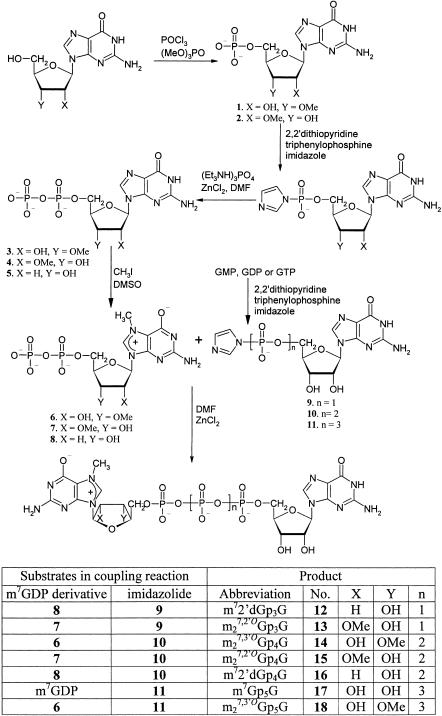
The two ARCAs synthesized previously, m73′dGp3G and m27,3′-OGp3G, behaved equivalently as either inhibitors of translation, when added as the free nucleotide, or as enhancers of translation, when incorporated at the 5′-terminus of mRNA (Stepinski et al. 2001). In the new series, we developed compounds with the same 3′-O-methyl ribose modification as one of these but with either four (Fig. 1 ▶, 14) or five (18) phosphate moieties. To serve as controls, we also synthesized conventional (non-ARCA) analogs with either four (Stepinski et al. 1995) or five (Fig. 1 ▶, compound 17) phosphate moieties. In addition, we synthesized analogs with two other types of ribose modification that might prevent reverse incorporation, 2′-O-methyl and 2′-deoxy, in either the three phosphate (12, 13) or four phosphate (15, 16) series.
FIGURE 1.
Scheme for organic synthesis of novel “anti-reverse” cap analogs.
Chemical synthesis of the new cap analogs (Fig. 1 ▶) was performed by a strategy similar to that developed previously (Stepinski et al. 2001). To obtain a 5′,5′-tetraphosphate linkage (14–16), the final coupling reaction involved modified 7-methylated nucleoside diphosphates and the imidazolide derivative of GDP (10). The coupling reaction in dimethylformamide (DMF) mediated by ZnCl2 occurred efficiently (75%–90%) without significant accumulation of by-products, which is extremely important because of purification problems common for this class of cap analogs. The methodology was extended to the synthesis of analogs with a 5′,5′-pentaphosphate bridge (17, 18). For this purpose, we developed a synthetic scheme for a novel compound, guanosine 5′-triphosphate P3-imidazolide (11). The structure of this imidazolide was confirmed indirectly after coupling with m7GMP because of its instability. The product was found to have the same retention time as an m7Gp4G standard (Stepinski et al. 1995) in both ion-exchange and reverse-phase HPLC systems. Intermediate 11 permitted synthesis of the unsymmetrical dinucleoside pentaphosphates 17 and 18 and even hexa- and heptaphosphates (J. Jemielity, unpubl.).
Solution conformations of the free cap analogs
The solution conformations assumed by the ribose moieties of the new cap analogs were determined from 1H NMR measurements (Table 1 ▶). The structural modifications alter the conformer populations only slightly from those of natural dinucleotide caps (Darzynkiewicz et al. 1990; Wieczorek et al. 1997) and previously synthesized ARCAs (Stepinski et al. 2001). Irrespective of the modifications in the m7Guo moiety, the populations of the Guo conformers in dynamic equilibrium are similar to one another (±5%) and close to the populations in 5′-GMP (Lassota et al. 1984). A characteristic increase in the amounts of the N (C3′-endo), +sc (gauche orientation of C5′ to O4′ and C3′), and ap (trans orientation of C4′ and Pα) conformers is observed upon methylation at N7 in all analogs. Methylation at C2′ and C3′ of m7Guo decreases the ribose N population only slightly (≤10%), but the 2′- and 3′-deoxy substitutions have a more profound effect, increasing the N population as much as 35%.
TABLE 1.
Conformer populations at 20°C in the dynamic N―S equilibria of the sugar ring (%N) and about C4′–C5′ (% + sc) and C5′–O5′ (%ap) bonds (±5%) derived by NMR, and populations of the stacked forms derived by fluorescence measurements (% stacking)
| Cap analog | %N | % + sc | %ap | % stacking | |
| m7Gp3G | m7Guo | 65a | 90a | 76a | 61.42 ± 0.06 |
| Guo | 34a | 55a | 63a | ||
| m27,3′-OGp3G | m27,3′-OGuo | 56b | 80b | 74b | 58.11 ± 0.07 |
| Guo | 36b | 54b | 66b | ||
| m27,2′-OGp3G (13) | m27,2′-OGuo | 65 | 90c | 80c | 56.85 ± 0.11 |
| Guo | 35 | 60c | 60c | ||
| m73′dGp3G | m73′dGuo | 100b | 80b | 72b | 62.73 ± 0.07 |
| Guo | 37b | 55b,c | 66b,c | ||
| m72′dGP3G (12) | m72′dGuo | 50 | 75c | 70c | 61.61 ± 0.07 |
| Guo | 37 | 55 | b | ||
| m7Gp4G | m7Guo | 61 | 85c | 75c | 48.45 ± 0.11 |
| Guo | 35 | 55c | 65c | ||
| m72′dGp4G (16) | m72′dGuo | 45c | 63 | 68 | 54.88 ± 0.07 |
| Guo | 32 | 55 | —d | ||
| m27,2′-OGp4G (15) | m27,2′-OGuo | 58 | —d | —d | 47.57 ± 0.08 |
| Guo | 33 | —d | —d | ||
| m27,3′-OGp4G (14) | m27,3′-OGuo | 51 | 77 | 74 | 50.02 ± 0.09 |
| Guo | 33 | 55c | —d | ||
| m7Gp5G (17) | m7Guo | 55 | 80c | 80c | 44.98 ± 0.10 |
| Guo | 29 | 60c | 75c | ||
| m27,3′-OGp5G (18) | m27,3′-OGuo | 51 | 75c | —d | 50.12 ± 0.08 |
| Guo | 30 | 62 | 68 | ||
aData from Darzynkiewicz et al. (1990).
bData from Stepinski et al. (2001).
cApproximate value.
dNot determined because of signal overlapping.
The populations of the stacked and unstacked forms in dynamic equilibrium were derived from fluorescence measurements (Table 1 ▶). Internal stacking was similar for all analogs and ranged from 45% to 63%. Stacking was less for dinucleotides with four- and five-member phosphate chains. This conformational feature may contribute to more efficient binding of the analogs to eIF4E (see the following section).
Binding affinity of cap analogs for eIF4E
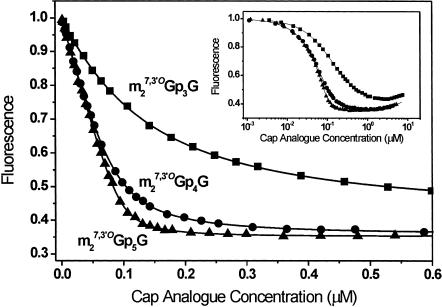
The binding affinity of the new cap analogs to mouse eIF4E (28–217) (i.e., missing the N-terminal 27 amino acid residues) was determined by quenching of intrinsic Trp fluorescence. Representative data are shown in Figure 2 ▶ for three cap analogs modified identically in the ribose ring of m7Guo but containing three (Stepinski et al. 2001), four (14), or five (18) phosphate moieties. The apparent plateaus arise from two effects that cancel each other out: a decrease of the intrinsic eIF4E fluorescence upon cap binding and an increase in the fluorescence of the free cap analog over the course of titration. The real plateaus cannot be detected graphically but must be calculated (Niedzwiecka et al. 2002). The calculated maximal eIF4E fluorescence quenching is approximately the same for each cap analog (~65%). Emission of the free cap does not influence the equilibrium association constant (KAS) values because it is explicitly taken into account in the fitting equation.
FIGURE 2.
Quenching of intrinsic Trp fluorescence in mouse eIF4E (28–217) with m27,3′-OGp3G (Stepinski et al. 2001), m27,3′-OGp4G (compound 14), and m27,3′-OGp5G (compound 18). Binding affinities for cap analogs to eIF4E were calculated as described in Materials and Methods.
The KAS values and free energy of binding of all new cap analogs, calculated from such data, are presented in Table 2 ▶ together with the same data for previously synthesized ARCAs (Stepinski et al. 2001) for comparison. Extension of the polyphosphate chain beyond the natural triphosphate results in a marked enhancement of binding. For instance, the dinucleoside tetraphosphate analogs in each ribose-modification subset bind eIF4E with higher affinity than their triphosphate counterparts (Table 2 ▶, m7Gp4G versus m7Gp3G, 8.9-fold; m27,2′-OGp4G versus m27,2′-OGp3G, 9.2-fold; etc.). Similarly, the pentaphosphate analogs bind eIF4E with higher affinity than their tetraphosphate counterparts (m7Gp5G versus m7Gp4G, 4.9-fold; m27,3′-OGp5G versus m27,3′-OGp4G, 3.5-fold). This has previously been reported for the tri- and tetraphosphate analogs with unmodified m7Guo moieties (Zuberek et al. 2003) but not for m27,2′-OGuo-, m27,3′-OGuo-, or m72′dGuo-containing analogs.
TABLE 2.
Equilibrium association constants (KAS) and binding free energies (ΔG°) for the binding of murine eIF4E (28–217) to dinucleoside tri-, tetra-, and pentaphosphate cap analogs, as determined by fluorescence quenching
| Cap analog | Number of assays | KAS × 10−6 (M−1)a | ΔG° (kcal/mole) |
| Triphosphate series | |||
| m7Gp3G | 9 | 12.5 ± 0.3 | 9.52 ± 0.01 |
| m27,2′-OGp3G (13) | 4 | 10.8 ± 0.3 | 9.43 ± 0.02 |
| m27,3′-OGp3G | 5 | 10.2 ± 0.3 | 9.40 ± 0.02 |
| m72′dGp3G (12) | 4 | 9.1 ± 0.5 | 9.33 ± 0.03 |
| m73′dGp3G | 4 | 13.1 ± 0.7 | 9.54 ± 0.03 |
| Tetraphosphate series | |||
| m7Gp4G | 8 | 110.6 ± 4.2 | 10.78 ± 0.02 |
| m27,2′-OGp4G (15) | 3 | 99.8 ± 6.0 | 10.73 ± 0.04 |
| m27,3′-OGp4G (14) | 3 | 84.5 ± 2.6 | 10.63 ± 0.03 |
| m72′dGp4G (16) | 3 | 88.0 ± 2.9 | 10.65 ± 0.03 |
| Pentaphosphate series | |||
| m7Gp5G (17) | 11 | 543 ± 55 | 11.71 ± 0.06 |
| m27,3′-OGp5G (18) | 8 | 299 ± 20 | 11.36 ± 0.04 |
aThe final KAS value for each cap analog was calculated as a weighted average of independent titrations.
In contrast, the affinities of C2′- and C3′-modified dinucleoside triphosphate analogs for eIF4E are similar to those of the parent m7Gp3G. The KAS of m73′dGp3G, m27,3′-OGp3G, m72′dGp3G, m27,2′-OGp3G, and m7Gp3G differ within only ±20% (Table 2 ▶). Similarly, no major difference is observed for the various ribose-modified forms within the tetraphosphate cap analogs (±15%). We therefore conclude that, unlike the phosphate chain extension, neither C2′ nor C3′ modifications of the m7Guo ribose moieties have much influence on cap analog affinity for eIF4E.
Novel cap analogs as inhibitors of cap-dependent translation
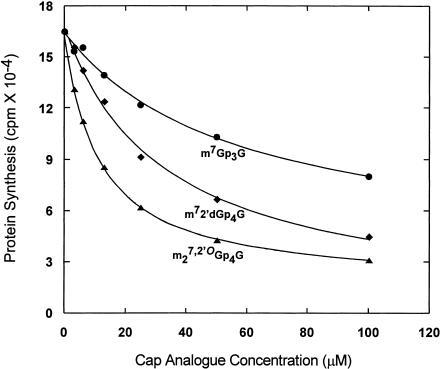
The ability of the new cap analogs to inhibit cap-dependent translation was assayed in a rabbit reticulocyte lysate system programmed with natural rabbit globin mRNA. Representative data for two analogs, m27,2′-OGp4G (15) and m72′dGp4G (16), are shown in Figure 3 ▶ in comparison with the physiological cap, m7Gp3G. Data for inhibition of translation were fit with a theoretical curve that describes cap-dependent translation as a function of a competitive inhibitor of mRNA binding (Cai et al. 1999). The curve fitting yields a value for KI, which is the cap analog concentration at which cap-dependent translation is inhibited 50%. Despite the fact that 15 and 16 bind to eIF4E with similar affinities (Table 2 ▶; KAS = 99.8 ± 6.0 versus 88.0 ± 2.9 × 106 M−1), 15 was considerably more inhibitory for cap-dependent translation than 16 (KI = 3.07 ± 0.31 versus 11.29 ± 0.85 μM).
FIGURE 3.
Inhibition of globin mRNA translation in rabbit reticulocyte lysate by m7Gp3G (•), m27,2′-OGp4G (compound 15; ▴), and m72′dGp4G (compound 16; ♦). Natural rabbit globin mRNA was translated at 5 μg/mL in a rabbit reticulocyte lysate system, and globin synthesis was detected by incorporation of [3H]Leu into protein as described in Materials and Methods.
KI values for all new cap analogs are shown in Table 3 ▶. Addition of a methyl group to either C2′ or C3′ improved the ability of cap analogs to inhibit cell-free translation, regardless of whether the series was triphosphates (m27,2′-O Gp3G or m27,3′-OGp3G versus m7Gp3G), tetraphosphates (m27,2′-OGp4G or m27,3′-OGp4G versus m7Gp4G), or pentaphosphates (m27,3′-OGp5G versus m7Gp5G). In contrast, substitution of H for OH at C2′ or C3′ produced either a poorer inhibitor, in the case of triphosphates (m72′dGp3G or m73′dGp3G versus m7Gp3G), or had no significant effect, in the case of tetraphosphates (m72′dGp4G versus m7Gp4G). The largest effect, however, was caused by the number of phosphates themselves. For ribose-unmodified m7Guo, the pentaphosphate (m7Gp5G) was more inhibitory than the tetraphosphate (m7Gp4G), which was in turn more inhibitory than the triphosphate (m7Gp3G). Likewise, for the 7,3′-O-dimethylguanosine series, the pentaphosphate (m27,3′-O Gp5G) was more inhibitory than the tetraphosphate (m27,3′-OGp4G), which was in turn more inhibitory than the triphosphate (m27,3′-OGp3G). In the 2′-deoxyguanosine series, the tetraphosphate (m72′dGp4G) was more inhibitory than the triphosphate (m72′dGp3G).
TABLE 3.
Inhibitory constants (KI) for inhibition of cap-dependent translation by dinucleoside tri-, tetra-, and pentaphosphate cap analogs in a rabbit reticulocyte lysate translation system
| Cap analog | Number of assays | KI (μM) |
| Triphosphate series | ||
| m7Gp3G | 13 | 17.1 ± 1.0a |
| m27,2′-OGp3G (13) | 3 | 14.1 ± 0.9 |
| m27,3′-OGp3G | 3 | 14.3 ± 1.3 |
| m72′dGp3G (12) | 2 | 27.2 ± 1.6 |
| m73′dGp3G | 3 | 27.8 ± 7.1b |
| Tetraphosphate series | ||
| m7Gp4G | 2 | 10.8 ± 0.8 |
| m27,2′-OGp4G (15) | 2 | 3.07 ± 0.31 |
| m27,3′-OGp4G (14) | 3 | 6.29 ± 0.93 |
| m72′dGp4G (16) | 3 | 11.3 ± 0.9 |
| Pentaphosphate series | ||
| m7Gp5G (17) | 3 | 4.91 ± 0.10 |
| m27,3′-OGp5G (18) | 3 | 3.49 ± 0.32 |
aData from Cai et al. (1999).
bData from Stepinski et al. (2001).
Cap dinucleotides modified at the 2′-O position of m7Guo are incorporated into RNA exclusively in the correct orientation
Because transcription by RNA polymerases in the presence of m7Gp3G is initiated with a nucleophilic attack by the 3′-OH of either m7Guo or Guo on the electrophilic α-phosphate of the first templated nucleoside triphosphate, we initially prevented reverse-orientation caps (Gp3m7GpN) by eliminating the 3′-OH group on m7Guo (Stepinski et al. 2001). Both m73′dGp3G and m27,3′-OGp3G were incorporated solely in the normal orientation, yielding the initial product of m7Gp3GpN. We reasoned that RNA polymerases are confronted with both ribonucleoside triphosphates and 2′-deoxyribonucleoside triphosphates in vivo, yet they incorporate only the former into RNA. We therefore tested compound 12, a cap dinucleotide that contains a 2′-deoxyribose in the m7Guo but not Guo moiety, as an initiator of RNA synthesis.
The strategy for determining incorporation of the cap analog was the same as described previously (Stepinski et al. 2001). Briefly, RNA was synthesized in the presence of cap dinucleotide, all four ribonucleoside triphosphates, and [α-32P]GTP. Any nucleotide on the 5′-side of a G residue acquires a 32P-labeled 3′-phosphate group by nearest neighbor transfer. Digestion of the product with RNase T2 yields two types of product, nucleoside-3′-monophosphates and cap structures containing a 3′-phosphate, both of which are labeled if located 5′ to a G residue. The DNA template used for in vitro transcription specified a G residue in the first templated position. Thus, for RNAs incorporating m7Gp3G in the normal orientation, the product of RNase T2 digestion is m7Gp3Gp* (where p* represents 32P), but for reverse orientation, it is Gp3m7Gp*. Additional digestion of the RNase T2 product with TAP yields two alternate labeled products, pGp* for normal orientation and pm7Gp* for reverse orientation. These nucleoside diphosphates differ in both charge and mass, because the m7 group confers a positive charge on G (Hendler et al. 1970), and are separable by ion-exchange HPLC.
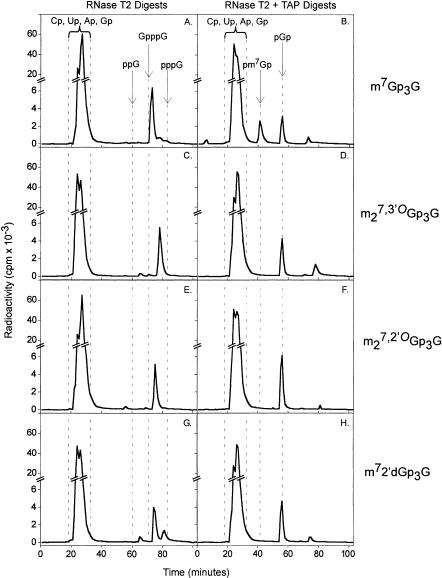
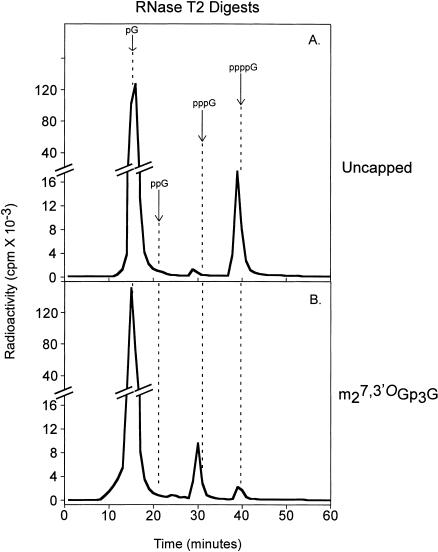
As shown in Figure 4 ▶, the RNA synthesized in the presence of the conventional cap m7Gp3G yielded a radioactive peak upon RNase T2 digestion that was retained slightly longer (73 min) than the chromatographic standard Gp3G (Fig. 4A ▶), which is consistent with the more negative charge of either m7Gp3Gp or Gp3m7Gp. Digestion with both RNase T2 and TAP converted this to two labeled compounds eluting with the chromatographic standards pGp (56 min) and pm7Gp (41 min; Fig. 4B ▶). The relative amounts of pGp* and pm7Gp* suggest that m7Gp3G had been incorporated in normal and reverse orientations to roughly the same extent. As shown previously (Stepinski et al. 2001), a cap analog in which the 3′-OH of m7Guo was blocked with a methyl group, m27,3′-OGp3G, was incorporated exclusively in the normal orientation, indicated by the absence of pm7Gp* (Fig. 4D ▶).
FIGURE 4.
Analysis of in vitro synthesized RNAs by enzymatic digestion with either RNase T2 (A,C,E,G) or RNase T2 plus tobacco acid pyrophosphatase (TAP; B,D,F,H) followed by anion-exchange HPLC on a Partisil 10SAX/25 column. Short mRNAs (see Materials and Methods) capped with (A,B) m7Gp3G, (C,D) m27,3′-OGp3G, (E,F) m27,2′-OGp3G (compound 13), and (G,H) m72′dGp3G (compound 12) were generated by in vitro transcription with T7 RNA polymerase in the presence of [α-32P]GTP. Fractions of 1 mL were collected, and the Cerenkov radiation was determined. The elution times of the following standard compounds, detected by UV absorption, are indicated with arrows: 3′-CMP (Cp), 3′-UMP (Up), 3′-AMP (Ap), 3′-GMP (Gp), 3′,5′-m7GDP (pm7Gp), 3′,5′-GDP (pGp), 5′-GDP (ppG), G(5′)p3(5′)G (GpppG), and 5′-GTP (pppG).
The novel analog m72′dGp3G (12) was also incorporated exclusively in the normal orientation (Fig. 4H ▶). This indicates that T7 polymerase discriminates between the two nucleotide moieties within the cap dinucleotide the same way it distinguishes between ribo- and 2′-deoxyribonucleotide triphosphates during RNA synthesis. We also tested m27,2′-OGp3G (13) in this assay, even though the modification at C2′ is not H, corresponding to the naturally occurring deoxyribonucleotide, but, rather, —OCH3. Interestingly, this analog was also incorporated exclusively in the normal orientation (Fig. 4F ▶).
Capping efficiency for novel ARCAs
As shown above, cap analogs of the tetraphosphate series bind eIF4E more avidly (Table 2 ▶) and inhibit in vitro translation more effectively (Table 3 ▶) than their triphosphate counterparts. Similarly, cap analogs of the pentaphosphate series bind eIF4E and inhibit translation more effectively than their tetraphosphate counterparts. Because mRNAs capped with ARCAs are translated more efficiently in vitro than those capped with unmodified cap analogs (Stepinski et al. 2001), we wished to test the effect of combining both of these modifications, that is, addition of more phosphate moieties together with the ARCA modification of m7Guo. However, it is not obvious that a cap structure that binds more tightly to eIF4E would necessarily lead to increased translational efficiency. In each round of protein synthesis initiation, the cap may first bind to eIF4E but then be released before the next round of initiation can occur (although this has not been experimentally demonstrated). Increased affinity of the cap for eIF4E could conceivably have either a positive or negative effect on translational efficiency.
Another consideration in assessing translational efficiency is the percentage of mRNAs capped. It is conceivable that some novel ARCAs compete with GTP more effectively than others during in vitro transcription, leading to a larger fraction of the mRNAs being capped. This would erroneously give the impression that these ARCA-capped mRNAs have a higher intrinsic translational efficiency. We therefore modified our chromatographic conditions to detect the highly charged guanosine-3′-[32P]phosphate-5′-triphosphate (p3Gp*) expected from RNase T2 digestion of uncapped mRNA. When uncapped mRNA was analyzed, a novel peak appeared (Fig. 5A ▶, fraction 39), very close to the OD standard of guanosine-5′-tetraphosphate (fraction 40), which should have a similar charge to p3Gp. When m27,3′-OGp3G-capped mRNA was analyzed (Fig. 5B ▶), the peak at fraction 39 was diminished and one at fraction 29 appeared. This was retained slightly longer than a peak at fraction 28 seen in some digests (e.g., Fig. 5A ▶) that is presumed to be trace amounts of the RNase T2-undigested NpNp*. The peak at fraction 29 runs at the correct charge for m27,3′-OGp3Gp* and was previously shown to be digested by TAP to pGp* (Stepinski et al. 2001). Based on all of these considerations, we believe that the peaks at fractions 29 and 39 represent the RNase T2 products of capped and uncapped mRNA, respectively. These results indicate that for m27,3′-OGp3G, the percent capping is 71.9%. Considering that the ratio of m27,3′-OGp3G to GTP was 10:1 during in vitro transcription (see Materials and Methods), this means that T7 polymerase discriminates against the cap analog in favor of GTP.
FIGURE 5.
Analysis of percent capping. Short mRNAs either (A) uncapped or (B) capped with m27,3′-OGp3G were generated by in vitro transcription with T7 RNA polymerase in the presence of [α-32P]GTP. The RNAs were digested with RNase T2 followed by anion-exchange HPLC as in Figure 4 ▶ except using a steeper phosphate gradient (see Materials and Methods). Fractions of 1 mL were collected, and the Cerenkov radiation was determined. The elution times of the following standard compounds, detected by UV absorption, are indicated with arrows: 5′-GMP (pG), 5′-GDP (ppG), 5′-GTP (pppG), and guanosine-5′-tetraphosphate (ppppG).
Percent capping was measured for the novel triphosphate and tetraphosphate ARCAs (Table 4 ▶). In the latter case, the RNase T2 digestion products, for example, m27,3′-OGp4Gp*, coeluted with the product from uncapped RNA, pppGp, under the gradient conditions shown in Figure 5 ▶. We therefore changed the pH of the eluting buffer from 3.5 to 4.5 and were able to achieve resolution (see Materials and Methods). Unfortunately, no chromatographic conditions were found to resolve the RNase T2 products of uncapped and pentaphosphate-capped mRNAs, and therefore we do not have percent capping results for the latter mRNAs.
TABLE 4.
Translational efficiency of luciferase mRNAs capped with dinucleoside tri-, tetra-, and pentaphosphate cap analogs in rabbit reticulocyte lysate system
| Cap analog | Overall translational efficiencyb (RLU/ng × 10−5) | Cap-dependent translational efficiencyc (RLU/ng × 10−5) | Relative translational efficiencyd | % cappingf |
| Triphosphate series | ||||
| Gp3G | 0.31 ± 0.03 | N/A | N/A | N/A |
| m7Gp3G | 4.61 ± 0.19 | 4.30 ± 0.19 | 1.00 | 66.9 |
| m27,2′-OGp3G (13) | 9.51 ± 0.45 | 9.21 ± 0.45 | 2.10 ± 0.15 | 62.3 |
| m27,3′-OGp3G | 8.52 ± 1.68 | 8.22 ± 1.68 | 1.88 ± 0.40 | 71.9 |
| m72′dGp3G (12) | 5.41 ± 0.49 | 5.12 ± 0.49 | 1.17 ± 0.13 | 83.1 |
| m73′dGp3G | 2.13 ± 0.5e | |||
| Tetraphosphate series | ||||
| Gp4G | 0.29 ± 0.03 | N/A | N/A | |
| m7Gp4G (1)a | 5.62 ± 0.24 | 5.38 ± 0.25 | 1.32 ± 0.08 | |
| m7Gp4G (2)a | 5.77 ± 0.23 | 5.52 ± 0.23 | 1.35 ± 0.07 | |
| m27,2′-OGp4G (15) | 10.71 ± 0.62 | 10.47 ± 0.62 | 2.56 ± 0.18 | 55.5 |
| m27,3′-OGp4G (14) | 10.14 ± 0.83 | 9.89 ± 0.83 | 2.42 ± 0.22 | 57.2 |
| m72′dGp4G (16) | 8.51 ± 0.74 | 8.13 ± 0.74 | 1.83 ± 0.18 | 61.7 |
| Pentaphosphate series | ||||
| m7Gp5G (17) | 5.03 ± 0.28 | 4.64 ± 0.29 | 1.05 ± 0.08 | |
| m27,3′-OGp5G (18) | 6.56 ± 0.65 | 6.18 ± 0.65 | 1.39 ± 0.16 | |
aTwo different organic syntheses of m7Gp4G were performed, and luciferase mRNA was made from each of them.
bRelative light units (RLU) per nanogram of luciferase mRNA were obtained as described in Figure 6 ▶. Because of the method of normalization, these results represent a single assay performed with three RNA concentrations (as in Fig. 6 ▶). However, between two and four syntheses of capped RNA were conducted for each cap analog, and between two and eight translation reactions were performed for each cap analog, with results comparable to those shown in the table.
cEach value for overall translational efficiency (second column) is corrected for cap-independent translation by subtracting the value for overall translational efficiency of mRNA capped with Gp3G, yielding cap-dependent translational efficiency (third column). Standard error is calculated from the error in overall translational efficiency as described in Blaedel and Meloche (1963).
dEach value for cap-dependent translational efficiency (third column) is normalized by dividing with the value for cap-dependent translational efficiency of mRNA capped with m7Gp3G. Standard error is calculated from the error in cap-dependent translational efficiency as described in Blaedel and Meloche (1963).
eData from Stepinski et al. (2001).
fData were obtained from experiments similar to that shown in Figure 5 ▶. The amount of product derived from a capped mRNA (the peak at fraction 29 of Fig. 5B ▶) is expressed as a percentage of total 5′-terminal structures (sum of the two peaks at fractions 29 and 39).
Translational efficiency of mRNAs capped with novel ARCAs
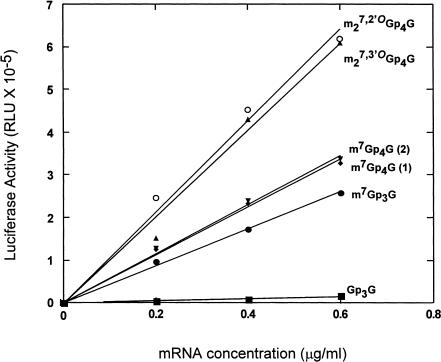
We incorporated the novel cap analogs into luciferase mRNA in vitro using T7 polymerase and translated the RNAs in a rabbit reticulocyte lysate system under conditions of linear production of luciferase with respect to both mRNA concentration and time of incubation. Figure 6 ▶ shows representative data in which m27,2′-OGp4G (15) and m27,3′-OGp4G (14) are compared to the conventional cap m7Gp3G. As a control for the additional phosphates in 14 and 15, we used the non-ARCA tetraphosphate m7Gp4G. To show the reproducibility of this assay, two independent organic syntheses of m7Gp4G were performed, and the resulting cap analogs were used to synthesize two separate preparations of RNA. As shown in Figure 6 ▶, the translational efficiencies of RNAs containing the two cap analogs [m7Gp4G (1) and m7Gp4G (2)] are nearly identical. The assay shows that the non-ARCA tetraphosphate m7Gp4G promotes translation slightly better than the non-ARCA triphosphate m7Gp3G, presumably because of its higher affinity for eIF4E. However, mRNAs capped with the two ARCA tetraphosphates, m27,2′-OGp4G and m27,3′-OGp4G, are considerably more active than any of the non-ARCAs.
FIGURE 6.
Translational efficiency of ARCA-capped mRNAs. Luciferase mRNAs were synthesized in vitro using T7 RNA polymerase and SmaI-digested pluc-A+ in the presence of all four NTPs and Gp3G (▪), m7Gp3G (•), m7Gp4G (first synthesis; ♦), m7Gp4G (second synthesis; ▾), m27,3′-OGp4G (compound 14; ▴), or m27,2′-OGp4G (compound 15; ○), as described in Materials and Methods. The RNAs were translated for 60 min in a rabbit reticulocyte lysate system, and luciferase activity was measured in triplicate in 1-μL aliquots by luminometry; (RLU) relative light units.
Translational efficiencies of mRNAs capped with each of the novel cap analogs are presented in Table 4 ▶. Two types of normalization were applied to the data. First, translation observed in vitro has both cap-dependent and cap-independent components, and the distribution between the two can be influenced by a variety of factors (salt concentration, pH, mRNA concentration, the particular lysate used, etc.; for review, see Rhoads 1985). The amount of cap-independent translation can be determined with a control mRNA capped with Gp3G, which is not recognized by eIF4E. Such a control was included in all translation assays. The luciferase activity per nanogram of mRNA (Table 4 ▶, second column) obtained with Gp3G-capped RNA was subtracted from the overall translational efficiency obtained with the various test cap analogs to yield cap-dependent translational efficiency (Table 4 ▶, third column). Second, there is a certain amount of day-to-day variability between in vitro translations using identical mRNAs and reticulocyte lysate preparations, for reasons that are only partially understood, for example, variable activation of HRI, an eIF2α kinase (Chen 2000). We corrected for this by including a standard mRNA capped with m7Gp3G in each assay. Relative translational efficiency (Table 4 ▶, fourth column) was calculated by dividing cap-dependent translational efficiency for each new cap analog by cap-dependent translational efficiency for m7Gp3G-capped mRNA.
The results indicate that the triphosphate ARCA previously characterized, m27,3′-OGp3G (Stepinski et al. 2001), had a relative translation of 1.88 ± 0.40. It is therefore twice as active as conventional m7Gp3G-capped mRNA (within experimental error), as would be predicted from the fact that approximately half of the caps are in reverse orientation (Fig. 4B ▶). In contrast, mRNA capped with the 2′deoxy analog, m72′dGp3G, had a relative translation of only 1.17 ± 0.13, that is, no better than m7Gp3G-capped mRNA. On the other hand, the 2′-O-methyl analog, m27,2′-OGp3G, had a relative translation of 2.10 ± 0.15, the same as m27,3′-OGp3G (within experimental error). This is consistent with the finding that m27,2′-OGp3G and m27,3′-OGp3G inhibit in vitro translation to the same extent, but m72′dGp3G is only half as effective (Table 3 ▶).
In the tetraphosphate series, the two different organic syntheses of m7Gp4G mentioned above (1 and 2) produced mRNAs with relative translational efficiency of 1.32 and 1.35, only marginally better than m7Gp3G-capped mRNA. However, the tetraphosphate ARCAs m27,3′-OGp4G and m27,2′-OGp4G stimulated translation 2.42- and 2.56-fold more than m7Gp3G, respectively. This is the highest translational efficiency promoted by any cap analog reported to date. Interestingly, the 2′-deoxy member of the tetraphosphate set, m72′dGp4G (16), stimulated translation more than m7Gp3G (relative translational efficiency of 1.83 ± 0.18), but not as much as the other ARCA tetraphosphates, in keeping with its diminished ability to inhibit in vitro translation relative to m27,2′-OGp4G (15) or m27,3′-OGp4G (14) (Table 3 ▶).
The effect on translational efficiency of the pentaphosphate cap analogs was contrary to that predicted by binding affinity to eIF4E (Table 2 ▶) or inhibition of in vitro protein synthesis (Table 3 ▶), in which pentaphosphate cap analogs were more effective than tri- or tetraphosphate analogs. The relative translational efficiencies of m7Gp5G and m27,3′-OGp5G were only 1.05 ± 0.08 and 1.39 ± 0.16, respectively (Table 4 ▶). This may indicate that the higher binding affinity interferes with release of the cap at the end of the initiation cycle and thereby inhibits subsequent initiation events.
mRNAs were also synthesized that contained a poly(A) tract of 31 nt in addition to a cap. The cap analogs used were Gp3G, m7Gp3G, m7Gp4G, m27,3′-OGp4G, m7Gp5G, and m27,3′-OGp5G. In each case, the differences in relative translational efficiency between mRNAs capped with control and novel cap analogs were similar to the differences with nonpolyadenylated mRNA (data not shown). mRNAs were also synthesized from a different plasmid using the new cap analogs and SP6 polymerase rather than T7 polymerase (see Materials and Methods). Again, the translation results were similar (data not shown).
Finally, we can rule out the hypothesis that novel ARCA-capped mRNAs exhibit higher translation efficiency because they have a higher percentage of capping. The results in Table 4 ▶ indicate that the mRNAs with the highest translational efficiencies, those produced from m27,2′-OGp4G (15) and m27,3′-OGp4G (14), actually had a lower percent capping than others in the series and than m7Gp3G. Thus, the theoretical relative translational efficiencies of mRNAs 100% capped with m27,2′-OGp4G (15) and m27,3′-OGp4G (14) are even higher than those represented in Table 4 ▶.
DISCUSSION
The series of new cap analogs 12–18 with modifications at C2′ and C3′ of m7Guo and in the length of oligophosphate bridge represents a search for more effective and more easily synthesized “anti-reverse” cap analogs. Although the strategy for the multistep synthesis was similar to that developed previously (Stepinski et al. 2001), it was necessary to create the novel reagent guanosine 5′-triphosphate-P3-imidazolide. This also represents the first demonstration of a coupling reaction involving the imidazole derivative of GDP and GTP in DMF in the presence of ZnCl2. mRNAs capped with two of the new ARCAs, m27,2′-OGp4G and m27,3′-OGp4G, are translated ~2.5-fold more efficiently than mRNA capped with the native m7Gp3G cap and ~1.4-fold more efficiently than the previously described m27,3′-OGp3G (Table 4 ▶). The cost and time required to synthesize all three compounds is similar, indicating that m27,2′-OGp4G and m27,3′-OGp4G are the preferred cap analogs for in vitro mRNA synthesis. m72′dGp4G, on the other hand, enhances translational efficiency as much as m27,3′-OGp3G (1.8-fold; Table 4 ▶) yet can be synthesized more easily, because 2′-deoxyguanosine is commercially available and inexpensive, compared with the laborious preparation of 3′- or 2′-O-methylguanosine.
The influence of structural modifications of the cap analogs on their interaction with eIF4E is most clearly seen from a comparison of the binding free energies (Table 2 ▶). The change of ΔG° for the triphosphate versus tetraphosphate series is ΔΔG° ≅ 1.3 kcal/mole, whereas for the tetraphosphate versus pentaphosphate series, it is ΔΔG° ≅ 0.8 kcal/mole. The energetic effect of the δ-phosphate is of the order of the typical H-bond or salt bridge (Pace et al. 1996). In the case of m7Gp4G binding to eIF4E, this can be explained in terms of an additional phosphate–protein-binding contact (enthalpy effect). In the latter case, the longer phosphate chain would move the hanging free Guo away from the remaining m7Gppp part of the cap, allowing it to fit more tightly with the protein (Niedzwiecka et al. 2002). Introduction of the subsequent ɛ-phosphate additionally lowers the binding free energy. If one takes into account some favorable effect of a smaller stacked population (Table 1 ▶) and the unfavorable entropic effect of the phosphate chain extension, which partially cancel each other out, the more negative ΔG° for the pentaphosphate might be caused by an additional stabilizing contact between the negatively charged chain and the positively charged protein surface, that is, Lys 159. It is clear that groups in capped mRNA “downstream” of the m7Guo interact with eIF4E, because (1) capped oligonucleotides bind more strongly than cap dinucleotides (Carberry et al. 1992), and (2) the Ado moiety in m7Gp3A is stabilized by two H-bonds to Thr 205 and Thr 211 and van der Walls contacts at the surface of full-length human eIF4E (Tomoo et al. 2002) and at the surface of yeast eIF4E (Matsuo et al. 2000). On the other hand, no such interactions were observed in the latter study for m7Gp3ACC. Also, the Guo in m7Gp3G does not bind to truncated murine eIF4E and is not visible in the crystallography electron density map (Niedzwiecka et al. 2002). Therefore, it is not entirely clear which groups in eIF4E interact with the δ- and ɛ-phosphates.
Changes in ΔG° upon structural modification at C2′ and C3′ do not exceed 0.35 kcal/mole (Table 2 ▶). This is consistent with the crystallographic studies of mammalian eIF4E complexes with m7GDP (Marcotrigiano et al. 1997), m7Gp3G (Niedzwiecka et al. 2002), m7GTP, and m7Gp3A (Tomoo et al. 2002), and with the NMR study of yeast eIF4E with m7GDP (Matsuo et al. 1997), which show that 2′-OH and 3′-OH of m7Guo are located outside the protein surface and do not interact directly with eIF4E.
There is a qualitative but not quantitative correlation between KAS (Table 2 ▶) and 1/KI (Table 3 ▶). Compounds with the same m7Guo ribose modification increase in both KAS and 1/KI as the number of phosphate moieties increases (m7Gp3G < m7Gp4G < m7Gp5G; m27,3′-OGp3G < m27,3′-OGp4G < m27,3′-OGp5G; etc.). However, the quantitative differences within these series are not the same. For instance, in the series of cap analogs with no ribose modifications, affinity for eIF4E increases 8.9-fold in going from tri- to tetraphosphate and an additional 4.9-fold from tetra- to pentaphosphate, yet the ability of these compounds to inhibit translation increases only 1.6- and 2.2-fold, respectively. For analogs with the same number of phosphate moieties but different m7Guo ribose modifications, even the qualitative correlations do not hold up. Thus, m27,2′-OGp3G is the best inhibitor in the triphosphate series, but m73′dGp3G has the highest affinity for eIF4E. Similarly, m27,2′-OGp4G is the best inhibitor in the tetraphosphate series, but m7Gp4G has the highest affinity for eIF4E. m27,2′-OGp4G is three- to fourfold more inhibitory to cap-dependent translation than would be predicted from its binding affinity to eIF4E, for reasons we do not understand. In the pentaphosphate series, m27,3′-OGp5G is a better inhibitor than m7Gp5G but has a lower affinity for eIF4E. A generalization that can be made about all three phosphate bridge series is that analogs that are 2′-O- or 3′-O-methylated in m7Guo are better inhibitors than ribose-unmodified dinucleotides, which are in turn better than 2′-deoxy or 3′-deoxy m7Guo-containing analogs.
One explanation for these qualitative and quantitative differences may be that the highly charged tetra- and pentaphosphate cap analogs bind to additional proteins in the reticulocyte lysate, lowering their effective concentrations and decreasing their binding to eIF4E, but this is not a consideration for fluorescence titration experiments (Fig. 2 ▶) that contain only eIF4E. It should also be borne in mind that KAS represents an equilibrium measurement whereas KI represents a kinetic measurement. Thus, as eIF4E becomes more inhibited, some step in the initiation of protein synthesis may become rate-limiting, making the addition of more cap analog less and less effective. Another consideration is that productive interaction between eIF4E and the mRNA cap occurs when eIF4E is bound to eIF4G (although the order of binding is not known), and it has been established that eIF4G binding increases the affinity of eIF4E for caps (Haghighat and Sonenberg 1997), and binding of the poly(A)-binding protein further enhances cap binding (Wei et al. 1998). The differences between tri-, tetra-, and pentaphosphate analogs for binding to the eIF4E–eIF4G–poly(A)-binding protein complex may not be as great as for binding to eIF4E alone.
In addition to the requirements of T7 RNA polymerase for an unaltered ribose moiety on m7Guo, the enzyme shows some specificity toward the nucleoside base, because the trimethylguanosine cap m32,2,7Gp3G is incorporated into RNA chains exclusively in the correct orientation (R.E. Rhoads, unpubl.). This observation may explain an earlier finding, that mRNA transcripts capped with m22,7Gp3G are translated in a rabbit reticulocyte system 1.5-fold more efficiently than m7Gp3G-capped mRNAs (Darzynkiewicz et al. 1988). Similarly, mRNAs containing the bulky benzyl group on N7 (bn7Gp3G) instead of methyl are translated 1.7-fold more efficiently than mRNAs capped with the natural structure (Darzynkiewicz et al. 1989). It is possible that m22,7Gp3G and bn7Gp3G behave as ARCAs, although we have not tested this.
The new ARCAs may be useful for the study of biochemical processes in addition to translation. For instance, synthetic capped pre-mRNAs are widely used in splicing experiments, and it would be interesting to test whether ARCA-capped pre-mRNAs are spliced more efficiently than conventional pre-mRNAs, and if so, at what stage this improvement is manifested. Similarly, ARCA-capped mRNAs may bind differently to the nuclear cap-binding complex (CBC 20/80), which plays a role in splicing (Izaurralde et al. 1994) and possibly also in the first, “pioneer” round of translation (Fortes et al. 2000). Recent crystallographic studies show that, in contrast to eIF4E, CBC requires the 2′-OH and 3′-OH of m7Guo for binding to the cap (Calero et al. 2002). The compounds described in the present study that are modified in either C2′ or C3′ of m7Guo may shed light on this interaction and provide differential inhibition of eIF4E versus CBC 20/80.
MATERIALS AND METHODS
Synthesis of cap analogs
Final products and intermediate nucleotides were isolated from appropriate reaction mixtures by column chromatography on DEAE-Sephadex (A-25, HCO3− form) using a linear gradient of triethylammonium bicarbonate (TEAB, water solution, at pH 7.5) at 4°C. Eluates were collected in an automated fraction collector, and products peaks (as monitored by UV absorption at 260 nm) were pooled and evaporated to dryness (bath temperature not exceeding 30°C, ethanol repeatedly added to remove the TEAB buffer). Thus, the products were obtained as TEA salts. Monitoring of purity of intermediates and products was performed at 260 nm by analytical HPLC using a Spectra-Physics SP8800 apparatus on a Supelcosil LC-18-T reverse-phase column (25 cm plus 2-cm guard) and a Supelcosil LC-SAX ion-exchange column (25 cm plus 2-cm guard). For reverse-phase chromatography, the mobile phase was a linear 0%–25% gradient of methanol in 0.05 M ammonium acetate (pH 5.9) over 15 min with a flow rate of 1.3 mL/min. For ion-exchange chromatography, the mobile phase was a linear gradient of 0.001–0.1 M KH2PO4 (pH 4.0) over 20 min with a flow rate of 1.0 mL/min. All compounds used for biophysical and biological studies exhibited purity higher than 95% in both chromatographic systems.
2′-O-Methylguanosine and 3′-O-methylguanosine were prepared as previously described (Kusmierek and Shugar 1978). A mixture of their 6-O-ethyl derivatives was separated by column chromatography using Dowex 1 × 2 Ion-Exchange Resin (200–400 mesh) and 25% methanol in water as eluent in an isocratic system.
3′-O-Methylguanosine 5′-monophosphate (1) was prepared by stirring 3′-O-methylguanosine (606 mg, 2.04 mmole) for 3 h with 18 mL of trimethyl phosphate and phosphorus oxychloride (0.95 mL, 10.2 mmole) at 0°C. The reaction was quenched with 20 mL of 1 M TEAB. DEAE Sephadex chromatography using a linear gradient of 0–0.7 M TEAB afforded 3 (yield: 980 mg as a TEA salt, 83%).
2′-O-Methylguanosine 5′-monophosphate (2) was synthesized in the same manner as 1 using 412 mg (1.39 mmole) of 2′-O-methylguanosine (yield: 620 mg as a TEA salt, 77%).
3′-O-Methylguanosine 5′-diphosphate (3) was synthesized by mixing compound 1 (695 mg, TEA salt, 1.2 mmole), imidazole (408 mg, 6.0 mmole), and 2,2′-dithiodipyridine (528 mg, 2.4 mmole) in anhydrous DMF (15 mL) and TEA (168 μL). Triphenylphosphine (629 mg, 2.4 mmole) was added, and the mixture was stirred at room temperature overnight. The reaction mixture was poured in a flask containing anhydrous sodium perchlorate (600 mg) dissolved in 72 mL of acetone. After cooling at 4°C for 2 h, the precipitate was filtered and dried in a vacuum desiccator or over P4O10. The imidazolide thus obtained was dissolved in DMF (15 mL), and 2.4 g of tris-(triethylammonium)phosphate was added. Finally, 960 mg of ZnCl2 was added, and the mixture was stirred at room temperature for 4 h (the reaction reached 80% HPLC yield after 30 min). The reaction mixture was poured into a beaker containing a solution of EDTA (3 g) in water (200 mL) and neutralized with 1 M NaHCO3. Chromatographic isolation (a linear gradient of 0–1 M TEAB) gave 3 (yield: 657 mg as a TEA salt, 72%).
2′-O-Methylguanosine 5′-diphosphate (4) was synthesized in the same manner as 3 using 579 mg (1.0 mmole) of 2′-O-methylguanosine 5′-monophosphate TEA salt (yield: 516 mg as a TEA salt, 68%).
2′-Deoxyguanosine 5′-diphosphate (5) was synthesized in the same manner as 3 starting with 175 mg (0.45 mmole) of 2′-deoxyguanosine 5′-monophosphate Na+ salt (purchased form Calbiochem), which was converted into the TEA salt before the synthesis using ion-exchange chromatography on Dowex 50WX1, TEA form (yield: 182 mg, 55.6%).
3′-O,7-Dimethylguanosine 5′-diphosphate (6) was synthesized by dissolving 3 (604 mg, TEA salt, 0.79 mmole) in 20 mL of DMSO, adding 1.4 mL of methyl iodide, and stirring the solution at room temperature for 3 h. Then 300 mL of cold water was added, and the solution was extracted with diethyl ether (3 × 100 mL). The aqueous phase, after neutralization with 1 M NaHCO3, was chromatographed on DEAE Sephadex and eluted with a linear 0–0.93 M gradient of TEAB (yield: 496 mg as a TEA salt, 81%).
2′-O,7-Dimethylguanosine 5′-diphosphate (7) was synthesized in the same manner as 6 using 516 mg (0.68 mmole) of 2′-O-methylguanosine 5′-diphosphate TEA salt (yield: 516 mg as a TEA salt, 74.2%).
2′-O,7-Dimethylguanosine 5′-diphosphate (8) was synthesized in the same manner as 6 using 182 mg (0.25 mmole) of 2′-O-methylguanosine 5′-diphosphate TEA salt (yield: 77.5 mg as a TEA salt, 40%).
Guanosine 5′-monophosphate imidazolide (9) was synthesized by changing guanosine 5′-monophosphate Na+ salt (407 mg, 1 mmole) into the TEA salt on Dowex 50WX1 resin and then mixing it with imidazole (340 mg, 5 mmole) and 2,2′-dithiodipyridine (440 mg, 2 mmole) in anhydrous DMF (12.5 mL) and TEA (140 μL). Triphenylphosphine (524 mg, 2 mmole) was added, and the mixture was stirred at room temperature overnight. The mixture was poured in a flask containing anhydrous sodium perchlorate (500 mg) dissolved in 60 mL of acetone. After cooling at 4°C for 2 h, the precipitate was filtered, washed twice with 15 mL of cold acetone, and dried in a vacuum desiccator over P4O10 (yield: 392 mg, 90%; HPLC purity: 97.4%).
Guanosine 5′-diphosphate imidazolide (10) was synthesized in the same manner as 9 using 509 mg (1 mmole) of guanosine 5′-diphosphate Na+ salt instead of GMP (yield: 462 mg, 86%; HPLC purity: 98.1%).
Guanosine 5′-triphosphate imidazolide (11) was synthesized in the same manner as 9 using 305.5 mg (0.5 mmole) of guanosine 5′-triphosphate Na+ salt instead of GMP (yield: 488 mg, 76.4%; HPLC purity: 93.3%).
Synthesis of tri-, tetra-, and pentaphosphate dinucleoside cap analogs was carried out by the same general procedure. The TEA salt of each 7-methylguanosine derivative (0.05 mmole) and imidazolide (0.06 mmole) were dissolved in 2 mL of anhydrous DMF, stirred for 5 min, and then ZnCl2 (0.4 mmole) was added. The mixture was stirred at room temperature for 4–8 h, poured into a beaker containing a solution of 170 mg of EDTA in 20 mL of water, and neutralized with 1 M NaHCO3. Chromatographic isolation on DEAE Sephadex using a linear gradient of TEAB (0–1.1 M for triphosphates; 0–1.2 M for tetraphosphates; 0–1.3 M for pentaphosphates) gave the product as the TEA salt. The final product was converted to its Na+ salt by ion-exchange chromatography (Dowex 50WX8, Na+ form). The eluate was evaporated to a small volume and the residue was precipitated with ethanol. Centrifugation gave an amorphous white powder that was dried in a vacuum desiccator over P4O10.
The following compounds were prepared using this general procedure. HPLC yield refers to content of product at the end of reaction in reaction mixture, whereas preparative yield refers to the yield of reaction after purification and conversion of product into the Na+ salt. P1-7-methyl-2′-deoxyguanosine-5′ P3-guanosine-5′ triphosphate (m72′dGp3G, 12) was prepared using 37.9 mg of 8 and 26.1 mg of 9 (HPLC yield: 76.6%; preparative yield: 23.9 mg, 56%). P1-2′-O,7-dimethylguanosine-5′ P3-guanosine-5′ triphosphate (m27,2′-OGp3G, 13) was prepared using 38.7 mg of 7 and 26.1 mg of 9 (HPLC yield: 85%; preparative yield: 29.1 mg, 66%). P1-3′-O,7-dimethylguanosine-5′ P4-guanosine-5′ tetraphosphate (m27,3′-OGp4G, 14) was prepared using 38.7 mg of 6 and 32.2 mg of 10 (HPLC yield: 88.2%; preparative yield: 25.1 mg, 51%). P1-2′-O,7-dimethylguanosine-5′ P4-guanosine-5′ tetraphosphate (m27,2′-OGp4G, 15) was prepared using 38.7 mg of 7 and 32.2 mg of 10 (HPLC yield: 81.7%; preparative yield: 13.2 mg, 26.8%). P1-7-methyl-2′-deoxyguanosine-5′ P4-guanosine-5′ tetraphosphate (m72′dGp4G, 16) was prepared using 37.9 mg of 8 and 32.2 mg of 10 (HPLC yield: 76.6%; preparative yield: 22.0 mg, 46%). P1-7-methylguanosine-5′ P5-guanosine-5′ pentaphosphate (m7Gp5G, 17) was prepared using 38.0 mg of 7-methylguanosine/TEA salt and 38.3 mg of 11 (HPLC yield: 74,1%; preparative yield: 28.1 mg, 52.4%). P1-3′-O,7-dimethylguanosine-5′ P5-guanosine-5′ pentaphosphate (m27,3′-OGp5G, 18) was prepared using 38.7 mg of 6 and 38.3 mg of 11 (HPLC yield: 84.6%; preparative yield: 34.8 mg, 64%).
The concentrations of dinucleotide cap analog solutions were measured by UV absorption at pH 7.0 using λ = 255 and ɛM = 22.6 × 10−3 M.
NMR spectroscopy
The structures (Table 5 ▶) and conformations (Table 1 ▶) of new cap analogs were confirmed by 1H and 31P spectra. Spectra were recorded on a Varian UNITYplus 500 MHz instrument in 2H2O at 20°C and at concentrations ranged from 2 to 3 mg/mL. Conformations of the sugar moieties were derived from the vicinal 1H–1H coupling constants (Haasnoot et al. 1980), and conformations of the α-phosphate groups about the C5′—O5′ bond from the vicinal 1H–31P coupling constants (Lankhorst et al. 1984).
TABLE 5.
1H NMR chemical shifts in parts per million (±0.001) versus internal sodium 3-trimethylsilyl-[2,2,3,3-2H4]- propionate and 31P NMR chemical shifts in parts per million (±0.01) versus external H3PO4, and 1H–1H, 1H–31P coupling constants in hertz (±0.2)
| m27,2′-OGp3G (13) | m72′dGp3G (12) | m72′Gp4G (16) | m27,2′-OGp4G (15) | m27,3′-OGp4G (14) | m7Gp5G (17) | m27,3′-OGp5G (18) | ||||||||
| m27,2′-OG | G | m72′dG | G | m72′dG | G | m27,2′-OG | G | m27,3′-O | G | m7G | G | m27,3′-OG | G | |
| H8 | a | 8.002 | a | 8.011 | a | 8.050 | a | 8.056 | a | 8.043 | a | 8.082 | a | 8.077 |
| H1′ | 5.923 | 5.784 | 6.177 | 5.797 | 6.258 | 5.839 | 6.002 | 5.829 | 5.923 | 5.819 | 5.996 | 5.854 | 5.972 | 5.842 |
| H2′ | 4.161 | 4.642 | 2.471 | 4.651 | 2.59b | 4.725 | 4.249 | 4.759 | 4.732 | 4.737 | 4.611 | 4.799 | 4.763 | 4.805 |
| H2″ | — | — | 2.549 | — | 2.59b | — | — | — | — | — | — | — | — | — |
| H3′ | 4.479 | 4.466 | 4.590 | 4.472 | 4.683 | 4.535 | 4.572 | 4.538 | 4.145 | 4.529 | 4.530 | 4.576 | 4.176 | 4.574 |
| H4′ | 4.29b | 4.331 | 4.217 | 4.343 | 4.25b | 4.354 | 4.32b | 4.34b | 4.459 | 4.352 | 4.395 | 4.362 | 4.488 | 4.366 |
| H5′ | 4.373 | 4.260 | 4.28b | 4.25b | 4.293 | 4.26b | 4.34b | 4.26b | 4.384 | 4.27b | 4.320 | 4.29b | 4.380 | 4.29b |
| H5″ | 4.29b | 4.236 | 4.25b | 4.25b | 4.233 | 4.26b | 4.31b | 4.27b | 4.242 | 4.27b | 4.29b | 4.222 | 4.26b | 4.238 |
| CH3 | 4.057 (N7) | — | 4.044 | — | 4.071 | — | 4.084 (N7) | — | 4.086 (N7) | — | 4.088 | — | 4.093 (N7) | — |
| 3.578 (O2′) | 3.559 (O2′) | 3.493 (O3′) | 3.508 (O3′) | |||||||||||
| Pα | −12.20 | −12.21 | −12.05 | −12.00 | −12.02 | −11.92 | −11.98 | |||||||
| Pβ | −23.79 | −23.76 | −23.81 | −23.74 | −23.79 | −23.58 | −23.64 | |||||||
| Pγ | −12.20 | −12.21 | −23.81 | −23.74 | −23.79 | −23.58 | −23.64 | |||||||
| Pδ | — | — | −12.05 | −12.00 | −12.02 | −23.58 | −23.64 | |||||||
| Pɛ | — | — | — | — | — | −11.92 | −11.98 | |||||||
| J(1′, 2′) | 3.2 | 6.4 | 5.3 | 6.0 | 6.2c | 6.4 | 3.7 | 6.7 | 4.4 | 6.4 | 4.0 | 6.8 | 4.4 | 6.6 |
| J(1′, 2″) | — | — | 6.4 | — | 6.2c | — | — | — | — | — | — | — | — | — |
| J(2′, 2″) | — | — | 14.3 | — | d | — | — | — | — | — | — | — | — | — |
| J(2′, 3′) | 4.8 | 4.9 | 6.4 | 5.2 | 5.6c | 5.4 | 5.0 | 5.0 | 5.0 | 5.3 | 5.2 | 5.2 | 4.9 | 5.3 |
| J(2", 3′) | — | — | 5.3 | — | 5.6c | — | — | — | — | — | — | — | — | — |
| J(3′, 4′) | 6.0 | 3.5 | 4.4 | 3.6 | 3.2 | 3.1 | 5.2 | 3.3 | 4.6 | 3.2 | 4.8 | 2.8 | 4.6 | 2.8 |
| J(4′, 5′) | 2.4 | 3.5 | 3.0c | 4.0c | 3.5 | 4.0c | d | d | 3.2 | 4.0 | 2.5 | 3.0b | 3.0 | 3.8 |
| J(4′, 5″) | 2.4b | 4 | 3.0c | 4.0c | 3.7 | 4.0c | d | d | 2.8 | 4.0b | 3.0b | 3.5 | d | 3.7 |
| J(5′, 5″) | 12.0 | 11.8 | d | d | 11.6 | d | d | d | 11.6 | 11.9 | 12.2 | 11.6 | 11.6 | 12.0 |
| J(4′, P) | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b | 1.0b |
| J(5′, P) | 4.0 | 6.0 | 5.5 | d | 6.7 | d | d | d | 4.6 | 6.6 | 4.5 | 5.5b | 4.4 | 6.4 |
| J(5", P) | 5.0b | 6.8 | d | d | 4.8 | d | d | d | 5.8 | d | 5.0b | 5.0 | d | 5.2 |
aDeuterated.
bApproximate value because of signal overlapping.
cMean value because of signal overlapping of the geminal protons H2′/H2" or H5′/H5".
dNot determined because of signal overlapping.
Expression and purification of eIF4E
Mouse eukaryotic initiation factor eIF4E (residues 28–217) was expressed in E. coli as inclusion bodies. The guanidinium-solubilized protein was renatured by one-step dialysis and purified by ion-exchange chromatography on a Mono SP column (Edery et al. 1988). The concentration of eIF4E was determined spectrophotometrically assuming ɛ280 = 53,400 M−1 cm−1.
Measurement of binding affinity by fluorescence quenching
Fluorescence time-synchronized titrations (Niedzwiecka et al. 2002) were performed on an LS-50B spectrofluorometer (Perkin Elmer Co.) in 50 mM HEPES/KOH (pH 7.2), 100 mM KCl, 0.5 mM EDTA, and 1 mM DTT at 20°C. Aliquots of the cap analog solution (1 μL) were added to 1400 μL of 0.1 μM or 0.05 μM eIF4E solution. Fluorescence intensities (excitation at 280 nm, detection at 337 nm) were corrected for the sample dilution (<4%), for the inner filter effect, and for the emission of the free cap analog. KAS was determined by fitting the theoretical curve for the fluorescence intensity as a function of total concentration of cap analog to the titration data points. The amount of active protein was fit exactly as a free parameter of the equilibrium equation (Niedzwiecka et al. 2002). The final KAS was calculated as a weighted average of five to 10 independent titrations, with the weights taken as the reciprocals of the numerical standard deviations squared. Numerical least-squares nonlinear regression analysis was performed using ORGIN 6.0 from Microcal Software Inc. The population of the stacked cap analog was determined from the temperature dependence of the fluorescence intensity in phosphate buffer at pH 5.2 (Wieczorek et al. 1997).
Synthesis of mRNAs
RNAs initiated with various cap analogs were synthesized by in vitro transcription as described previously (Stepinski et al. 2001) except that T7 polymerase replaced SP6 polymerase in some cases and incubation was for 45 rather than 60 min. Short RNAs were generated using [α-32P]GTP for analysis of cap orientation and capping efficiency by digesting the plasmid pluc-A+, which contains the entire firefly luciferase mRNA sequence in pGEM4 (Promega) plus a 3′-terminal 31-nt poly(A) tract, with NcoI to produce a T7 RNA polymerase template that yields a 48-nt RNA exclusive of the cap. Long RNAs were generated using [α-32P]ATP for measurement of translational efficiency from one of two plasmids. The first, pSP-luc+ (Promega), was digested with EcoRI to produce an SP6 RNA polymerase template that yields an RNA product of 1711 nt exclusive of the cap containing the entire firefly luciferase coding region but lacking a poly(A) tract (Stepinski et al. 2001). The second plasmid, pluc-A+, was digested with either HpaI, to produce a T7 RNA polymerase template for an RNA of 1742 nt exclusive of the cap containing the luciferase coding region plus a 3′-terminal 31-nt poly(A) tract, or SmaI, to produce a similar RNA but lacking the poly(A) tract.
Reaction mixtures were extracted with phenol and chloroform, and then RNA was separated from free nucleotides using one of three methods. The ethanol precipitation method was carried out as described previously (Stepinski et al. 2001). In the column chromatography method, reaction mixtures (50–75 μL) were passed over 5-mL columns of Sephadex G-50 (fine; Pharmacia), made RNase-free by treatment with 0.1% aqueous diethylpyrocarbonate followed by equilibration with TE buffer (10 mM Tris, 1 mM EDTA at pH 8.0). In the spin-column method, NucAway Spin Columns (Ambion) were filled with 650 μL of RNase-free water, vortexed, hydrated at room temperature for 15 min, and centrifuged at 750g for 2 min to remove the excess interstitial fluid. Then 50 μL of reaction mixture was loaded, and the spin column was centrifuged as before, the effluent being used as the source of RNA. In all three cases, concentrations of the resultant mRNAs were determined by Cerenkov counting, with specific radioactivity calculated from the transcription reaction mixture. All three methods were comparable, but the spin-column method was the simplest, quickest, and gave the most reproducible results.
Analysis of cap orientation and percentage capping in mRNAs
Short RNAs (see above) were digested with either RNase T2 (Invitrogen) alone or TAP (Epicentre Technologies) followed by RNase T2 and analyzed by anion-exchange HPLC as described previously (Stepinski et al. 2001). To determine the relative amounts of capped and uncapped mRNAs, labeled RNAs were digested with RNase T2 and analyzed by a modified anion-exchange HPLC protocol. The gradient consisted of water for the first 5 min, a linear gradient of 0–1.5 M KH2PO4 at pH 3.5 (for the RNase T2-digested triphosphate analogs) or pH 4.5 (for the RNase T2-digested tetraphosphate analogs) for 40 min, and isocratic elution at 1.5 M KH2PO4 for 2 min, all at a flow rate of 1 mL/min. The radioactivity in each peak was then summed to obtain the relative amounts of RNase T2 digestion products resulting from capped (Fig. 5B ▶, 29-min peak) and uncapped (39-min peak) mRNAs.
In vitro translation
Inhibition of globin mRNA translation with the novel cap analogs and measurement of translational efficiency using luciferase mRNAs capped with the novel cap analogs were performed in a rabbit reticulocyte lysate system as described previously (Cai et al. 1999; Stepinski et al. 2001) except that 3 mM glucose was included. Normalization of KI data based on inhibition by the standard m7Gp3G included in the same assay was performed as described previously (Cai et al. 1999). Luciferase activity was determined by adding 1 μL of translation reaction mixture to 360 μL of the following freshly made solution: 25 mM glycylglycine (pH 7.8), 15 mM MgSO4, 4 mM EGTA, 15 mM KH2PO4 (pH 7.8), 1 mM DTT, and 2 mM ATP. This mixture was placed in a Monolight 2010 luminometer programmed to inject 100 μL of a freshly made solution containing 25 mM glycylglycine (pH 7.8), 15 mM MgSO4, 4 mM EGTA, 4 mM DTT, and 0.225 mg/mL beetle luciferin (Promega). After a 2-sec delay, relative light units (RLU) of the samples were read over a 10-sec interval. Each translation reaction mixture was assayed for luciferase activity in triplicate. Normalization of translational efficiency data was performed as described in the legend to Table 4 ▶ using Gp3G- and m7Gp3G-capped luciferase mRNA as standards. Optimal cap-dependent translation was achieved at 100 mM potassium acetate and 1.4 mM magnesium chloride.
Acknowledgments
This project was supported by Grant No. GM20818 from the National Institute of General Medical Sciences, NIH, and Grant Nos. PBZ-KBN-059/T09/10, KBN 6 P04A 055 17, BST 763/BF, and BW1565/BF from the Polish Committee for Scientific Research (KBN). The authors express their gratitude to Nahum Sonenberg for the mouse eIF4E (28–217) plasmid and to Dorota Haber for her excellent chemical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Adams, B.L., Morgan, M., Muthukrishnan, S., Hecht, S.M., and Shatkin, A.J. 1978. The effect of “cap” analogs on reovirus mRNA binding to wheat germ ribosomes. J. Biol. Chem. 253: 2589–2595. [PubMed] [Google Scholar]
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., and Parker, R. 1998. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646. [DOI] [PubMed] [Google Scholar]
- Blaedel, W.J. and Meloche, V.W. 1963. Elementary quantitative analysis: Theory and practice, p. 640. Harper & Row, New York.
- Cai, A., Jankowska-Anyszka, M., Centers, A., Chlebicka, L., Stepinski, J., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 1999. Quantitative assessment of mRNA cap analogs as inhibitors of in vitro translation. Biochemistry 38: 8538–8547. [DOI] [PubMed] [Google Scholar]
- Calero, G., Wilson, K.F., Ly, T., Rios-Steiner, J.L., Clardy, J.C., and Cerione, R.A. 2002. Structural basis of m7GpppG binding to the nuclear cap-binding protein complex. Nat. Struct. Biol. 9: 912–917. [DOI] [PubMed] [Google Scholar]
- Carberry, S.E., Friedland, D.E., Rhoads, R.E., and Goss, D.J. 1992. Binding of protein synthesis initiation factor 4E to oligoribonucleotides: Effects of cap accessibility and secondary structure. Biochemistry 31: 1427–1432. [DOI] [PubMed] [Google Scholar]
- Chen, J.-J. 2000. Heme-regulated eIF2α kinase. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 529–546. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Contreras, R., Cheroutre, H., Degrave, W., and Fiers, W. 1982. Simple, efficient in vitro synthesis of capped RNA useful for direct expression of cloned eukaryotic genes. Nucleic Acids Res. 10: 6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Ekiel, I., Tahara, S.M., Seliger, L.S., and Shatkin, A. 1985. Chemical synthesis and characterization of 7-methylguanosine cap analogues. Biochemistry 24: 1701–1707. [Google Scholar]
- Darzynkiewicz, E., Ekiel, I., Lassota, P., and Tahara, S.M. 1987. Inhibition of eukaryotic translation by analogues of messenger RNA 5′-cap: Chemical and biological consequences of 5′-phosphate modifications of 7-methylguanosine 5′-monophosphate. Biochemistry 26: 4372–4380. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Stepinski, J., Ekiel, I., Jin, Y., Haber, D., Sijuwade, T., and Tahara, S.M. 1988. β-Globin mRNAs capped with m7G, m2,2G, or m2,2,7G differ in intrinsic translation efficiency. Nucleic Acids Res. 16: 8953–8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Stepinski, J., Ekiel, I., Goyer, C., Sonenberg, N., Temeriusz, A., Jin, Y., Sijuwade, T., Haber, D., and Tahara, S.M. 1989. Inhibition of eukaryotic translation by nucleoside 5′-monophosphate analogues of mRNA 5′-cap: Changes in N7 substituent affect analogue activity. Biochemistry 28: 4771–4778. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz, E., Stepinski, J., Tahara, S.M., Stolarski, R., Ekiel, I., Haber, D., Neuvonen, K., Lehikoinen, P., Labadi, I., and Lönnberg, H. 1990. Synthesis, conformation and hydrolytic stability of 1,P3-dinucleoside triphosphates related to mRNA 5′-cap, and comparative kinetic studies on their nucleoside and nucleoside monophosphate analogs. Nucleosides & Nucleotides 9: 599–618. [Google Scholar]
- Edery, I., Altmann, M., and Sonenberg, N. 1988. High-level synthesis in Escherichia coli of functional cap-binding eukaryotic initiation factor eIF-4E and affinity purification using a simplified cap-analog resin. Gene 74: 517–525. [DOI] [PubMed] [Google Scholar]
- Fischer, U., Darzynkiewicz, E., Tahara, S.M., Dathan, N.A., Luhrmann, R., and Mattaj, I.W. 1991. Diversity in the signals required for nuclear accumulation of U snRNPs and variety in the pathways of nuclear transport. J. Cell Biol. 113: 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes, P., Inada, T., Preiss, T., Hentze, M.W., Mattaj, I.W., and Sachs, A.B. 2000. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell 6: 191–196. [PubMed] [Google Scholar]
- Furuichi, Y. and Shatkin, A.J. 2000. Viral and cellular mRNA capping: Past and prospects. Adv. Virus Res. 55: 135–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.-C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Ann. Rev. Biochem. 68: 913–963. [DOI] [PubMed] [Google Scholar]
- Haasnoot, C.A.G., deLeeuw, F.A.A.M., and Altona, C. 1980. The relationship between proton–proton NMR coupling constants and substituent electronegativities. Tetrahedron 36: 2783–2792. [Google Scholar]
- Haghighat, A. and Sonenberg, N. 1997. eIF4G dramatically enhances the binding of eIF4E to the mRNA 5′-cap structure. J. Biol. Chem. 272: 21677–21680. [DOI] [PubMed] [Google Scholar]
- Hamm, J., Darzynkiewicz, E., Tahara, S.M., and Mattaj, I.W. 1990. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell 62: 569–577. [DOI] [PubMed] [Google Scholar]
- Hendler, S., Fürer, E., and Srinivasan, P.R. 1970. Synthesis and chemical properties of monomers and polymers containing 7-methylguanine and an investigation of their substrate or template properties for bacterial DNA or RNA polymerases. Biochemistry 9: 4141–4153. [DOI] [PubMed] [Google Scholar]
- Izaurralde, E., Stepinski, J., Darzynkiewicz, E., and Mattaj, I.W. 1992. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol. 118: 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde, E., Lewis, J., McGuigan, C., Jankowska, M., Darzynkiewicz, E., and Mattaj, I.W. 1994. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78: 657–668. [DOI] [PubMed] [Google Scholar]
- Jankowska-Anyszka, M., Lamphear, B.J., Aamodt, E.J., Harrington, T., Darzynkiewicz, E., Stolarski, R., and Rhoads, R.E. 1998. Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in C. elegans can distinguish between mono- and trimethylated mRNA cap structures. J. Biol. Chem. 273: 10538–10542. [DOI] [PubMed] [Google Scholar]
- Keiper, B.D., Lamphear, B.J., Deshpande, A.M., Jankowska-Anyszka, M., Aamodt, E.J., Blumenthal, T., and Rhoads, R.E. 2000. Functional characterization of five eIF4E isoforms in Caenorhabditis elegans. J. Biol. Chem. 275: 10590–10596. [DOI] [PubMed] [Google Scholar]
- Konarska, M.M., Padgett, R.A., and Sharp, P.A. 1984. Recognition of a cap structure in splicing in vitro of mRNA precursors. Cell 38: 731–736. [DOI] [PubMed] [Google Scholar]
- Kusmierek, J. and Shugar, D. 1978. A new route to 2′(3′)-O-alkyl purine nucleosides. Nucleic Acids Res. Special Publ. No. 4: s73–s77. [Google Scholar]
- Lankhorst, P.L., Haasnoot, C.A.G., Erkelens, C., and Altona, C. 1984. Carbon-13 NMR in conformational analysis of nucleic acid fragments. Reparametrization of the Karplus equation for vicinal NMR coupling constants in COOP and HCOP fragments. J. Biomol. Struct. Dyn. 1: 1387–1405. [DOI] [PubMed] [Google Scholar]
- Lassota, P., Stolarski, R., and Shugar, D. 1984. Conformation about the glycosidic bond and susceptibility to 5′-nucleotidase of 8-substituted analogues of 5′-GMP. Z. Naturforsch. 39c: 55–63. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano, J., Gingras, A.-C., Sonenberg, N., and Burley, S.K. 1997. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell 89: 951–961. [DOI] [PubMed] [Google Scholar]
- Maroney, P.A., Denker, J.A., Darzynkiewicz, E., Laneve, R., and Nilsen, T.W. 1995. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: A role for spliced leader addition in translational efficiency. RNA 1: 714–723. [PMC free article] [PubMed] [Google Scholar]
- Matsuo, H., Li, H., McGuire, A.M., Fletcher, C.M., Gingras, A.-C., Sonenberg, N., and Wagner, G. 1997. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat. Struct. Biol. 4: 717–724. [DOI] [PubMed] [Google Scholar]
- Matsuo, H., Moriguchi, T., Takagi, T., Kusakabe, T., Buratowski, S., Sekine, M., Kyogoku, Y., and Wagner, G. 2000. Efficient synthesis of 13C,15N-labeled RNA containing the cap structure m7Gp3A. J. Am. Chem. Soc. 122: 2417–2421. [Google Scholar]
- Mattaj, I.W. 1986. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 46: 905–911. [DOI] [PubMed] [Google Scholar]
- Miyoshi, H., Dwyer, D.S., Keiper, B.D., Jankowska-Anyszka, M., Darzynkiewicz, E., and Rhoads, R.E. 2002. Discrimination between mono- and trimethylated cap structures by two isoforms of Caenorhabditis elegans eIF4E. EMBO J. 21: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedzwiecka, A., Marcotrigiano, J., Stepinski, J., Jankowska-Anyszka, M., Wyslouch-Cieszynska, A., Dadlez, M., Gingras, A.-C., Mak, P., Darzynkiewicz, E., Sonenberg, N., et al. 2002. Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319: 615–635. [DOI] [PubMed] [Google Scholar]
- Pace, C.N., Shirley, B.A., McNutt, M., and Gajiwala, K. 1996. Forces contributing to the conformational stability of the proteins. FASEB J. 10: 75–83. [DOI] [PubMed] [Google Scholar]
- Pasquinelli, A.E., Dahlberg, J.E., and Lund, E. 1995. Reverse 5′ caps in RNAs made in vitro by phage RNA polymerases. RNA 1: 957–967. [PMC free article] [PubMed] [Google Scholar]
- Peng, Z.-H., Sharma, V., Singleton, S.F., and Gershon, P.D. 2002. Synthesis and application of a chain-terminating dinucleotide mRNA cap analog. Organic Letters 4: 161–164. [DOI] [PubMed] [Google Scholar]
- Rhoads, R.E. 1985. The cap structure of eukaryotic messenger RNA and its interaction with cap-binding protein. Prog. Mol. Subcell. Biol. 9: 104–155. [Google Scholar]
- ———. 1999. Minireview: Signal transduction pathways that regulate eukaryotic protein synthesis. J. Biol. Chem. 274: 30337–30340. [DOI] [PubMed] [Google Scholar]
- Stachelska, A., Wieczorek, Z., Ruszczynska, K., Stolarski, R., Pietrzak, M., Lamphear, B.J., Rhoads, R.E., Darzynkiewicz, E., and Jankowska-Anyszka, M. 2002. Interaction of three Caenorhabditis elegans isoforms of translation initiation factor eIF4E with mono- and trimethylated mRNA 5′ cap analogues. Acta Biochim. Polon. 49: 671–682. [PubMed] [Google Scholar]
- Stepinski, J., Bretner, M., Jankowska, M., Felczak, K., Stolarski, R., Wieczorek, Z., Cai, A.-L., Rhoads, R.E., Temeriusz, A., Haber, D., et al. 1995. Synthesis and properties of P1,P2,P3- and P1,P4-dinucleoside di-, tri- and tetraphosphate mRNA 5′-cap analogues. Nucleosides & Nucleotides 14: 717–721. [Google Scholar]
- Stepinski, J., Waddell, C., Stolarski, R., Darzynkiewicz, E., and Rhoads, R.E. 2001. Synthesis and properties of mRNAs containing the novel “anti-reverse” cap analogues 7-methyl(3′-O-methyl)GpppG and 7-methyl(3′-deoxy)GpppG. RNA 7: 1486–1495. [PMC free article] [PubMed] [Google Scholar]
- Tomoo, K., Shen, X., Okabe, K., Nozoe, Y., Fukuhara, S., Morino, S., Ishida, T., Taniguchi, T., Hasegawa, H., Terashima, A., et al. 2002. Crystal structure of 7-methylguanosine 5′-triphosphate (m7GTP)- and P1–7-methylguanosine-P3-adenosine-5′,5′-triphosphate (m7GpppA)-bound human full-length eukaryotic initiation factor 4E: Biological importance of the C-terminal flexible region. Biochem. J. 362: 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, C.C., Balasta, M.L., Ren, J.H., and Goss, D.J. 1998. Wheat germ poly(A)-binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry 37: 1910–1916. [DOI] [PubMed] [Google Scholar]
- Wieczorek, Z., Zdanowski, K., Chlebicka, L., Stepinski, J., Jankowska, M., Kierdaszuk, B., Temeriusz, A., Darzynkiewicz, E., and Stolarski, R. 1997. Fluorescence and NMR studies of intramolecular stacking of mRNA cap-analogues. Biochim. Biophys. Acta 1354: 145–152. [DOI] [PubMed] [Google Scholar]
- Yisraeli, J.K. and Melton, D.A. 1989. Synthesis of long, capped transcripts in vitro by SP6 and T7 RNA polymerases. In Methods in enzymology (eds. J.E. Dahlberg and J.N. Abelson), Vol. 180, pp. 42–50. Academic Press, San Diego. [DOI] [PubMed]
- Zuberek, J., Wyslouch-Cieszynska, A., Niedzwiecka, A., Dadlez, M., Stepinski, J., Augustyniak, W., Gingras, A.-C., Zhang, Z., Burley, S.K., Sonenberg, N., et al. 2003. Phosphorylation of eIF4E attenuates its interaction with mRNA cap analogs by electrostatic repulsion: Intein-mediated protein ligation strategy to obtain phosphorylated protein. RNA 9: 52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]