Abstract
Regulation of decapping is a critical determinant of mRNA stability. We recently identified hDcp2 as a human decapping enzyme with intrinsic decapping activity. This activity is specific to N7-methylated guanosine containing RNA. The hDcp2 enzyme does not function on the cap structure alone and is not sensitive to competition by cap analog, suggesting that hDcp2 requires the RNA for cap recognition. We now demonstrate that hDcp2 is an RNA-binding protein and its recognition and hydrolysis of the cap substrate is dependent on an initial interaction with the RNA moiety. A biochemical characterization of hDcp2 revealed that a 163 amino acid region containing two evolutionarily conserved regions, the Nudix fold hydrolase domain and the adjacent Box B region contained methyl-cap-specific hydrolysis activity. Maximum decapping activity for wild-type as well as truncation mutants of hDcp2 required Mn2+ as a divalent cation. The demonstration that hDcp2 is an RNA-binding protein with an RNA-dependent decapping activity will now provide new approaches to identify specific mRNAs that are regulated by this decapping enzyme as well as provide novel avenues to control mRNA decapping and turnover by influencing the RNA-binding property of hDcp2.
Keywords: mRNA stability, in vitro mRNA decay, RNA-binding protein, Nudix motif
INTRODUCTION
A common theme among all eukaryotic mRNAs is the presence of an N7 methyl cap structure at the 5′ end. The cap structure with its unusual 5′-5′ pyrophosphate linkage is resistant to most ribonucleases, and confers stability onto the 5′ end of an mRNA. A subset of specific nucleases have been identified that can efficiently hydrolyze the pyrophosphate linkage of the methylated cap structure. To date, at least two different decapping enzymes with distinct activities have been identified. The first enzyme utilizes capped RNA as a substrate and hydrolyzes the cap to release m7GDP. In yeast, this is carried out by the Dcp1p/Dcp2p complex (Hsu and Stevens 1993; Beelman et al. 1996; Steiger et al. 2003) and the human Dcp2 (hDcp2) protein also contains decapping activity (Lykke-Andersen 2002; Van Dijk et al. 2002; Wang et al. 2002). The second enzyme has a scavenger decapping activity that hydrolyzes the resulting cap structure following 3′ to 5′ mRNA decay (Wang and Kiledjian 2001). This activity is carried out by DcpS in humans and Dcs1p in yeast (Liu et al. 2002).
Decapping is a critical step in the turnover of the mRNA. Depending on the pathway of mRNA decay, the cap structure is hydrolyzed either at the onset or following the decay of the mRNA body. By virtue of their substrate specificity, each decapping enzyme functions in one of the two major mRNA decay pathways. A major pathway in mammals involves sequential decay steps from the 3′ end proceeding with removal of the poly(A) tail, degradation of the RNA body, and hydrolysis of the resulting cap structure by DcpS (Liu et al. 2002; Rodgers et al. 2002; Wang and Kiledjian 2000). The DcpS homolog, Dcs1p, carries out the same function in Saccharomyces cerevisiae (Liu et al. 2002). Interestingly, a similar protein was reported recently in Schizosaccharomyces pombe, and it hydrolyzes the cap of an intact mRNA substrate (Salehi et al. 2002).
An alternate and most likely regulated pathway for mRNA decay in mammals involves decapping of the mRNA following the initial deadenylation step. This deadenylation-dependent decapping step is carried out by hDcp2 or an hDcp2-like activity and effectively results in the removal of the terminal structures that protect the ends of the mRNA. Following decapping, the mRNA is degraded, yet the directionality of decay is currently unclear in mammals. In yeast, the Dcp1p/Dcp2p decapping enzyme complex can hydrolyze the cap structure following deadenylation, and the mRNA is subsequently degraded from the 5′ end by Xrn1 (Larimer et al. 1992; Hsu and Stevens 1993; Decker and Parker 1994; Muhlrad et al., 1994; Beelman et al. 1996).
Dcp2p was originally isolated as a high-copy suppressor of a temperature-sensitive dcp1 allele and shown to be required for decapping (Dunckley and Parker 1999). More recently, both the hDcp2 protein (Lykke-Andersen 2002; Van Dijk et al. 2002; Wang et al. 2002) and yeast Dcp2p (Van Dijk et al. 2002; Steiger et al. 2003) were shown to contain intrinsic decapping activity. The Dcp2 proteins are members of the Nudix (nucleotide diphosphate linked to an X moiety) hydrolase family of proteins, which consist of a conserved 23 amino acid sequence core Nudix motif, GX5EX7REUXEEXGU, in which X can be any amino acid, and U is a hydrophobic amino acid (Koonin 1993; Mejean et al. 1994). The evolutionarily conserved Nudix motif is found in a large number of proteins having a diverse spectrum of substrates including nucleoside triphosphates, dinucleotide triphosphates, coenzymes, and nucleotide sugars (Bessman et al. 1996). The Nudix motif is contained within a larger Nudix fold, which consists of two β-sheets sandwiched between α-helices (Gabelli et al. 2001). Residues in the Nudix fold are critical for catalysis, and crystallographic analysis has confirmed the role of key residues in the coordination of metal ion in the MutT and ADP-Ribose proteins (Abeygunawardana et al. 1995; Gabelli et al. 2001).
The hDcp2 protein is cytoplasmic (Wang et al. 2002) and concentrated in distinct foci (Ingelfinger et al. 2002; Lykke-Andersen 2002; Van Dijk et al. 2002). In addition to the Nudix fold, hDcp2 also contains at least two additional motifs conserved within diverse organisms (Wang et al. 2002). The first is Box A that precedes the Nudix fold in all the Dcp2 proteins examined, and the second is Box B that is just carboxy-terminal to the Nudix fold. A fourth region, which is mainly conserved among yeast and human Dcp2 proteins, is located carboxy-terminal to Box B (Lykke-Andersen 2002). Other than the Nudix fold, the functional significance for the conserved domains is not known.
To further investigate the mechanism of mRNA decapping mediated by the hDcp2 protein, we have initiated a biochemical characterization of the hDcp2 protein. We demonstrate that hDcp2 is an RNA-binding protein that utilizes RNA binding to recognize the cap. It preferentially utilizes Mn2+ as a cofactor and the minimal region of the protein that retains methyl cap-specific mRNA decapping activity is constituted by the Nudix fold and Box B domains.
RESULTS
hDcp2 preferentially utilizes Mn2+ as a cofactor for efficient decapping activity
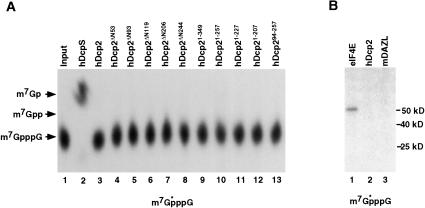
In an initial effort to characterize the hDcp2 protein, we examined the requirement and ability of hDcp2 to utilize various divalent cations for the decapping activity. Our initial studies used Mg2+ exclusively as the cation source that yielded low levels of activity (Wang et al. 2002). Decapping reactions were carried out with equivalent concentrations of different divalent cations. The pcDNA3 polylinker RNA containing a G-track of 16 guanosine residues at the 3′ end (pcP-G16) was utilized as a decapping substrate. We demonstrated previously that the presence of the G-track has no detectable affect on decapping activity, but minimizes potential nonspecific nuclease decay from the 3′ end (Wang and Kiledjian 2001). The decapping products were resolved by polyethyleneimine-cellulose (PEI)-thin layer chromatography (TLC) developed in 0.75 M LiCl as described previously (Wang et al. 2002). As shown in Figure 1 ▶, hDcp2 decapping is dependent upon the presence of a divalent cation, as its omission is inhibitory to cap hydrolysis (Fig. 1 ▶, lane 2). Consistent with our previous findings, m7GDP decapping product was detected in the presence of Mg2+ (Fig. 1 ▶, lane 3). Decapping was not detected or minimally detected with the use of several different metal ions tested except for Mn2+. A significant increase in the detected decapping activity was observed with Mn2+ compared with Mg2+ (Fig. 1 ▶, cf. lanes 3 and 9). Similar stimulation is also detected with the endogenous hDcp2-like decapping activity in mammalian extract (data not shown) as well as yeast Dcp2p (Steiger et al. 2003). We conclude that hDcp2 preferentially uses manganese as a cofactor in vitro. Optimal decapping activity was found to occur with 0.5 mM Mn2+, as higher concentrations did not increase decapping efficiency (data not shown). Addition of Mg2+ to the Mn2+-containing reactions did not have any detectable consequence on decapping efficiency (data not shown). Therefore, all subsequent reactions were carried out in our normal in vitro decay buffer that contains 2 mM Mg2+ supplemented with 0.5 mM Mn2+.
FIGURE 1.
Recombinant hDcp2 utilizes Mn2+ for optimal decapping activity. In vitro decay assays were carried out by incubating 0.5 μg of histidine-tagged recombinant hDcp2 protein at 37°C for 30 min with cap-labeled pcP-G16 RNA. Reactions were carried out in IVDA-2 buffer lacking divalent cation and supplemented with 2 mM of the indicated cations. Products were resolved on PEI-TLC in 0.75 M LiCl. Migration of unlabeled m7GDP standard was visualized by UV-shadowing and is indicated at right.
Identification of the minimal decapping domain in hDcp2
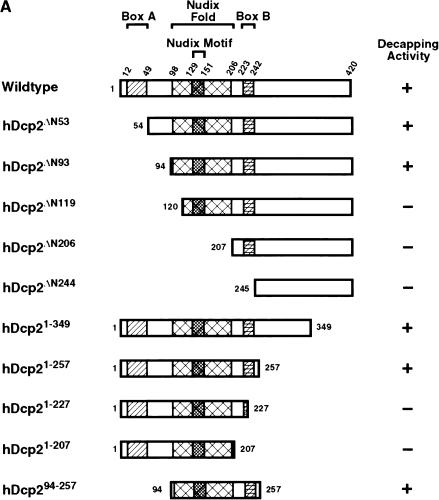
A systematic analysis was undertaken to identify the critical regions in hDcp2 sufficient for decapping. There are at least three motifs that are conserved among the Dcp2 proteins from different organisms: Box A, the Nudix fold, and Box B (Wang et al. 2002). The functional significance of Boxes A and B are unknown, whereas the Nudix fold is predicted to contain the hydrolysis active site. A series of hDcp2 proteins with increasing truncations at the amino terminus and carboxyl terminus were generated. The proteins are represented schematically in Figure 2A ▶ and the corresponding purified recombinant proteins are shown in Figure 2B ▶. The ability of each mutant protein to hydrolyze the cap on an RNA was tested simultaneously. The individual recombinant proteins were incubated with cap-labeled pcP-G16 RNA for 30 min, and the products were resolved by TLC. The ability of each truncation protein to hydrolyze the cap is shown in Figure 2C ▶ and summarized in Figure 2A ▶. Recombinant hDcp2 proteins that truncated regions of the amino terminus removing Box A, but containing an intact Nudix fold, still retained the ability to decap (Fig. 2C ▶, lanes 3,4). Curiously, the protein with a smaller amino-terminal truncation that removes up to and including Box A (hDcp2ΔN53) exhibited reduced activity compared with the protein that removed a larger portion of the amino terminus (hDcp2ΔN93) (Fig. 2C ▶, cf. lanes 3 and 4). An amino-terminal truncation that extended into the Nudix fold (hDcp2ΔN119) resulted in an inactive protein, demonstrating the significance of the Nudix fold for decapping and showing that the amino-terminal 93 amino acids are not required for this function.
FIGURE 2.
Identification of a minimal hDcp2 domain sufficient for cap hydrolysis. (A) The hDcp2 protein and the various truncation proteins are shown schematically. The three regions highly conserved in hDcp2 from diverse organisms are denoted as the 37 amino acid Box A, the 109 amino acid Nudix fold with the central 23 amino acid Nudix motif, and the 19 amino acid Box B segment. The numbers above and on either side of the schematic diagrams represent amino acid positions. The decapping activity for each protein is summarized at right by a + or − for those that can and cannot decap, respectively. (B) Purified recombinant protein was resolved on a 12.5% SDS-PAGE and detected with Coomassie blue. (C) Decapping assays of hDcp2 and hDcp2 mutant proteins. The assays were carried out using 20 pmoles of each protein with cap-labeled pcP-G16 substrate, and the reaction products were resolved by TLC. The quantitation for the decapping efficiency of each protein is presented as the percentage decapping using ImageQuant 5.2 software. Migration of the standards are shown at right. (D) The minimal hDcp294–257 protein retains methyl cap specificity and efficiently utilizes different RNA substrates. Decapping assays were carried out as described above using the 3′ UTR sequences of the indicated mRNAs (α-globin, β-globin, c-myc) and pcP. N-7 methyl-containing capped pcP RNA is denoted as m7G—RNA, whereas the unmethylated is represented as G—RNA. Migration of the standards are indicated on the right.
Mutant hDcp2 proteins containing carboxyl terminus truncations were used to identify the carboxy-terminal border for an active hDcp2 protein. Removal of carboxy-terminal sequences up to Box B had no deleterious consequence on decapping (Fig. 2C ▶, lanes 8,9), whereas larger truncation removing all or part of Box B abrogated the ability of the protein to decap (Fig. 2C ▶, lanes 10,11). These data suggest that the carboxy-terminal boundary for hDcp2 protein decapping activity extends to Box B.
The above analysis indicates that sequences from amino acid 94 to 257 are necessary for decapping. We next determined whether this region was also sufficient for decapping. An hDcp2 recombinant protein from amino acids 94 to 257 was generated (hDcp294–257), which spanned from the start of the Nudix fold to the end of Box B. Figure 2C ▶ demonstrates that this minimal 163 amino acid domain can efficiently hydrolyze the cap-labeled pcP-G16 RNA substrate (Fig. 2C ▶, lane 12) and contains both the substrate recognition and hydrolysis activities. Similar to the full-length hDcp2 protein, the minimal hDcp294–257 protein retained the specificity for utilizing methylated cap as substrate and was able to hydrolyze multiple RNA substrates (Fig. 2D ▶).
Interestingly, the data in Figure 2 ▶, C and D also demonstrate that, in addition to the expected m7GDP product, m7GMP was also generated by the proteins that lacked Box A, (Fig. 2C ▶, lanes 3,4,12, and D, lanes 2,6–8). These results suggest that Box A is essential to maintain enzyme fidelity to hydrolyze the correct phosphate within the cap pyrophosphate linkage.
hDcp2 is an RNA-binding protein
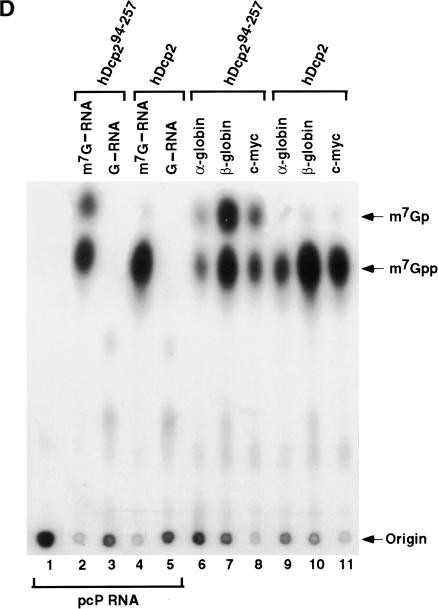
An interesting property of hDcp2 is its ability to hydrolyze a capped RNA but not a cap structure lacking an RNA moiety (Van Dijk et al. 2002; Wang et al. 2002). This observation suggests that either the hydrolysis activity of hDcp2 is activated upon RNA binding, or that hDcp2 is unable to recognize the cap structure directly and is an RNA-binding protein that detects its substrate through association with the RNA moiety. To distinguish between these two possibilities, the hDcp2 truncation mutants listed in Figure 2A ▶ were tested for their ability to hydrolyze 32P-labeled methylated cap structure (m7GpppG). We reasoned that if a regulatory region that masked decapping activity was present, the decapping and regulation could be uncoupled. Figure 3A ▶ shows that none of the mutants were capable of decapping the 32P-labeled m7GpppG cap structure substrate. Furthermore, addition of unlabeled RNA competitor to labeled cap analog in trans also did not stimulate the ability of the hDcp2 proteins to hydrolyze the cap structure (data not shown). These results indicate that the requirement for an RNA moiety is not to unmask an inhibitory domain or to activate the cap hydrolysis activity of the hDcp2 enzyme. The data suggest that hDcp2 is unable to bind the cap structure directly. Consistent with this hypothesis, under conditions in which efficient ultraviolet (UV) cross-linking of the eIF4E cap-binding protein was detected to 32P-labeled cap structure substrate (Fig. 3B ▶, lane 1), we were unable to detect cross-linking of hDcp2 to the cap structure (Fig. 3B ▶, lane 2). These data are consistent with a model whereby hDcp2 is an RNA-binding protein, and its ability to recognize and hydrolyze the cap structure requires prior association with the RNA body.
FIGURE 3.
hDcp2 does not utilize the cap structure as substrate. (A) hDcp2 is unable to hydrolyze the cap structure in the absence of the RNA body. 32P-labeled cap analog was incubated with 10 pmole of each hDcp2 protein and the products resolved by TLC. The scavenger decapping activity DcpS, which can hydrolyze the cap structure, was used as the positive control (lane 2). Standards are indicated at left. (B) hDcp2 is unable to cross-link to cap analog. 32P-labeled cap analog was UV cross-linked with 20 pmole of the indicated proteins and the products resolved by 12.5% SDS-PAGE. GST fusion protein of the cap-binding protein, eIF4E (lane 1), was used as the positive control. Histidine-tagged hDcp2 protein in lane 2, and an unrelated RNA-binding protein control, GST-mDAZL (Jiao et al. 2002), was used in lane 3. Molecular weight standards are indicated at right. The cap analog substrate is shown at bottom, and the star denotes the labeled phosphate.
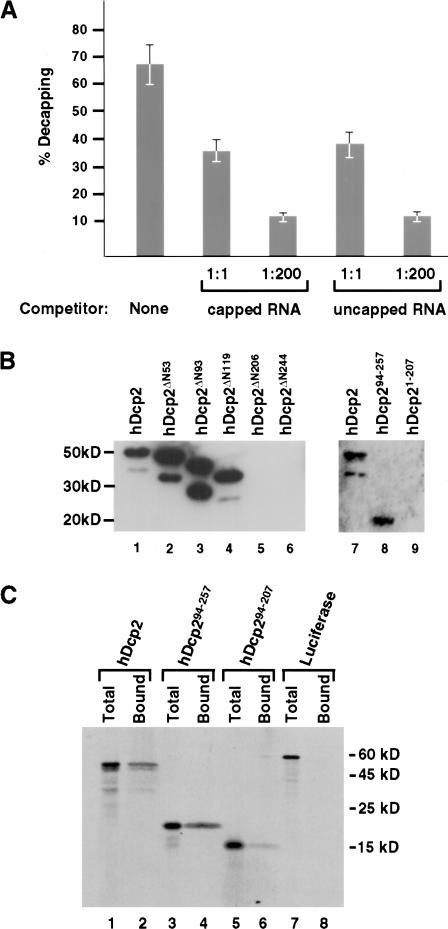
The ability of uncapped RNA to compete for hDcp2 decapping was next tested. Decapping assays were carried out with hDcp2 protein and a 32P-cap labeled pcP-G16 RNA substrate in the presence of unlabeled RNA competitor. As shown in Figure 4A ▶, hDcp2-mediated decapping was inhibited by the addition of capped RNA competitor. Surprisingly, a similar reduction of decapping was also detected when uncapped RNA was used as competitor. The ability of uncapped RNA to compete for hDcp2 decapping was indistinguishable from capped RNA competitor even at 200-fold molar excess, which would suggest the difference is not due to rapid hydrolysis of the capped RNA competitor. These data suggest that hDcp2 has an intrinsic RNA recognition property that is independent of the cap structure, and RNA binding is the primary means by which hDcp2 can recognize its capped RNA substrate for hydrolysis.
FIGURE 4.
hDcp2 is an RNA-binding protein. (A) RNA can compete for hDcp2-mediated decapping. Capped and uncapped pcP-G16 RNAs were used as competitor. In vitro decay assays were carried out with 20 pmole of hDcp2 with no competitor (None), 1:1 or 1:200 molar fold excess competitor RNA as indicated. The decapping products were resolved by TLC and the percent decapping is plotted. Both RNAs were found to compete equally for hDcp2 activity. (B) Binding of hDcp2 to RNA by Northwestern analysis. Six μg of each protein was resolved by SDS-PAGE, transferred to nitrocellulose, and probed with uniform-labeled, uncapped pcP RNA. The proteins are shown schematically in Figure 2 ▶ and their size on SDS-PAGE corresponds to the following molecular weights as follows: hDcp2, 48kD; hDcp2ΔN53, 43 kD; hDcp2ΔN93, 38 kD; hDcp2ΔN119, 35 kD; hDcp2ΔN206, 25 kD; hDcp2ΔN244, 20 kD; hDcp294–257, 19 kD; hDcp21–207, 23 kD. (C) Binding of hDcp2 to ssDNA. [35S]methionine in vitro-translated proteins were bound to ssDNA-agarose beads and resolved by SDS-PAGE. The total lanes contain 25% of the input protein and the Bound lanes contain the protein bound to ssDNA. The hDcp2 and hDcp294–257 proteins are as shown in Figure 2 ▶ and hDcp294–207 contains only the Nudix fold and lacks Box B. The Firefly Luciferase control protein is designated as Luciferase.
To directly determine whether hDcp2 is an RNA-binding protein, Northwestern analysis was carried out. hDcp2 truncation proteins were resolved (SDS-PAGE; 12.5%), transferred to nitrocellulose, and probed with uniform-labeled, uncapped pcP RNA. Figure 4B ▶ shows that recombinant hDcp2 was capable of binding RNA, as the 32P-labeled RNA was retained by the protein (Fig. 4B ▶, lane 1). In an attempt to map the region of hDcp2 required for RNA binding, a subset of the truncation mutants were next tested for their ability to bind RNA. The ability of hDcp2ΔN119 to bind RNA (Fig. 4B ▶, lane 4) indicates that the presence of an intact Nudix fold containing hydrolysis activity is not essential for RNA binding. Therefore, RNA binding is not dependent on a functional cap hydrolysis activity. Larger truncations from the amino terminus that removed the entire Nudix fold were unable to bind RNA (Fig. 4B ▶, lanes 5,6) demonstrating that at least part of the Nudix fold is necessary for RNA binding. As expected, the hDcp294–257 minimal decapping protein was also capable of binding RNA (Fig. 4B ▶, lane 8), yet an hDcp2 protein lacking the carboxy-terminal region including Box B was unable to bind RNA (Fig. 4B ▶, lane 9).
The significance of Box B for nucleotide binding was further reinforced with the use of ssDNA-binding analysis. The hDcp2 and hDcp294–257 proteins were translated in vitro in the presence of [35S]methionine, and incubated with ssDNA agarose beads. Bound proteins were isolated and resolved by SDS-PAGE. Consistent with the Northwestern results, both proteins were able to bind ssDNA (Fig. 4C ▶, lanes 2,4). A third protein, hDcp294–207, which contains only the Nudix motif was significantly compromised in its ability to bind. Collectively, these data suggest that hDcp2 is an RNA-binding protein and the carboxy-terminal region of the Nudix fold as well as Box B are necessary for RNA binding.
RNA binding is essential for cap binding
To determine the correlation between cap recognition and RNA binding, we tested the ability of hDcp2 to bind the cap. hDcp2 was incubated with cap-labeled pcP-G16 RNA on ice to minimize hydrolysis, and the ability to associate with the cap was tested by UV cross-linking. Contrary to the lack of hDcp2 cross-linking to the cap structure without an RNA moiety shown in Figure 3B ▶, hDcp2 can cross-link to the cap structure of pcP-G16 RNA (Fig. 5 ▶, lane 2), demonstrating that hDcp2 requires the presence of the RNA moiety to gain access to the cap. The detected cross-linking is most likely an underestimation, as the capped RNA is also a substrate for the hDcp2 hydrolysis reaction.
FIGURE 5.
Cap binding of hDcp2 is RNA dependent. (A) hDcp2 can be UV cross-linked to cap-labeled RNA. An autoradiograph of an SDS-PAGE showing 10 pmole of the indicated protein cross-linked to 32P-cap-labeled pcP-G16 RNA. Cross-linking of the eIF4E cap-binding protein (GST–eIF4E, 50 kD; lanes 1,4) was used as a positive control. An unrelated GST–mDAZL RNA-binding protein (68 kD; lane 3), or purification of a His-tag empty vector (lane 9) are shown as negative controls. The sizes of the hDcp2 proteins and truncations are as described in the legend to Figure 4 ▶. The capped RNA substrate is shown at bottom, where the line represents the RNA and the star denotes the labeled phosphate.
The ability of the mutant proteins to cross-link the cap-labeled pcP-G16 RNAs was also tested. As expected, the truncation mutant proteins hDcp2ΔN93 and hDcp294–257, which were able to bind RNA and contained decapping activity, bound to the cap structure of the cap-labeled RNA (Fig. 5 ▶, lanes 5,7). The increased cross-linking of hDcp2ΔN93 was reproducible and could reflect more optimal positioning of the protein for UV cross-linking. The hDcp21–207 mutant protein, which has an intact Nudix fold, does not bind RNA and is inactive for decapping, was also unable to cross-link to the cap (Fig. 5 ▶, lane 8). However, the most telling result was with the cross-linking of hDcp2Δ119N (Fig. 5 ▶, lane 6). This mutant protein is unable to hydrolyze the cap, yet is able to bind RNA (Fig. 4 ▶) and is also capable of cross-linking to the cap of the pcP-G16 RNA, albeit at lower efficiency (Fig. 5 ▶, lane 6). Our data indicates that the hDcp2 protein cap recognition requires prior RNA-binding and that hDcp2 has RNA-dependent decapping activity.
DISCUSSION
In this study, we have presented a biochemical characterization of the hDcp2 decapping enzyme. Maximal decapping activity by hDcp2 was found to require Mn2+ as a cofactor. A minimal 163 amino acid domain including the Nudix hydrolase fold and the evolutionarily conserved Box B region were found to retain all of the properties for hydrolysis of the capped RNA. Last, we demonstrate that hDcp2 is an RNA-binding protein that recognizes the capped RNA substrate by an association with the RNA moiety prior to recognition and hydrolysis of the cap.
Most Nudix proteins characterized thus far appear to preferentially utilize Mg2+ as a cofactor. Exceptions include the Escherichia coli GDP-mannose mannosyl hydrolase, which is equally efficient with both Mg2+ and Mn2+ (Legler et al. 2002) and the Arabidopsis thaliana NADH hydrolase protein, with a cofactor preference of Mn2+ over Mg2+ (Dobrzanska et al. 2002). hDcp2 is the second example of a Nudix protein whose activity is greatly enhanced with Mn2+ as the divalent cation. The activity of the yeast Dcp2p enzyme is also more efficient with Mn2+ (Steiger et al. 2003), implying a structural conservation in the catalytic core of the two Dcp2 proteins. How Mn2+ promotes more efficient decapping by hDcp2 is currently unknown. The demonstration that Mn2+ decreases the Km of the Nudix-containing MutT enzyme for its dGTP substrate (Frick et al. 1994) suggests that Mn2+ could similarly decrease the Km of the hDcp2 cap-substrate complex. Further analyses are required to determine which kinetic parameter of the decapping reaction is specifically affected by Mn2+.
We noted previously the presence of two conserved motifs in addition to the Nudix hydrolase domain within hDcp2. A region termed Box A is present at the amino terminus of the protein and a Box B is positioned carboxy-terminal to the Nudix fold (Wang et al. 2002). Our current data indicate that a minimal domain, spanning amino acids 94–257, which encompasses the Nudix fold and the conserved Box B region is competent to decap an mRNA. Therefore, both the substrate recognition and hydrolysis domains are present within this 163 amino acid region. The truncation analysis demonstrated that Box B is essential for the RNA-binding activity, as carboxy-terminal truncations that lacked Box B were unable to bind RNA (Fig. 4B ▶). More significantly, removal of Box B from the minimal domain protein also prevented nucleic acid binding (Fig. 4C ▶). However, despite the requirement of this region for RNA binding, it is not sufficient to function as an autonomous RNA-binding domain, as the hDcp2ΔN206 protein, which retains an intact Box B, did not bind RNA with this assay system. Sequences at the carboxyl terminus of the Nudix fold in addition to Box B were also required.
The significance of Box A for hDcp2 protein decapping is less obvious. Truncations that remove this region of the protein are still capable of hydrolyzing the cap, however, the fidelity of the enzyme to cleave at the appropriate phosphate is compromised. hDcp2 normally hydrolyzes the pyrophosphate linkage of the cap between the β- and γ-phosphate to produce the m7GDP product. The mutant proteins that lacked Box A also generated m7GMP in addition to m7GDP. Therefore, although Box A is not essential for cap hydrolysis, it could be involved in maintaining the structural integrity of the Nudix fold to enable decapping at the appropriate phosphate linkage. Ongoing structural analysis of hDcp2 will help elucidate the requirement of Box A for correct phosphate bond cleavage. It is also possible that Box A could be a potential regulatory site to influence the decapping product of hDcp2 and enable production of m7GMP under certain physiological conditions.
hDcp2 is an RNA-binding protein with an RNA-dependent decapping activity. Several lines of evidence suggest that hDcp2 is unable to recognize the cap directly and requires prior binding to the RNA to detect and hydrolyze the cap substrate. First, unlike the DcpS decapping enzyme, which is competed by cap analog competitor (Liu et al. 2002), hDcp2 decapping is not inhibited by cap analog competitor (Van Dijk et al. 2002; Wang et al. 2002), suggesting that it is unable to directly recognize the cap structure. Furthermore, it is also unable to hydrolyze the cap structure lacking an RNA moiety (Fig. 3A ▶), requires a capped RNA containing 23 nucleotides or greater as a substrate for decapping (Van Dijk et al. 2002; Wang et al. 2002), and is equally competed by both capped and uncapped RNA (Fig. 4A ▶). Collectively, these observations suggest that hDcp2 detects its cap substrate by RNA binding, and the recognition of the 5′ cap is strictly dependent on the presence of the RNA moiety. Furthermore, we have observed that the addition of uncapped RNA does not promote hDcp2 hydrolysis of cap structure in trans (C. Piccirillo and M. Kiledjian, unpubl.). This result suggests RNA binding is not sufficient to facilitate hDcp2 cap recognition. Instead, RNA binding may serve to position the catalytic center of the enzyme in proximity to the cap structure, while in turn promoting hydrolysis.
Decapping by hDcp2 can be regulated at multiple levels. One level of regulation involves modulation by the cap-binding proteins. The eIF4E cap-binding protein has been demonstrated to inhibit the yeast decapping enzyme (Schwartz and Parker 1999, 2000; Vilela et al. 2000) as well as the human enzyme (R. Khanna and M. Kiledjian, unpubl.). It is likely that other proteins with cap-binding potential can also regulate decapping. Second, in addition to control at the cap-binding step, regulation could occur at the initial hDcp2 RNA-binding step. The sequence and/or structure at the 5′ end of an mRNA, as well as proteins that could potentially influence the accessibility of the mRNA sequences at the 5′ end, will greatly impact decapping efficiency. For example, the stimulatory affect of decapping by the putative helicase protein Dhh1p (Coller et al. 2001; Fischer and Weis 2002) could be to relieve potential secondary structures present at the 5′ end of the mRNA to enable hDcp2 binding. Similarly, the Edc1p and Edc2p proteins, which are RNA-binding proteins that stimulate decapping (Schwartz et al. 2003) could function by maintaining the RNA in an open conformation to enable more efficient hDcp2 binding. The opposite can also be envisioned, in which RNA-binding proteins can prevent access of hDcp2 to the 5′ end of an mRNA independent of the cap-binding proteins as well as proteins that can directly prevent the RNA-binding activity of hDcp2.
We propose a model by which hDcp2 functions to recognize and hydrolyze the cap. The RNA-binding property of hDcp2 anchors the enzyme onto the capped RNA substrate adjacent to the cap structure and positions the Nudix motif for cap hydrolysis. The requirement of an accessible RNA at the 5′ end is also true for the yeast Dcp2p activity, in which an antisense oligonucleotide at the 5′ end adjacent to the cap inhibits decapping (Steiger et al. 2003). Whether hDcp2 detection of the cap structure occurs by random binding events on the RNA or by an active 3′ to 5′ scanning mechanism to search for the cap substrate is currently unknown. All RNA substrates we have thus far tested are effectively decapped by hDcp2 (Fig. 2D ▶; C. Piccirillo and M. Kiledjian, unpubl.). It is unclear whether the observed differences in decapping efficiency of the different RNA substrates in Figure 2D ▶ are a consequence of local structures at the cap influencing the accessibility of hDcp2, or a reflection of sequence preference. However, most RNA-binding proteins have both a basal level of low-affinity nonspecific RNA-binding activity as well as high-affinity sequence-specific binding activity. Whether high-affinity hDcp2-binding sites that are capable of targeting mRNA for rapid decapping exist, remains to be determined.
The demonstration that hDcp2 is an RNA-binding protein will now permit investigations into novel mechanisms regulating mRNA decapping. Factors that modulate the RNA-binding efficiency of hDcp2 can provide a new mode of decapping regulation. Identification of high-affinity binding sites for hDcp2 and specific RNA substrates containing these sites will greatly expedite our understanding of targeted mRNA turnover and provide novel avenues to control gene expression by mRNA stability.
MATERIALS AND METHODS
Plasmids and expression of recombinant protein
The pET-hDcp2 plasmid expressing the wild-type hDcp2 protein with a histidine tag and the pET-hDcp21–349 encoding hDcp2 amino acids 1–349 have been described previously (Wang et al. 2002). All hDcp2 truncations were generated by PCR with primers that introduce a BamHI site on the 5′ end and an XhoI site on the 3′ end and were cloned into the same sites in pET28a (Novagen). The plasmid pET-hDcp2ΔN53 encodes sequences corresponding to hDcp2 amino acids 54–420 and was generated using a 5′ primer (5′-TAATAGGATCCCCAGGATTACCTCAGTGTG-3′) and the 3′ hDcp2 primer (5′-CAATACTCGAGCTGCTATCAAAGGTC CAAG-3′). pET-hDcp2ΔN93 was similarly constructed with a 5′ primer (5′-TAAAAGGATCCATGGGAGTACCAACATATG-3′) and the 3′ hDcp2 primer and encodes amino acids 94–420. pET-hDcp2ΔN119 was generated with the 5′ primer (5′-TAATAGGA TCCGCAAAATCAGGCTGGG GA-3′) and the 3′ hDcp2 primer, and removes the amino-terminal 119 amino acids of hDcp2. The plasmid pET-hDcp2ΔN206 was PCR amplified with the 3′ hDcp2 primer and the 5′ primer (5′-TAATAGGATCCTCATAGAAAT GATATGAC-3′). This truncation corresponded to a deletion of the amino-terminal-most 206 amino acids. pET-hDcp2ΔN244 removes the amino-terminal 244 amino acids and was constructed using a 5′ primer (5′-TAATAGGATCCTTCCTCAGACAGTGAC AA-3′) and the 3′ hDcp2 primer. pET-hDcp21–257 encodes hDcp2 amino acids 1–257 and was generated with the 5′ primer (5′-TAATA GGATCCCCAGGATTACCTCAGTGTG-3′) and a 3′ primer (5′CAATACTCGAGGCGTGCTACCAGTTGAGGA-3′). pET-hDcp21–227 encompassing amino acids 1–227 was generated by digesting pGEMT-hDcp2 (Wang et al. 2002) with MscI and BamHI and inserted directionally into the BamHI site and blunt-ended XhoI site. pET-hDcp21–207 encodes hDcp2 sequences from the amino terminus to amino acid 207, was PCR amplified with the 5′ primer (5′-TAATAGGATCCCCAGGATTACCTCAGTGTG-3′) and 3′ primer (5′-AAGCTTCTCGAGGTCATATCATTTCTATGACAAG G-3′). The plasmid pET-hDcp294–257 encodes amino acids 94–257 and was PCR amplified using a 5′ primer (5′-TAAAAGGATC CATGGGAGTACCAACATATG-3′) and a 3′ primer (5′-CAAT ACTCGAGGCGTGCTACCAGTTGAGGA-3′). The recombinant proteins were expressed in E. coli BL21DE3 cells (Novagen) and purified on a nickel column under denaturing conditions with 4 M urea as described by the manufacturer. Purified protein was dialyzed overnight in Renaturation Buffer (50 mM Tris-HCl, 500 mM L-Arginine, 5 mM EDTA, 0.4% PEG 4000, 5 mM reduced Glutathione, oxidized 1 mM Glutathione) to renature the protein and remove urea. Stepwise dialysis was subsequently performed to replace the Renaturation Buffer with Storage Buffer (10 mM Tris, 100 mM NaCl, 2 mM Mg-Ac, 1 mM DTT, 10% Glycerol).
RNA production
RNAs corresponding to the 121-nucleotide pcDNA3 polylinker containing a polyG track at the 3′ end (pcP-G16) were transcribed with SP6 RNA Polymerase from a template generated by using the SP6 promoter sequence as the 5′ primer and an antisense T7 promoter primer, followed by 16 cytosines for the 3′ primer. The 105-nucleotide pcP RNA was transcribed similar to pcP-G16 RNA, except the 3′ primer lacked the cytosines. RNA corresponding to α-globin 3′UTR was generated as described previously (Wang et. al. 1999). The c-myc 3′UTR RNA was generated by PCR amplification using a 5′ primer containing a T7 promoter (5′-CGTAAT ACGACTCACTATAGGGGACTTTGGGCATAAAAG-3′) and a 3′ primer (5′-CTGCTATAAACGTTTTAT-3′). The β-globin 3′UTR RNA was PCR amplified using a T7 promoter containing 5′ primer (5′-CGTAATACGACTCACTATAGGGGCTCGCTTTCTT GCTGTCC-3′) and a 3′ primer (5′-CAATGAAAATAAATGTTT TTTATTAGGCAGAATCC-3′). The c-myc and β-globin RNAs were transcribed using T7 RNA Polymerase. Cap-labeled RNAs were generated as described previously (Wang et al. 2001), and all RNAs were gel purified prior to use (Wang et al. 1999). 32P-labeled cap analog was generated with RNase P1 digestion of cap-labeled RNA as described previously (Wang and Kiledjian 2001).
In vitro RNA decay assay
In vitro RNA decay assays were carried out essentially as described previously (Wang et al. 1999) with minor modifications. The indicated amount of extract or recombinant protein was incubated with cap-labeled pcP-G16 RNA (10,000 cpm) at 37°C for 30 min in IVDA-2 buffer (10 mM Tris:HCl at pH 7.5, 100 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM MnCl2 2 mM dithiothreitol, and 0.1 mM Spermine). An aliquot of each sample was resolved by PEI-TLC developed in 0.75 M LiCl. In Figure 1 ▶, the magnesium acetate and MnCl2 were omitted from the buffer and 2 mM of the indicated cation was used. Unlabeled TLC standards were visualized by UV-shadowing, and 32P-labeled signals were detected by autoradiography.
In vitro translation and ssDNA-agarose bead-binding assay
The TNT T7-Coupled Reticulocyte Lysate Transcription/Translation System (Promega) was used to generate in vitro-translated [35S]methionine hDcp2 proteins according to the manufacturers’ protocol (Promega). Single-stranded DNA (ssDNA)-binding assays were carried out with ssDNA-agarose beads (GIBCO-BRL), washed four times with RBB (150 mM KCl, 10 mM Tris-HCl at pH 8.0, 1.5 mM MgCl2, 0.5 mM DTT, 0.5% Triton X-100) prior to use. Binding reactions were carried out in 450μL reaction volume containing RBB and the indicated in vitro-translated hDcp2 proteins (~150,000 cpm trichloroacetic acid precipitable protein) at 4°C for 1 h on a nutator. Beads containing the bound protein were pelleted with a brief spin and washed four times with RBB. Bound proteins were eluted by boiling with 50μL of SDS-polyacrylamide sample buffer. The proteins were resolved on a 12.5% SDS-PAGE, dried and exposed to Kodak BioMax film, and detected by autoradiography.
Northwestern RNA-binding assay
Northwestern analysis was carried out as described previously (Kiledjian et al. 1999). Briefly, 6 μg of histidine-tagged recombinant hDcp2 or truncation mutant proteins were resolved by 12.5% SDS-PAGE. The proteins were electroblotted onto nitrocellulose membrane and renatured with a 1-h incubation on a shaking platform in Northwestern (NW) buffer (10 mM Tris-HCl at pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 1× Denhardt’s solution) at room temperature. The blot was probed with uniform-labeled, pcP RNA lacking a poly(A) tail or cap, in NW buffer containing 10 mg/mL E. coli tRNA for 1 h at room temperature, washed twice for 30 min with NW buffer containing 1 mg/mL Heparin, exposed to X-ray film, and detected by autoradiography.
UV cross-linking
Binding of hDcp2 or hDcp2 truncation mutant proteins to capped RNA or cap structure were carried out in Eppendorf tube caps placed on ice. Binding reactions were in IVDA-2 buffer lacking MnCl2 in the presence of 3 pmole of 32P cap-labeled pcP-G16 RNA or 32P-labeled cap structure (m7GpppG) incubated with 40 pmole of the indicated protein for 15 min on ice. Cross-linking of protein to RNA was carried out with Ultraviolet (UV) light irradiation for 10 min with a germicidal lamp positioned 5 cm above the caps. The RNA was subsequently digested with an RNase cocktail containing 0.1 mg/mL each of RNase A and U2 for 30 min at 37°C, and the cross-linked proteins were resolved on 12.5% SDS-PAGE, followed by detection by autoradiography.
Acknowledgments
We thank A.C. Gingras and N. Sonenberg (McGill University) for providing the pGEX-meIF4E plasmid, A. Carr-Schmid for critical reading of the manuscript, and members of the Kiledjian lab for helpful discussion. This work was supported by funds from the National Institutes of Health to M.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Abeygunawardana, C., Weber, D.J., Gittis, A.G., Frick, D.N., Lin, J., Miller, A.F., Bessman, M.J., and Mildvan, A.S. 1995. Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase. Biochemistry 34: 14997–15005. [DOI] [PubMed] [Google Scholar]
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., and Parker, R. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646. [DOI] [PubMed] [Google Scholar]
- Bessman, M.J., Frick, D.N., and O’Handley, S.F. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271: 25059–25062. [DOI] [PubMed] [Google Scholar]
- Coller, J.M., Tucker, M., Sheth, U., Valencia-Sanchez, M.A., and Parker, R. 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7: 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J. and Parker, R. 1994. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem. Sci. 19: 336–340. [DOI] [PubMed] [Google Scholar]
- Dobrzanska, M., Szurmak, B., Wyslouch-Cieszynska, A., and Kraszewska, E. 2002. Cloning and characterization of the first member of the Nudix family from Arabidopsis thaliana. J. Biol. Chem. 277: 50482–50486. [DOI] [PubMed] [Google Scholar]
- Dunckley, T. and Parker, R. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18: 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, N. and Weis, K. 2002. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 21: 2788–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick, D.N., Weber, D.J., Gillespie, J.R., Bessman, M.J., and Mildvan, A.S. 1994. Dual divalent cation requirement of the MutT dGTPase. Kinetic and magnetic resonance studies of the metal and substrate complexes. J. Biol. Chem. 269: 1794–1803. [PubMed] [Google Scholar]
- Gabelli, S.B., Bianchet, M.A., Bessman, M.J., and Amzel, L.M. 2001. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat. Struct. Biol. 8: 467–472. [DOI] [PubMed] [Google Scholar]
- Hsu, C.L. and Stevens, A. 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13: 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger, D., Arndt-Jovin, D.J., Luhrmann, R., and Achsel, T. 2002. The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA 8: 1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Jiao, X., Trifillis, P., and Kiledjian, M. 2002. Identification of target messenger RNA substrates for the murine deleted in azoospermia-like RNA-binding protein. Biol. Reprod. 66: 475–485. [DOI] [PubMed] [Google Scholar]
- Kiledjian, M., Day, N., and Trifillis, P. 1999. Purification and RNA binding properties of the polycytidylate-binding proteins αCP1 and αCP2. Methods 17: 84–91. [DOI] [PubMed] [Google Scholar]
- Koonin, E.V. 1993. A highly conserved sequence motif defining the family of MutT-related proteins from eubacteria, eukaryotes and viruses. Nucleic Acids Res. 21: 4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimer, F.W., Hsu, C.L., Maupin, M.K., and Stevens, A. 1992. Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: Sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene 120: 51–57. [DOI] [PubMed] [Google Scholar]
- Legler, P.M., Lee, H.C., Peisach, J., and Mildvan, A.S. 2002. Kinetic and magnetic resonance studies of the role of metal ions in the mechanism of Escherichia coli GDP-mannose mannosyl hydrolase, an unusual nudix enzyme. Biochemistry 41: 4655–4668. [DOI] [PubMed] [Google Scholar]
- Liu, H., Rodgers, N.D., Jiao, X., and Kiledjian, M. 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 21: 4699–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejean, V., Salles, C., Bullions, L.C., Bessman, M.J., and Claverys, J.P. 1994. Characterization of the mutX gene of Streptococcus pneumoniae as a homologue of Escherichia coli mutT, and tentative definition of a catalytic domain of the dGTP pyrophosphohydrolases. Mol. Microbiol. 11: 323–330. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C.J., and Parker, R. 1994. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes & Dev. 8: 855–866. [DOI] [PubMed] [Google Scholar]
- Rodgers, N.D., Wang, Z., and Kiledjian, M. 2002. Characterization and purification of a mammalian endoribonuclease specific for the α-globin mRNA. J. Biol. Chem. 277: 2597–2604. [DOI] [PubMed] [Google Scholar]
- Salehi, Z., Geffers, L., Vilela, C., Birkenhager, R., Ptushkina, M., Berthelot, K., Ferro, M., Gaskell, S., Hagan, I., Stapley, B., et al. 2002. A nuclear protein in Schizosaccharomyces pombe with homology to the human tumour suppressor Fhit has decapping activity. Mol. Microbiol. 46: 49–62. [DOI] [PubMed] [Google Scholar]
- Schwartz, D., Decker, C.J., and Parker, R. 2003. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA 9: 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D.C. and Parker, R. 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 5247–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 20: 7933–7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger, M., Carr-Schmid, A., Schwartz, D.C., Kiledjian, M., and Parker, R. 2003. Analysis of recombinant yeast decapping enzyme. RNA 9: 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., and Seraphin, B. 2002. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21: 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela, C., Velasco, C., Ptushkina, M., and McCarthy, J.E. 2000. The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J. 19: 4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. and Kiledjian, M. 2000. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol. Cell. Biol. 20: 6334–6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751–762. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Day, N., Trifillis, P., and Kiledjian, M. 1999. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol. Cell. Biol. 19: 4552–4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Jiao, X., Carr-Schmid, A., and Kiledjian, M. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. 99: 12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]