Abstract
Transcription antitermination of the Bacillus subtilis glyQS gene, a member of the T box gene regulation family, can be induced during in vitro transcription in a minimal system using purified B. subtilis RNA polymerase by the addition of unmodified T7 RNA polymerase-transcribed tRNAGly. Antitermination was previously shown to depend on base-pairing between the glyQS leader and the tRNA at the anticodon and acceptor ends. In this study, variants of tRNAGly were generated to identify additional tRNA elements required for antitermination activity, and to determine the effect of structural changes in the tRNA. We find that additions to the 3′ end of the tRNA blocked antitermination, in agreement with the prediction that uncharged tRNA is the effector in vivo, whereas insertion of 1 nucleotide between the acceptor stem and the 3′ UCCA residues had no effect. Disruption of the D-loop/T-loop tertiary interaction inhibited antitermination function, as was previously demonstrated for tRNATyr-directed antitermination of the B. subtilis tyrS gene in vivo. Insertion of a single base pair in the anticodon stem was tolerated, whereas further insertions abolished antitermination. However, we find that major alterations in the length of the acceptor stem are tolerated, and the insertions exhibited a pattern of periodicity suggesting that there is face-of-the-helix dependence in the positioning of the unpaired UCCA residues at the 3′ end of the tRNA for interaction with the antiterminator bulge and antitermination.
Keywords: tRNA, gene regulation, Gram-positive, transcription termination, attenuation, RNA structure
INTRODUCTION
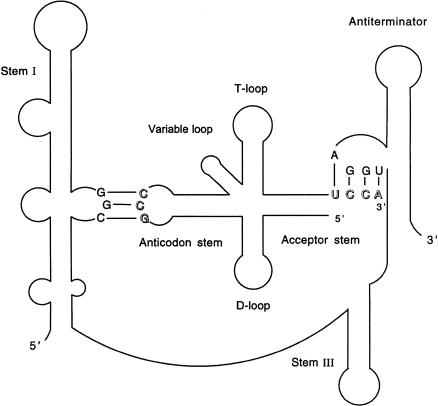
Regulation of gene expression at the level of premature termination of transcription has emerged as a common mechanism in bacteria (Henkin 2000; Henkin and Yanofsky 2002). Synthesis of the full-length mRNA is controlled by modulation of the activity of a termination signal located in the leader region of the target gene, upstream of the start of the coding region. The activity of the terminator can be controlled by modification of the activity of RNA polymerase (RNAP), or by interaction of regulatory components with the leader RNA to direct folding of the RNA into the terminator structure or an alternate antiterminator structure. The T box antitermination system of Gram-positive bacteria is unique in that formation of the antiterminator structure, and expression of the target gene is dependent on interaction of the leader RNA with the cognate uncharged tRNA (Grundy and Henkin 1993; Grundy et al. 1994). Genes in this family exhibit a complex set of conserved structural and sequence elements within the leader region. The specificity of the leader RNA–tRNA interaction is determined at least in part by pairing of the tRNA anticodon with a single codon (the “specifier sequence”) in the leader, whereas stabilization of the antiterminator and prevention of formation of the competing terminator helix is dependent on base-pairing of the four free nucleotides (5′-NCCA-3′) at the 3′ end of the tRNA with complementary residues (5′-UGGN-3′) in a 7-nt bulge of the antiterminator (Fig. 1 ▶). This second interaction is blocked by attachment of the amino acid to the tRNA.
FIGURE 1.
Interaction of the glyQS leader in the antiterminator conformation with tRNAGly. The anticodon of tRNAGly (5′-GCC-3′) pairs with the specifier sequence (5′-GGC-3′) in Stem I; the acceptor end of tRNA (5′-UCCA-3′) pairs with four residues (5′-UGGA-3′) in the antiterminator bulge. Stabilization of the antiterminator structure sequesters sequences required for formation of the leader region transcription terminator, causing synthesis of the full-length transcript.
Although these two base-pairing interactions have been shown to be necessary for antitermination both in vivo and in vitro (Grundy et al. 1994, 1997, 2002; Putzer et al. 2002), it appears that other elements of the tRNA and leader are also required. Studies in which the specifier sequence and antiterminator bulge elements of the Bacillus subtilis tyrS leader were varied to match noncognate tRNA species demonstrated that although some classes of tRNAs could direct antitermination of an appropriately altered tyrS leader RNA, others could not, and no noncognate tRNA was as efficient as the cognate tRNATyr (Grundy et al. 1997). Similar results have been observed with other T box family genes (Putzer et al. 1995; Marta et al. 1996; Luo et al. 1997). Mutational analysis of tRNATyr-directed antitermination of tyrS in vivo showed that the sequence in the helical regions of the tRNA could be varied, and the long variable arm was dispensable for antitermination, and could tolerate a large insertion; however, mutations that disrupted tRNA tertiary structure abolished antitermination function (Grundy et al. 2000).
The in vivo studies of tRNATyr variants were designed to generate tRNA molecules independent of the normal tRNA processing pathways. Nevertheless, these studies were limited by the fact that certain tRNA variants are subject to repair within the cell (Deutscher 1990). The development of an in vitro antitermination assay using the B. subtilis glyQS gene, encoding glycyl-tRNA synthetase, and unmodified tRNAGly generated by in vitro transcription by T7 RNAP (Grundy et al. 2002) permits testing of any tRNA variant that is generated by manipulation of the template DNA. For example, we were able to generate tRNA variants with extra residues at the 3′ end of the tRNA, which was not possible in vivo due to posttranscriptional 3′ end processing of the tRNA. Because it was previously demonstrated for tRNATyr-directed antitermination that only the isoacceptor tRNA with an exact match to the specifier sequence was able to promote antitermination (Grundy et al. 1994), we used the tRNAGly isoacceptor whose anticodon matched the specifier sequence of the glyQS leader. In this study, we show that the tertiary structure of the tRNA is required for antitermination in vitro, consistent with previous in vivo analyses. Insertion of a full turn of the RNA helix in the acceptor stem was allowed, whereas a partial turn abolished function, indicating a remarkable tolerance for an increase in distance between the ends of the tRNA, in conjunction with constraints on the angle of presentation of the 3′ residues for interaction with the antiterminator bulge.
RESULTS
Requirement for the tRNAGly D-loop/T-loop interaction
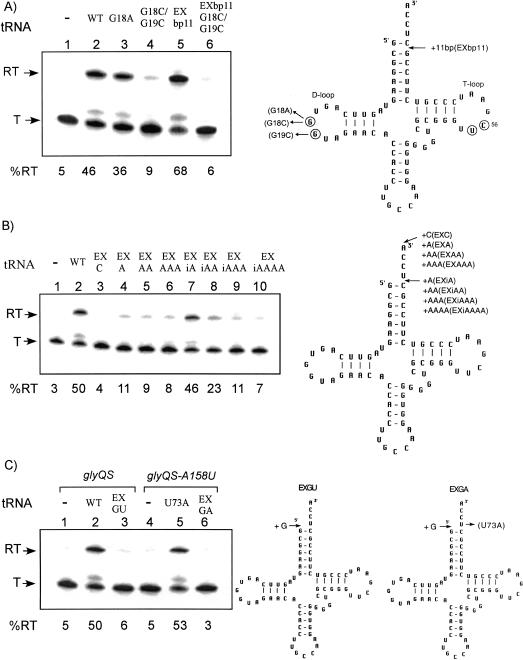
Interaction between the highly conserved G18 and G19 residues in the D-loop with residues U55 and C56 in the T-loop is an important component of tRNA tertiary structure (Giege et al. 1993). Characterization of tRNATyr variants that function in tyrS antitermination in vivo indicated that formation of the D-loop/T-loop interaction was required (Grundy et al. 2000). Changes in the G18 and G19 residues were therefore introduced into tRNAGly to test whether glyQS antitermination in vitro exhibited the same requirements. The G18A mutation of tRNAGly did not interfere with efficient readthrough (Fig. 2A ▶, lane 3), in contrast to the results for tRNATyr, where the equivalent allele abolished tyrS expression (Grundy et al. 2000). An alteration of tRNALeu corresponding to G18A allows antitermination of the B. subtilis ilv-leu operon in vivo (Garrity and Zahler 1994), indicating that this substitution is tolerated for antitermination directed by some tRNAs, including tRNALeu and tRNAGly, but not for others, including tRNATyr. A more drastic alteration (G18C/G19C) in tRNAGly, which was predicted to destroy the D-loop/T-loop interaction, resulted in complete loss of antitermination activity (Fig. 2A ▶, lane 4). The loss of antitermination function with the G18C/G19C substitutions indicates that the D-loop/T-loop interaction in tRNAGly is essential for interaction with the glyQS leader region under the conditions used in the in vitro transcription assay, in agreement with the analysis of tyrS antitermination in vivo. However, there appears to be more flexibility in the nature of this interaction than was observed for tRNATyr.
FIGURE 2.
Effects of D-loop mutations and 5′ and 3′ extensions of tRNAGly. (Left) The in vitro transcription of glyQS templates; (right) the cloverleaf models of B. subtilis tRNAGly variants. (T) Terminated transcript; (RT) readthrough transcript; (WT) wild-type tRNAGly. Percent readthrough is indicated at the bottom of each lane. Arrows indicate positions of substituted or added (+) bases or base pairs. Residues involved in the D-loop/T-loop tertiary interaction are circled. (A) D-loop mutations. (B) Additions to the unpaired region of the acceptor end of tRNAGly. (C) Addition to the 5′ end of tRNAGly. (Lanes 1–3) Wild-type glyQS template DNA (A158 antiterminator); (lanes 4–6) glyQS-A158U template DNA (A158U antiterminator).
Additions to the unpaired region of the acceptor end of tRNAGly
The 3′ unpaired residues of tRNAGly (5′-UCCA-3′) are predicted to pair with the 5′-UGGA-3′ residues of the antiterminator bulge, based on mutational and biochemical analyses (Grundy et al. 1994, 2002; Gerdeman et al. 2002). Charging of the tRNA, which occurs by addition of an amino acid to the 3′ terminal A residue, results in loss of antitermination activity both in vivo and in vitro (Grundy et al. 1994; Putzer et al. 2002). We therefore tested the effect of adding of 1–3 residues at the 3′ end of the tRNA, which was predicted to mimic the effect of tRNA charging. These additions resulted in complete loss of antitermination activity (Fig. 2B ▶, lanes 3–6), consistent with the model that charging of the tRNA blocks antitermination by steric hindrance, such that the 3′end of the tRNA can no longer make the proper interaction with the antiterminator bulge. In contrast, a tRNAGly variant with one residue inserted just before the 5′-UCCA-3′ was able to stimulate antitermination as efficiently as wild-type tRNAGly (Fig. 2B ▶, lane 7). Insertion of two residues at this position led to a twofold decrease in antitermination, whereas readthrough was lost after insertion of three or four extra residues (Fig. 2B ▶, lanes 8–10). These results indicate that some flexibility in the position of the terminal UCCA residues relative to the acceptor helix is tolerated.
Addition of an extra residue at the 5′ end of the tRNA was also tested. In the context of the wild-type glyQS template, this was predicted to allow formation of a G-U wobble pair at the top of the acceptor stem, by pairing of the introduced G with the U73 residue that normally pairs with A158 in the leader. This variant resulted in complete loss of antitermination (Fig. 2C ▶, lane 3). We also generated a second tRNAGly variant in which the extra G at the 5′ end was combined with the U73A substitution, which was previously shown to promote antitermination of a glyQS template having a compensatory A158U substitution in the antiterminator bulge (Grundy et al. 2002). In this case, the extra G was not predicted to pair with A73. This variant exhibited no antitermination activity (Fig 2C ▶, lanes 4–6), indicating that an extra residue at the 5′ end blocks the proper interaction of the 3′ residues with the antiterminator, independent of pairing with position 73 of the tRNA.
Alteration of tRNAGly acceptor stem length
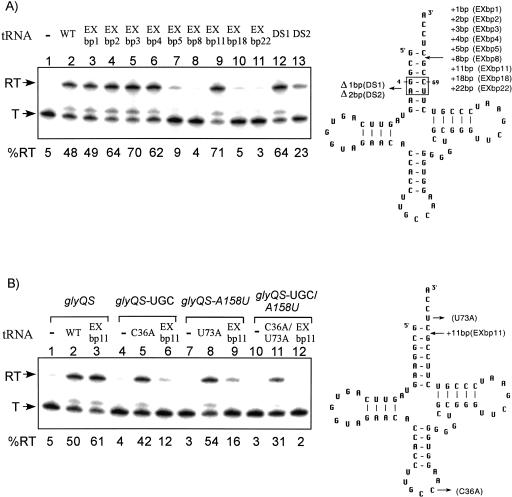
Because addition of residues between the acceptor helix and the terminal UCCA was tolerated, extension of the acceptor helix was also tested. Insertion of 1–4 bp one position from the top of the acceptor stem helix allowed efficient readthrough (Fig. 3A ▶, lanes 3–6), whereas insertion of 5 or 8 bp resulted in loss of antitermination function (Fig. 3A ▶, lanes 7,8), suggesting that a 4-bp insertion was the maximum allowed. We then examined a tRNAGly variant with an insertion of 11 bp (tRNAGly EXbp11), which comprises a complete turn in the RNA helix; remarkably, antitermination function was fully restored (Fig. 3A ▶, lane 9). Constructs with insertions of 2, 3, 4, or 11 bp exhibited increased antitermination efficiency relative to that observed with wild-type tRNAGly. Further extension of the acceptor stem by insertion of 18 or 22 bp resulted in loss of antitermination activity (Fig. 3A ▶, lanes 10,11). Deletion of 1 bp (residues 4, 69) within the acceptor helix allowed efficient antitermination, whereas deletion of 2 bp (residues 4, 5, 68, 69) resulted in a significant decrease in readthrough (Fig. 3A ▶, lanes 12,13); loss of activity for the 2-bp deletion variant could be due to altered positioning of the acceptor end or decreased stability of the helix. Overall, these results indicate that alterations in the length of the acceptor helix can be tolerated, within limits, suggesting that there is flexibility in the relative positioning of the specifier sequence and antiterminator elements of the leader RNA. The observation that addition of 5 or 8 bp destroyed function, whereas function was restored with an 11-bp insertion, suggests that the orientation of the 5′-UCCA-3′ residues relative to the antiterminator bulge is crucial.
FIGURE 3.
Effects of tRNAGly acceptor stem insertions and deletions. (Left) The in vitro transcription of glyQS templates; (right) the cloverleaf models of B. subtilis tRNAGly variants. (T) terminated transcript; (RT) readthrough transcript; (WT) wild-type tRNAGly. Percent readthrough is indicated at the bottom of each lane. Arrows indicate positions of substituted, added (+), or deleted (Δ) residues or base pairs. (A) Alteration of tRNAGly acceptor stem length. (B) Specificity of tRNAGly EXbp11-directed antitermination. (Lanes 1–3) Wild-type glyQS template DNA (GGC specifier sequence, A158 antiterminator); (lanes 4–6) glyQS-UGC template DNA (UGC cysteine specifier sequence); (lanes 7–9) glyQS-A158U template DNA (A158U antiterminator); (lanes 10–12) glyQS-UGC/A158U template DNA (UGC cysteine specifier sequence, A158U antiterminator).
One possible explanation for efficient antitermination by tRNAGly EXbp11 is that the extended helix nonspecifically stabilizes the antiterminator, independent of a functional interaction with the complete leader. To address this issue, we tested the ability of this variant to direct antitermination of leader variants that do not match the tRNA at the anticodon (glyQS leader with cysteine specifier sequence; glyQS-UGC), position 73 (glyQS leader with A158U change at the antiterminator bulge; glyQS-A158U), or both (glyQS leader with cysteine specifier sequence and A158U change at the antiterminator bulge; glyQS-UGC/A158U). These alterations prevent antitermination directed by wild-type tRNAGly, but allow readthrough directed by tRNAGly variants with compensatory changes (Grundy et al. 2002). As shown in Figure 3B ▶, tRNAGly EXbp11 exhibited efficient readthrough only for a template with matching sequence at both the specifier sequence and the antiterminator variable position, indicating dependence on normal base-pairing at both positions. We also tested the effect of introduction of the G18C/G19C alterations, which destroyed the antitermination activity of wild-type tRNAGly, into that variant. The G18C/G19C alterations resulted in complete loss of activity (Fig. 2A ▶, lane 6), providing further evidence that interaction of tRNAGly EXbp11 variant with the leader region follows the pattern observed for wild-type tRNAGly.
Extension of the tRNAGly anticodon stem
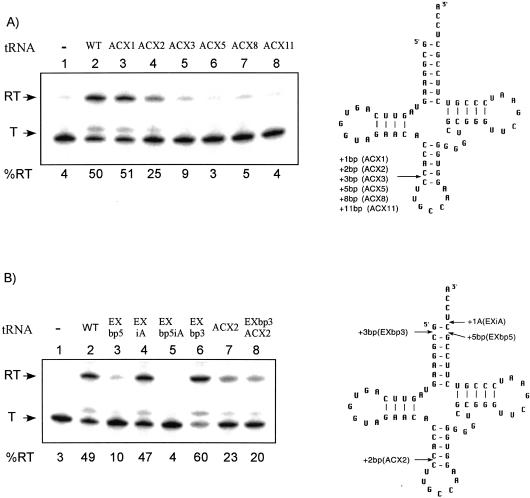
Because extension of the acceptor helix was well tolerated, we tested the effect of inserting extra base pairs into the anticodon stem of tRNAGly. Variants with 1, 2, 3, 5, 8, or 11 bp inserted into the anticodon helix were generated (Fig. 4A ▶). Antitermination activity similar to that of wild-type tRNAGly was observed for the variant containing a 1-bp insertion, whereas insertion of 2 bp resulted in a twofold decrease in antitermination activity (Fig. 4A ▶, lanes 3,4), and further extension of the helix resulted in loss of antitermination function. These results indicate that the anticodon stem is much more sensitive to extension than is the acceptor stem.
FIGURE 4.
Effects of tRNAGly anticodon stem mutations. (Left) The in vitro transcription of glyQS templates;, (right) the cloverleaf models of B. subtilis tRNAGly variants. (T) terminated transcript; (RT) readthrough transcript; (WT) wild-type tRNAGly. Percent readthrough is indicated at the bottom of each lane. Arrows indicate positions of insertions. (A) Anticodon stem insertions. (B) Combination of acceptor end and anticodon stem alterations. EXbp5iA has both EXbp5 and EXiA; EXbp3ACX2 has both EXbp3 and ACX2.
Combination of acceptor end and anticodon stem alterations
Insertion of 5 or 8 bp in the tRNAGly acceptor stem blocked antitermination function, whereas insertion of 4 or 11 bp was tolerated. One explanation for this effect is that the 5- and 8-bp insertions resulted in improper positioning of the 5′-UCCA-3′ relative to the antiterminator bulge. We therefore generated a tRNA variant with both the 5-bp insertion in the acceptor helix and an extra base inserted just before 5′-UCCA-3′ residues, to determine whether the extra unpaired base, which alone allowed efficient antitermination, would provide added flexibility to compensate for the extended acceptor helix. This tRNA variant was unable to promote antitermination (Fig. 4B ▶, lane 5), indicating that improper positioning of acceptor end could not be suppressed in this way.
The acceptor and anticodon ends of the tRNA exhibited differential sensitivity to insertions. It was possible that the two domains operate independently in their interaction with the leader. Alternatively, insertions at both ends of the tRNA could function additively, therefore the effect of combinations of added base pairs at the two ends would mimic the effect of insertion of the total number of base pairs at one end. Alterations in the acceptor and anticodon helices of the tRNA were therefore tested in combination. A tRNAGly variant with 3 extra bp in the acceptor stem and 2 extra bp in the anticodon stem exhibited antitermination activity similar to that of the tRNA with the anticodon stem insertion alone (Fig. 4B ▶, lanes 6–8). Because this activity is greater than that observed for insertion of 5 bp into the acceptor stem (Fig. 4B ▶, lane 3), it appears that the insertions do not act additively, but rather that the overall ability of the tRNA variant to promote antitermination is dictated by the domain with the greatest defect in interaction with the leader.
DISCUSSION
The tRNA-directed antitermination of T box family genes is dependent on a complex set of determinants in both the leader RNA and its tRNA partner that are required for a functional RNA–RNA interaction. Phylogenetic and mutational analyses provided insight into the leader RNA structure and its requirements for antitermination (Rollins et al. 1997; Delorme et al. 1999; Winkler et al. 2001; F.J. Grundy, S.M. Rollins, W.C. Winkler, and T.M. Henkin, unpubl.), and structural mapping of the B. subtilis thrS leader gave results consistent with most of the features of the model (Luo et al. 1998). The NMR structure of a small RNA modeled on the antiterminator domain revealed a complex tertiary arrangement overlaying the known secondary structure features (Gerdeman et al. 2003). Overall, little is known about the tertiary structure of the leader RNA, and the arrangement of leader domains relative to the tRNA. Because the tRNA structure is well characterized and two positions of leader–tRNA interaction have been established, we chose to further analyze the structural limitations of the tRNA for antitermination in vitro.
The tRNA structure is generally highly conserved; however, certain features vary, often by amino acid class. These variable features, including the size of the variable arm, nucleotide identity at position 73 (which is designated the discriminator base), and the structure of the acceptor stem, often serve in conjunction with the anticodon as identity elements for correct aminoacylation (Labouze and Bedouelle 1989; Sherman et al. 1992a,b; Giege et al. 1993; McClain et al. 1999). We previously showed that substitutions in the variable arm, anticodon and position 73 of the tRNA, did not block antitermination activity, as long as base-pairing at the specifier sequence and antiterminator variable position (position 158 in glyQS) was maintained (Grundy et al. 2000, 2002). In addition, the sequence within the helical regions of the tRNA does not appear to be important. The work presented here demonstrates that there is significant flexibility in the length of the acceptor helix, as well as moderate flexibility in the anticodon helix and in the positioning of the terminal unpaired residues relative to the acceptor helix. We also show that the D-loop/T-loop tertiary interaction in the tRNA is required in vitro, consistent with what was previously shown in vivo (Grundy et al. 2000).
The anticodon loop of the tRNA is responsible for the essential codon–anticodon pairing at the specifier sequence within Stem I of the leader. This region of the tRNA is highly constrained in natural tRNAs, and is often heavily modified in vivo. Because only unmodified tRNAGly variants were examined, the possible effect of tRNA modification is not known. Insertion of 1 or 2 bp into the anticodon stem of yeast pre-tRNAPhe yielded a precursor that is recognized as a substrate for splicing (Reyes and Abelson 1988). Here we show that extension of the anticodon helix by 1 bp is tolerated in the context of unmodified tRNAGly, whereas a 2-bp insertion significantly reduced antitermination function, and larger insertions abolished activity. It therefore appears that the structural arrangement of the anticodon domain relative to the remainder of the tRNA molecule is an important component of the tRNA–leader interaction.
The acceptor end of the tRNA makes the second known interaction with the leader, by pairing of the terminal four unpaired residues with four residues in the antiterminator bulge (Grundy et al. 1994). This interaction has been demonstrated to occur with a small model RNA based on the antiterminator domain alone (Gerdeman et al. 2002). In the NMR structure of the antiterminator domain, these bulge residues (UGGA in glyQS) are not highly constrained, whereas the remaining three residues in the bulge (ACC) are stacked onto the helix below the bulge, suggesting that the residues required for pairing are mobile to provide access to the tRNA (Gerdeman et al. 2003). Insertion of 1–2 residues between the top of the acceptor helix and the terminal UCCA residues was well tolerated, indicating that the terminal residues remained available for pairing with the antiterminator bulge. In contrast, addition of residues at the 3′ end of the tRNA completely blocked function, consistent with the model that the terminal A must fit precisely into the junction between the bulge and the adjacent helix, providing discrimination between charged and uncharged tRNA. Extension of the 5′ end of the tRNA also blocked antitermination, suggesting that the positioning of the 3′ terminal residues for pairing with the bulge is disrupted by the extra 5′ residue.
It was previously demonstrated that extension of the acceptor stem of Escherichia coli pre-tRNATyr Su3 by 3 bp resulted in a precursor that could be cleaved by Rnase P (Svard and Kirsebom 1993). In this study we show that for antitermination activity, the acceptor stem of tRNAGly is highly tolerant of alteration in length. Deletion of 1 bp or insertion of 1–4 bp did not decrease the efficiency of antitermination. Deletion of 2 bp reduced activity, whereas insertion of 5 or 8 bp abolished activity. Surprisingly, insertion of 11 bp resulted in complete restoration of activity in the antitermination assay. Because 11 bp constitutes one full turn of the RNA helix, the periodicity of this effect suggests that there is face-of-the-helix dependence in the positioning of the 3′ end of the tRNA relative to the antiterminator, such that the bases are available for pairing only if presented on the appropriate face of the tRNA (Fig. 5 ▶). The increase in antitermination activity observed for constructs with longer acceptor stems suggests that stacking of the acceptor helix onto the paired residues at the antiterminator bulge may contribute to the stability of the antiterminator–tRNA complex. This interaction is still dependent on leader RNA–tRNA pairing at both the specifier sequence and the antiterminator, and on formation of the D-loop/T-loop interaction in the tRNA, indicating that the extended helix must be presented in the appropriate context. Tolerance of the addition of one full turn of the helix, but not a half-turn, suggests that there is flexibility in the distance between the specifier loop and antiterminator domains in the leader RNA, but that twisting of the acceptor end of the tRNA relative to the remainder of the tRNA can prevent its interaction with the antiterminator. Extension of the acceptor helix by two full turns did not allow function, indicating that there is a limit to the distance between the specifier sequence and antiterminator. Because the structural arrangement of the Stem I domain is highly conserved, the unpaired regions between Stem I and Stem III or Stem III and the antiterminator (Fig. 1 ▶) may serve as a hinge for the proper positioning of the specifier loop and antiterminator, and accommodation of the longer tRNA.
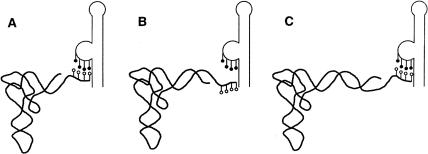
FIGURE 5.
Face-of-the-helix dependent effect of tRNAGly acceptor end insertions on interaction with the antiterminator bulge. White circles indicate the 3′ unpaired residues of tRNAGly (5′-UCCA-3′); black circles indicate the 5′-UGGA-3′ residues of the antiterminator bulge. (A) Wild-type tRNAGly. (B) tRNAGly variant with its acceptor stem extended by a half-turn of the RNA helix. (C) tRNAGly variant with its acceptor stem extended by a full turn of the RNA helix.
Pairing of tRNA anticodons with mRNA codons normally occurs in the context of the ribosome. The glyQS in vitro antitermination system is unique in that codon–anticodon recognition is mediated by the RNA itself, in the absence of the ribosome or any other protein factor. The tighter constraints on the structure of the anticodon end of the tRNA may reflect constraints on the codon–anticodon interaction, perhaps contributed by additional interactions of the tRNA with other domains of Stem I or other elements of the leader RNA. Our ability to assay the activity of tRNAGly in antitermination, independent of other tRNA functions or possible processing within the cell, allowed us to investigate extreme variations in tRNA structure. The apparent face-of-the-helix dependence of the presentation of the 3′ end of the tRNA provides new insight into the three-dimensional constraints on the acceptor end–antiterminator bulge interaction, and extends our analysis of the overall leader RNA–tRNA complex.
MATERIALS AND METHODS
In vitro transcription assays
The B. subtilis strain BR151MA (lys-3 trpC2) was used as the source of chromosomal DNA for amplification by PCR. Leader region variants (Grundy et al. 2002) were wild-type glyQS (GGC glycine specifier sequence, A158 antiterminator bulge), glyQS-UGC (UGC cysteine specifier sequence), glyQS-A158U (A158U antiterminator bulge), and glyQS-UGC/A158U (UGC cysteine specifier sequence, A158U antiterminator bulge). The templates used were 440-bp PCR fragments containing sequences from 135 bp upstream of the transcription start-site to position 305 of the transcript; the termination site is at position 220.
In vitro transcription of glyQS templates was carried out with B. subtilis RNAP as described by Grundy et al. (2002). The dinucleotide ApU (Sigma) was used at 150 μM to initiate transcription. The initiation reaction included template DNA at 10 nM, B. subtilis RNAP at 6 nM, ATP and GTP at 2.5 μM, UTP at 0.75 μM, and [α-32P]UTP (800 Ci/mmol; 1 Ci = 37 GBq) at 0.25 μM; omission of CTP from the initiation reaction resulted in a halt at position +17. The initiation mixtures were incubated at 37°C for 15 min, heparin (25 nM) was added to block reinitiation, tRNA was added to the reaction mixture at 200 nM, and elongation was triggered by the addition of NTPs to 10 μM final concentration. Transcription reactions were terminated by extraction with phenol, and the products were resolved by denaturing 6% polyacrylamide gel electrophoresis and visualized by PhosphorImager analysis. Readthrough and termination bands were quantified using ImageQuant 5.2, and the readthrough percentage was calculated as the ratio of readthrough product to total transcript. The band just above the termination product is reproducibly observed in reactions in which efficient tRNA-dependent readthrough is occurring (data not shown), but was not included in the quantification. The results obtained had a margin of error of ± 3%.
Generation of tRNA variants
The tRNAGly variants were prepared by T7 RNAP transcription of PCR products containing a T7 promoter positioned so that the first base of the transcript corresponds to the 5′ position of the tRNA and the final residue of the PCR product corresponds to the 3′ position of the tRNA (Grundy et al. 2002). Alterations in the 5′ or 3′ ends were introduced into the template by PCR using oligonucleotide primers containing the desired sequence change. The tRNAGly variants with alterations in the anticodon stem or D-loop were generated using three or four pairs of complementary oligonucleotides; each pair was designed to have a 4-nt 3′ overhang complementary to the overhang of the next pair. The 5′ ends of all internal oligonucleotides were phosphorylated with T4 polynucleotide kinase (United States Biochemicals) using the conditions recommended by the manufacturer. Complementary pairs of oligonucleotides (60 pmole in 70 mM Tris-HCl at pH 7.6, 10 mM MgCl2, 5 mM DTT, 50 μM rATP) were mixed, heated to 95°C for 5 min, and slowly cooled to room temperature for 90 min. Sets of oligonucleotides that formed the template for each tRNAGly variant (10 pmole each) were then mixed, incubated at 45°C for 5 min, and allowed to cool to room temperature for 30 min. T4 DNA ligase (800 U, New England BioLabs) was added and the mixture was incubated overnight at 16°C. The resulting DNA was used as a template for PCR amplification using the outside oligonucleotides as primers. PCR products were purified by a Qiagen PCR clean up kit, and sequence variations were verified by DNA sequencing by the 3700 DNA Analyzer (Applied Biosystems, Inc., Ohio State University Plant Microbe Genomics Facility). Sequence additions in different tRNAGly variants are summarized in Table 1 ▶.
TABLE 1.
Sequences added to tRNA variants
| tRNA variant | Insertion position | Added residues or base pairs | |||||||||||||||||||||
| EXC | 3′ end | C | |||||||||||||||||||||
| EXA | A | ||||||||||||||||||||||
| EXAA | A | A | |||||||||||||||||||||
| EXAAA | A | A | A | ||||||||||||||||||||
| EXiA | −72 | A | |||||||||||||||||||||
| EXiAA | A | A | |||||||||||||||||||||
| EXiAAA | A | A | A | ||||||||||||||||||||
| EXiAAAA | A | A | A | A | |||||||||||||||||||
| EXGU | −1 | G | |||||||||||||||||||||
| EXGA | G | ||||||||||||||||||||||
| EXbp1 | 2/71 | G | |||||||||||||||||||||
| | | |||||||||||||||||||||||
| C | |||||||||||||||||||||||
| EXbp2 | G | G | |||||||||||||||||||||
| | | | | ||||||||||||||||||||||
| C | C | ||||||||||||||||||||||
| EXbp3 | G | G | C | ||||||||||||||||||||
| | | | | | | |||||||||||||||||||||
| C | C | G | |||||||||||||||||||||
| EXbp4 | G | G | C | A | |||||||||||||||||||
| | | | | | | | | ||||||||||||||||||||
| C | C | G | U | ||||||||||||||||||||
| EXbp5 | G | G | C | A | A | ||||||||||||||||||
| | | | | | | | | | | |||||||||||||||||||
| C | C | G | U | U | |||||||||||||||||||
| EXbp8 | G | G | C | A | A | G | G | A | |||||||||||||||
| | | | | | | | | | | | | | | | | ||||||||||||||||
| C | C | G | U | U | C | C | U | ||||||||||||||||
| EXbp11 | G | G | C | A | A | G | G | A | A | U | A | ||||||||||||
| | | | | | | | | | | | | | | | | | | | | | | |||||||||||||
| C | C | G | U | U | C | C | U | U | A | U | |||||||||||||
| EXbp18 | G | G | C | A | A | G | G | A | A | U | A | G | G | C | A | A | G | G | |||||
| | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | ||||||
| C | C | G | U | U | C | C | U | U | A | U | C | C | G | U | U | C | C | ||||||
| EXbp22 | G | G | C | A | A | G | G | A | A | U | A | G | G | C | A | A | G | G | A | A | U | A | |
| | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | | ||
| C | C | G | U | U | C | C | U | U | A | U | C | C | G | U | U | C | C | U | U | A | U | ||
| ACX1 | 31/39 | G | |||||||||||||||||||||
| | | |||||||||||||||||||||||
| C | |||||||||||||||||||||||
| ACX2 | G | G | |||||||||||||||||||||
| | | | | ||||||||||||||||||||||
| C | C | ||||||||||||||||||||||
| ACX3 | G | G | C | ||||||||||||||||||||
| | | | | | | |||||||||||||||||||||
| C | C | G | |||||||||||||||||||||
| ACX5 | G | G | C | A | A | ||||||||||||||||||
| | | | | | | | | | | |||||||||||||||||||
| C | C | G | U | U | |||||||||||||||||||
| ACX8 | G | G | C | A | A | G | G | A | |||||||||||||||
| | | | | | | | | | | | | | | | | ||||||||||||||||
| C | C | G | U | U | C | C | U | ||||||||||||||||
| ACX11 | G | G | C | A | A | G | G | A | A | U | A | ||||||||||||
| | | | | | | | | | | | | | | | | | | | | | | |||||||||||||
| C | C | G | U | U | C | C | U | U | A | U | |||||||||||||
The T7 RNAP transcription was carried out using a MEGAshortscript T7 high yield transcription kit (Ambion). The tRNA transcripts were purified on a 6% denaturing polyacrylamide gel, visualized by UV shadowing, and eluted in 300 mM NaOAC (pH 4.5), 1 mM EDTA. The tRNA variants were purified by phenol–chloroform extraction, precipitated with ethanol, and suspended in water. The resulting tRNA was refolded by incubation at 80°C for 2 min and slow cooling to room temperature (Grundy et al. 2002). The tRNAs were quantified by measuring absorbance at 260 nm.
Acknowledgments
We thank Jennifer Hines and Michael Ibba for helpful discussions on RNA and tRNA structure. This work was supported by National Institutes of Health Grant GM47823.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Delorme, C., Ehrlich, S.D., and Renault, P. 1999. Regulation of expression of the Lactococcus lactis histidine operon. J. Bacteriol. 181: 2026–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher, M.P. 1990. Ribonucleases, tRNA nucleotidyltransferase and the 3′ processing of tRNA. Prog. Nucl. Acid Res. Mol. Biol. 39: 209–240. [DOI] [PubMed] [Google Scholar]
- Garrity, D.B. and Zahler, S.A. 1994. Mutations in the gene for a tRNA that functions as a regulator of a transcriptional attenuator in Bacillus subtilis. Genetics 137: 627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdeman, M.S., Henkin, T.M., and Hines, J.V. 2002. In vitro structure–function studies of the Bacillus subtilis tyrS mRNA antiterminator: Evidence for factor-independent tRNA acceptor stem binding specificity. Nucleic Acids Res. 30: 1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2003. Solution structure of the Bacillus subtilis T-box antiterminator RNA: Seven nucleotide bulge characterized by stacking and flexibility. J. Mol. Biol. 326: 189–201. [DOI] [PubMed] [Google Scholar]
- Giege, R., Puglisi, J.D., and Florentz, C. 1993. tRNA structure and aminoacylation efficiency. Prog. Nucl. Acid Res. Mol. Biol. 45: 129–206. [DOI] [PubMed] [Google Scholar]
- Grundy, F.J. and Henkin, T.M. 1993. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74: 475–482. [DOI] [PubMed] [Google Scholar]
- Grundy, F.J., Rollins, S.M., and Henkin, T.M. 1994. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: A new role for the discriminator base. J. Bacteriol. 176: 4518–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, F.J., Hodil, S.E., Rollins, S.M., and Henkin, T.M. 1997. Specificity of tRNA–mRNA interactions in Bacillus subtilis tyrS antitermination. J. Bacteriol. 179: 2587–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, F.J., Collins, J.A., Rollins, S.M., and Henkin, T.M. 2000. tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene. RNA 6: 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, F.J., Winkler, W.C., and Henkin, T.M. 2002. tRNA-mediated transcription antitermination in vitro: Codon–anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. 99: 11121–11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin, T.M. 2000. Transcription termination control in bacteria. Curr. Opin. Microbiol. 3: 149–153. [DOI] [PubMed] [Google Scholar]
- Henkin, T.M. and Yanofsky, C. 2002. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. Bioessays 24: 700–707. [DOI] [PubMed] [Google Scholar]
- Labouze, E. and Bedouelle, H. 1989. Structural and kinetic bases for the recognition of tRNATyr by tyrosyl-tRNA synthetase. J. Mol. Biol. 205: 729–735. [DOI] [PubMed] [Google Scholar]
- Luo, D., Leautey, J., Grunberg-Manago, M., and Putzer, H. 1997. Structure and regulation of the expression of the Bacillus subtilis valyl-tRNA synthetase gene. J. Bacteriol. 179: 2472–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998. In vitro and in vivo secondary structure probing of the thrS leader in Bacillus subtilis. Nucleic Acids Res. 26: 5379–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marta, P.T., Ladner, R.D., and Grandoni, J.A. 1996. A CUC triplet confers leucine-dependent regulation of the Bacillus subtilis ilv-leu operon. J. Bacteriol. 178: 2150–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain, W.H., Gabriel, K., Bhattacharya, S., Jou, Y.-Y., and Schneider, J. 1999. Functional compensation by particular nucleotide substitutions of a critical G • U wobble base-pair during aminoacylation of transfer RNA. J. Mol. Biol. 286: 1025–1032. [DOI] [PubMed] [Google Scholar]
- Putzer, H., Laalami, S., Brakhage, A.A., Condon, C., and Grunberg-Manago, M. 1995. Aminoacyl-tRNA synthetase gene regulation in Bacillus subtilis: Induction, repression, and growth-rate regulation. Mol. Microbiol. 16: 709–718. [DOI] [PubMed] [Google Scholar]
- Putzer, H., Condon, C., Brechemier-Baey, D., Brito, R., and Grunberg-Manago, M. 2002. Transfer RNA-mediated antitermination in vitro. Nucleic Acids Res. 30: 3026–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, V.M. and Abelson, J. 1988. Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55: 719–730. [DOI] [PubMed] [Google Scholar]
- Rollins, S.M., Grundy, F.J., and Henkin, T.M. 1997. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol. Microbiol. 25: 411–421. [DOI] [PubMed] [Google Scholar]
- Sherman, J.M., Rogers, M.J., and Soll, D. 1992a. Competition of aminoacyl-tRNA synthetases for tRNA ensures the accuracy of aminoacylation. Nucleic Acids Res. 20: 2847–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, J.M., Rogers, K., Rogers, M.J., and Soll, D. 1992b. Synthetase competition and tRNA context determine the in vivo identity of tRNA discriminator mutants. J. Mol. Biol. 228: 1055–1062. [DOI] [PubMed] [Google Scholar]
- Svard, S.G. and Kirsebom, L.A. 1993. Determinants of Escherichia coli RNase P cleavage site selection: A detailed in vitro and in vivo analysis. Nucleic Acids Res. 21 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, W.C., Grundy, F.J., Murphy, B.A., and Henkin, T.M. 2001. The GA motif: An RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA 7: 1165–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]