Abstract
β-globin mRNA bearing a nonsense codon is degraded in the cytoplasm of erythroid cells by endonuclease cleavage, preferentially at UG dinucleotides. An endonuclease activity in polysomes of MEL cells cleaved β-globin and albumin mRNA in vitro at many of the same sites as PMR1, an mRNA endonuclease purified from Xenopus liver. Stable transfection of MEL cells expressing normal human β-globin mRNA with a plasmid vector expressing the catalytically active form of PMR1 reduced the half-life of β-globin mRNA from 12 to 1–2 h without altering GAPDH mRNA decay. The reduced stability of β-globin mRNA in these cells was accompanied by an increase in the production of mRNA decay products corresponding to those seen in the degradation of nonsense-containing β-globin mRNA. Therefore, β-globin mRNA is cleaved in vivo by an endonuclease with properties similar to PMR1. Inhibiting translation with cycloheximide stabilized nonsense-containing β-globin mRNA, resulting in a fivefold increase in its steady-state level. Taken together, our results indicate that the surveillance of nonsense-containing β-globin mRNA in erythroid cells is a cytoplasmic process that functions on translating mRNA, and endonucleolytic cleavage constitutes one step in the process of β-globin mRNA decay.
Keywords: β-globin mRNA, nonsense codon, endonuclease, PMR1, mRNA decay
INTRODUCTION
Control of the rate of mRNA decay has a significant impact on overall gene expression patterns, and significant strides have been made in recent years in identifying cis-acting sequence elements and cognate-binding proteins that regulate mRNA decay rates. In the case of the AU-rich mRNA instability elements found on unstable mRNAs like c-fos mRNA, both a direct interaction with the exosome (Mukherjee et al. 2002), or a secondary interaction of the exosome through ARE-binding proteins (Chen et al. 2001) have been reported. These findings suggest that one pathway of mRNA degradation involves this complex of 3′–5′ exonucleases. In yeast, the mRNA body is primarily degraded in a 5′–3′ manner by Xrn1p following decapping (Tucker and Parker 2000). Mammals express orthologs to yeast Xrn1p (Bashkirov et al. 1997), Dcp1p and Dcp2p (Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002), raising the possibility of an analogous degradation pathway in vertebrates.
mRNAs are also subject to endonuclease-mediated decay (Schoenberg and Chernokalskaya 1997; Tourriere et al. 2002), and in many cases, this occurs in response to extracellular or developmental stimuli. Some mRNAs that have been shown to undergo endonuclease-mediated mRNA decay include transferrin receptor (Binder et al. 1994), c-myc (Herrick and Ross 1994; Gallouzi et al. 1998; Lee et al. 1998; Hanson and Schoenberg 2001), insulin-like growth factor II (van Dijk et al. 2000), serum albumin (Chernokalskaya et al. 1997; Hanson et al. 2001) and vitellogenin mRNA (Cunningham et al. 2000). Work by Brawerman and coworkers in the 1990’s (Bandyopadhyay et al. 1990) identified a mammalian endonuclease activity in mouse sarcoma 180 cells that displayed selectivity toward β-globin mRNA. This endonuclease activity preferentially cleaved β-globin mRNA at UG dinucleotides, but could also cleave at UC dinucleotides. In addition, it cosedimented with polysomes, but could be released into a more slowly sedimenting RNP fraction following polysome dissociation with EDTA. An endonuclease activity that behaved similarly was identified in RNP complexes from K562 cells (Konstantinova et al. 1999).
Estrogen stimulates the generalized destabilization of most serum protein mRNAs in Xenopus (Pastori et al. 1991a) through the activation of a polysome-associated endonuclease termed PMR1 (polysomal ribonuclease 1) (Cunningham et al. 2001b). This enzyme is unique in that it lacks signature RNase motifs and is closely related to the peroxidase gene family (Chernokalskaya et al. 1998). Similar to the endonuclease activities noted above, polysome-bound PMR1 is released following EDTA treatment as a component of a >680-kD RNP particle (Cunningham et al. 2001b). Both the crude enzyme present in polysome extracts and the purified endonuclease preferentially cleave single-stranded RNA at AYUGA elements, as well as other UG dinucleotides within albumin mRNA (Chernokalskaya et al. 1997). Degradation intermediates resulting from cleavage within these sequences can be observed in vivo using a sensitive ligation-mediated RT–PCR assay (Hanson and Schoenberg 2001), indicating that crude polysome-based mRNA decay systems can accurately reflect the in vivo process of endonuclease-mediated mRNA degradation.
In the early 1990s, Maquat and coworkers (Lim et al. 1989, 1992; Lim and Maquat 1992) developed mice expressing human β-globin transgenes corresponding to several β° thalassemias caused by the presence of premature termination codons (PTCs). mRNA expressed from the nonsense-containing transgene was selectively lost from the cytoplasmic fraction (Lim et al. 1989, 1992; Lim and Maquat 1992). In addition, the loss of nonsense-containing β-globin mRNA was associated with the appearance of polyadenylated decay intermediates that lacked sequences from the 5′ end (Lim and Maquat 1992). To date, this is the only case in which decay intermediates have been associated with NMD.
Previous work showed that the cytoplasmic degradation of nonsense-containing human β-globin mRNA could be recapitulated in murine erythroleukemia (MEL) cells that were stably transfected with the same transgenes used in the earlier work with transgenic mice (Stevens et al. 2002). S1 nuclease protection mapping using 5′ and 3′-labeled probes showed that the metastable decay intermediates of nonsense-containing human β-globin mRNA were generated by endonuclease cleavage. Furthermore, detailed S1 nuclease protection and primer extension experiments mapped the primary cleavage sites to UG dinucleotides, and also showed that normal β-globin mRNA is subject to the same endonucleolytic degradation pathway. Just as in our earlier work with Xenopus PMR1, the in vivo endonucleolytic degradation of β-globin mRNA could be accurately reproduced in vitro using polysome extracts from both control and DMSO-induced MEL cells (Stevens et al. 2002).
The endonuclease activity present in the above in vitro decay system displayed properties that were strikingly similar to those observed previously for Xenopus PMR1. Results presented here show that the polysome-associated endonuclease activity identified in MEL cells shares similar biochemical properties with Xenopus PMR1. The in vitro degradation patterns for β-globin and albumin mRNA by the polysome-associated endonuclease were almost indistinguishable from those observed using PMR1 purified from Xenopus liver. In addition, an antibody to PMR1 cross-reacts with a single 67-kD protein in MEL polysome extracts. To determine whether PMR1 could selectively destabilize β-globin mRNA in vivo, MEL cells expressing the normal human β-globin allele were stably transfected with plasmid vectors expressing the catalytically active 60-kD form of Xenopus PMR1. This resulted in a selective reduction in the steady-state level and half-life of β-globin mRNA. Furthermore, only those intermediates associated with β-globin mRNA decay were detected by S1 nuclease protection experiments, indicating that the exogenous endonuclease cleaved β-globin mRNA at the same sites as the endogenous enzyme. A characteristic feature of nonsense-mediated mRNA decay is its link to translation (Maquat 2000), and inhibiting translation with cycloheximide resulted in significant stabilization of nonsense-containing β-globin mRNA. A similar stabilization was not observed in cells expressing exogenous PMR1, in which β-globin mRNA was reduced to the level approximating that of the nonsense-containing mRNA. Our results suggest that the cytoplasmic degradation of nonsense-containing β-globin mRNA in erythroid cells is a specialized form of NMD, in which both surveillance and degradation are cytoplasmic, and endonucleolytic cleavage functions perhaps as the initiating step in mRNA decay.
RESULTS
Similarities in the in vitro cleavage specificity of the β-globin mRNA endonuclease and PMR1
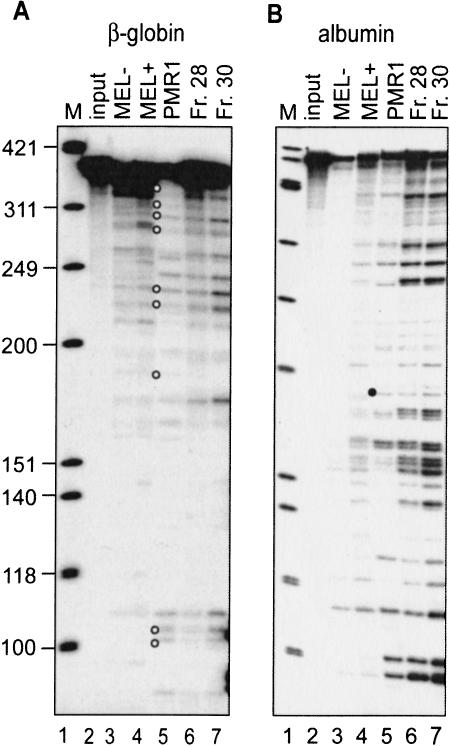
In vitro mRNA decay systems have been applied successfully to evaluate endonuclease-mediated mRNA decay, where they generate products that correspond to sites of in vivo endonuclease cleavage as well as nonspecific products resulting from the use of naked RNA substrates and the action of other RNases present in crude mixtures (Cunningham et al. 2001a). Previous work showed that MEL cells polysomes contained an endonuclease activity that cleaved β-globin mRNA in vitro at many of the same sites as seen in vivo for both normal and nonsense-containing human β-globin mRNA (Stevens et al. 2002). In addition, that study showed that endonuclease activity could be enriched by fractionation on a Mono S column. In the experiment in Figure 1 ▶, the activity of the crude and partially purified endonuclease activity present in MEL cells polysomes was compared with that of PMR1 purified to homogeneity from Xenopus liver. Accurate identification of endonuclease-generated decay products was obtained by using 5′ 32P-labeled substrate transcripts (Cunningham et al. 2001a). In the experiment in Figure 1A ▶, β-globin mRNA was digested as described previously (Stevens et al. 2002) with polysome extracts from control (MEL−, Fig. 1A ▶, lane 3) or DMSO-induced MEL cells (MEL+, Fig. 1A ▶, lane 4), purified PMR1 (Fig. 1A ▶, lane 5), or the endonuclease activity following partial purification of polysome extract from DMSO-induced cells on Mono S (Fig. 1A ▶, lanes 6,7). The products corresponding to in vivo β-globin mRNA endonuclease cleavage sites are identified by open circles. PMR1 generated a pattern of cleavage products that was similar to that generated by either crude or partially purified MEL cell endonuclease activity.
FIGURE 1.
In vitro cleavage of β-globin and albumin mRNA by PMR1 and the β-globin mRNA endonuclease. (A) A 5′ 32P-labeled transcript of the 5′-most 347 nucleotides of human β-globin mRNA was incubated for 30 min at 37°C with no added protein (lane 2, input), polysome extract from uninduced (MEL−, lane 3) or 48-h DMSO-induced MEL cells (MEL+, lane 4), or two fractions containing β-globin mRNA endonuclease activity recovered from Mono S fractionation of the polysome extract (lanes 6,7). Alternatively, the transcript was incubated for 20 min at 25°C with 20 U of purified Xenopus PMR1 (PMR1, lane 5). The reaction products were separated on a denaturing 6% polyacrylamide/urea gel and visualized by PhosphorImager. Lane 1 contains a marker consisting of φX174 DNA HinfI fragments and the open circles identify products with counterparts in vivo (Stevens et al. 2002). (B) An in vitro-synthesized, 5′ 32P-labeled transcript consisting of the 5′-most 420 nucleotide of Xenopus albumin mRNA was incubated as described in A. The characteristic PMR1 cleavage at overlapping APyrUGA elements is identified by a solid circle.
PMR1 was identified on the basis of its substrate selectivity for cleaving Xenopus albumin mRNA (Pastori et al. 1991b; Dompenciel et al. 1995). To explore further the possible relationship between the MEL cell polysome-associated endonuclease and PMR1, the endonuclease cleavage assay was performed as above using a 5′ end-labeled albumin mRNA (Fig. 1B ▶). Again, similar cleavage patterns were generated by both purified PMR1 and the MEL cell endonuclease. In addition, the polysome-associated endonuclease from MEL cells shows the same pH range for activity, resistance to inhibition by EDTA, resistance to placental ribonuclease inhibitor, and salt sensitivity as described previously (Dompenciel et al. 1995) for PMR1 (data not shown). These results raise the possibility that the polysomal endonuclease is either a mammalian homolog of Xenopus PMR1, or is a paralog that degrades the same mRNAs in a similar manner as PMR1.
Expression of Xenopus PMR1 reduces the half-life of β-globin mRNA in transfected MEL cells
Previous work described the development of two cell lines used to investigate the mechanisms of nonsense codon-induced destabilization of human β-globin mRNA (Stevens et al. 2002). The Norm2 line consists of MEL cells stably transfected with a normal human β-globin gene, and the Thal10 cell line is identical except for a single nucleotide deletion within codon 44 of human β-globin mRNA that introduces a premature termination codon (PTC) between codons 60 and 61. Both human β-globin genes were inserted downstream of β-globin locus control region of the GSC1417 β-globin gene-expression plasmid (Antoniou and Grosveld 1990; Collis et al. 1990). These same genes were used in previous studies on nonsense-mediated decay of β-globin mRNA in transgenic mice (Lim et al. 1989, 1992; Lim and Maquat 1992). We reported previously that in DMSO-induced cells, the steady-state level of full-length nonsense-containing cytoplasmic human β-globin mRNA was approximately sevenfold lower than that of the normal allele, but the quantity of cytoplasmic mRNA decay intermediates relative to full-length mRNA was 5- to 10-fold greater (Stevens et al. 2002). The latter finding provided evidence that both normal and nonsense-containing β-globin mRNA are substrates for endonucleolytic cleavage.
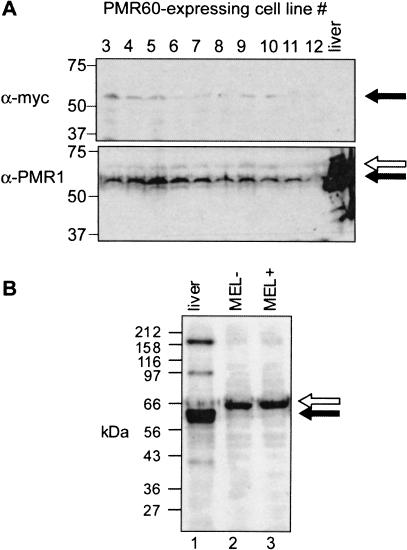
If the degradation of normal and nonsense-containing β-globin mRNA involves a PMR1-like endonuclease, transfecting Norm2 cells (which express the normal β-globin allele) with a vector expressing catalytically active Xenopus PMR1 should result in a selective decrease in the half-life of human β-globin mRNA. Norm2 cells were stably transfected with a vector expressing the catalytically active 60-kD form of Xenopus PMR1 (Chernokalskaya et al. 1998) bearing an amino-terminal myc epitope tag. Twelve cell lines expressing myc-PMR60 were recovered following selection in hygromycin-containing medium. The expression of myc-PMR60 in 10 of 12 surviving cell lines is shown in Figure 2A ▶. Low-level expression of a 60-kD protein was observed in each cell line by Western blot with a monoclonal antibody to the myc epitope. A second gel bearing the same samples plus a positive control of Xenopus liver polysome extract was analyzed by Western blot with a polyclonal antibody to Xenopus PMR1. A 60-kD band that comigrated with Xenopus PMR1 was seen in each of the transfected cell lines (solid arrow). The relative amount of this generally corresponded to that seen with the myc antibody. In addition, a somewhat larger endogenous protein was observed (open arrow) that reacted weakly with the PMR1 antiserum. The nature of this cross-reacting protein was examined more carefully in the experiment in Figure 2B ▶. Here, 15 μg of polysome extract from control (MEL−, Fig. 2B ▶, lane 2) or DMSO-induced (MEL+, Fig. 2B ▶, lane 3) MEL cells was compared with a polysome extract from Xenopus liver (Fig. 2B ▶, lane 1). Both the major ~60-kD form of PMR1 and the smaller 40-kD breakdown product (Dompenciel et al. 1995) are seen in lane 1, along with several cross-reacting bands that are larger than the 80-kD PMR1 precursor. Notably, only a single 67-kD peptide cross-reacts in both samples from MEL cells. The quantity of this cross-reacting material corresponded to the elution profile of the β-globin mRNA endonuclease from a Mono S column (data not shown), suggesting a relationship between the cross-reacting protein and enzymatic activity. The PMR1 antibodies currently available neutralize its activity, and experiments to test the catalytic activity of immunoprecipitated protein were inconclusive.
FIGURE 2.
Expression of myc-PMR60 in transfected Norm2 cells. (A) Norm2 cells were stably transfected with a plasmid expressing the catalytically active 60-kD form of Xenopus PMR1 bearing an amino-terminal myc epitope tag. Equal amounts of cytoplasmic extract from 10 stable cell lines recovered following selection were applied to duplicate 10% SDS-PAGE gels. In addition, one blot contained a sample of Xenopus liver polysome extract. Western blots of the duplicate gels were probed with a monoclonal antibody to the myc epitope (top) or a polyclonal antibody to Xenopus PMR1 (Dompenciel et al. 1995) (bottom). The solid arrow corresponds to a 60-kD PMR1, and the open arrow to a larger cross-reacting protein seen in all cultures. (B) Polysomes from Xenopus liver (lane 2), uninduced MEL cells (lane 2, MEL−), or DMSO-induced MEL cells (lane 3, MEL+) were analyzed as above by Western blotting using a polyclonal antibody to Xenopus PMR1.
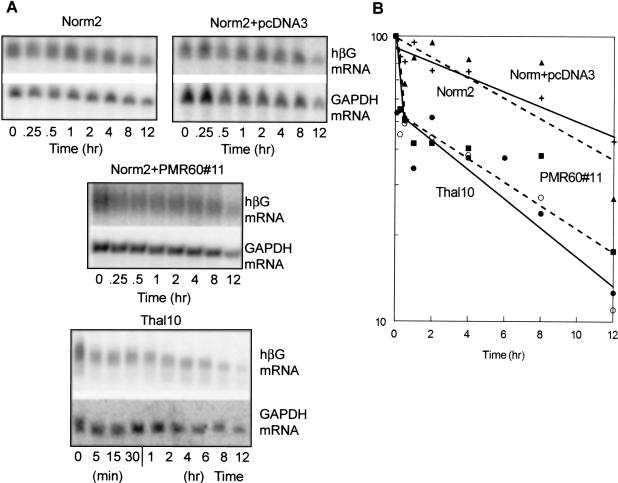
The impact of PMR1 expression on mRNA half-life was determined by Northern blots of RNA extracted from the individual stable cell lines following addition of actinomycin D to cultures that were induced previously for 48 h with DMSO. Selectivity of the endonuclease for β-globin mRNA was determined by comparing β-globin mRNA decay with that of GAPDH mRNA in the same preparation. Northern blots for a typical series of decay experiments are shown in Figure 3A ▶, and decay curves determined by PhosphorImager analysis are shown in Figure 3B ▶. In Norm2 and Norm2+pcDNA3 cells, the decay of wild-type β-globin mRNA could be fit to a single line, consistent with the pseudo-first order kinetics normally associated with mRNA decay. Results from three independent experiments resulted in an average half-life of 12 h, which is similar to the half-life obtained previously using erythroid cells from transgenic mice (Maquat et al. 1981). In all of the cell lines, the half-life of GAPDH mRNA was also 12 h.
FIGURE 3.
β-globin mRNA turnover in Norm2 cells, Thal10 cells, or Norm2 cells transfected with vector or PMR1. (A) Norm2 cells expressing wild-type β-globin mRNA (Norm2), Thal10 cells expressing nonsense-containing β-globin mRNA (Thal10), Norm2 cells that were stably transfected with empty vector (Norm2+pcDNA3), or a line of Norm2 cells stably transfected with vector expressing PMR1 (Norm2+PMR60#11) were induced for 48 h with DMSO prior to addition of actinomycin D. Cytoplasmic RNA was isolated at the indicated times and analyzed by Northern blot for human β-globin mRNA (hβG, top, each set) or GAPDH mRNA (bottom, each set). Shown are typical results from three independent experiments. The results in A were quantified by PhosphorImager analysis and are presented graphically in B. With the exception of Thal10 cells (•, ▪, solid line) the datapoints represent the decay of the mRNAs in A (Norm2 +, solid line; Norm2+pcDNA3 ▴, broken line; Norm2+PMR60#11 ○, broken line. The data points for Thal10 cells consist of those derived from the blot in A plus an independent experiment (not shown).
A distinctly different pattern of decay was observed for either nonsense-containing β-globin mRNA in Thal10 cells or for wild-type β-globin mRNA in cells expressing PMR1. In both cell lines, the amount of β-globin mRNA dropped within 15 min of adding Actinomycin D to the medium. Approximately 50% of each mRNA decayed within 90–100 min, followed by a slower rate of degradation as evidenced by the biphasic decay curve. The decay of nonsense-containing β-globin mRNA in Thal10 cells was also accompanied by the appearance of weakly hybridizing decay intermediates that were most evident during the slower step in decay beginning 2 h after Actinomycin D addition.
Xenopus PMR1 generates similar in vivo decay products in transfected MEL cells
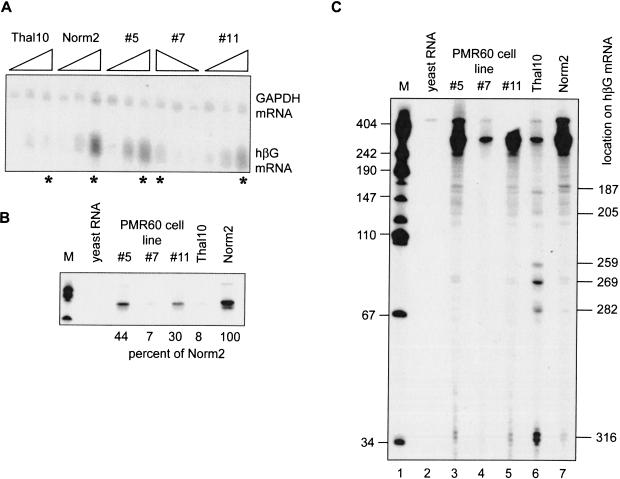
Previously, we showed that the same decay products were generated in vivo by endonuclease cleavage of both normal and nonsense-containing β-globin mRNA (Stevens et al. 2002). The latter were more abundant, because both the nonsense-containing mRNA is more rapidly degraded, and because these decay products contain a 5′ cap-like modification (Lim and Maquat 1992) that may render them more stable to subsequent degradation than those generated from the normal allele. The experiment in Figure 4 ▶ compared the impact of exogenous PMR1 expression on the pattern of degradation intermediates in PMR1-transfected cells with that of Norm2 and Thal10 cells. RNA was isolated from Norm2 cells, Thal10 cells, and the PMR60-expressing lines #5, #7, and #11 that were cultured without DMSO or induced for 48 or 96 h (Fig. 4A ▶). Northern blot analysis shows that DMSO treatment induced β-globin mRNA in each of the cell lines, but had little impact on the amount of GAPDH mRNA.
FIGURE 4.
Identification of β-globin mRNA decay intermediates in Norm2 cells expressing Xenopus PMR1. Cytoplasmic RNA was isolated at time 0, 48, and 96 h following DMSO-induction from Norm2, Thal10, or three lines of Norm2 cells stably transfected with plasmids expressing myc-PMR60. (A) Each of the RNA samples was analyzed by Northern blot that was hybridized to a mixed probe for β-globin and GAPDH mRNA. The RNAs were applied in the order indicated above with the exception of cell line #7, in which they were applied in reverse order. (B) Equal amounts of the 96-h samples identified with by an asterisk (*) in A were analyzed by S1 nuclease protection using a NaeI–BamH1 probe, and the protected products were analyzed as described previously (Stevens et al. 2002). An 18-h exposure of the portion of the gel corresponding to the fully protected probe is shown in B. The marker (M) in lane 1 consists of HpaII fragments of pUC19 DNA. (Lane 2) Yeast RNA was added to the initial hybridization reaction; (lanes 3–5) S1 nuclease protection was performed on RNA obtained from three independent clones of Norm2 cells stably transfected with myc-PMR60; (lanes 6,7) S1 nuclease protection was performed on RNAs from Thal10 and parental Norm2 cells. The relative quantity of each fully protected product was determined by scanning densitometry, and the percent of the signal for β-globin mRNA from Norm2 cells is shown beneath the gel. (C) The gel was exposed for 96 h to visualize β-globin mRNA decay intermediates. Human β-globin mRNA decay products characterized previously (Stevens et al. 2002) are identified on the right side of the autoradiogram.
In addition, the overall level of induced β-globin mRNA was lower in each of the PMR60 transfectants than the parental Norm2 cells, and in the case of line #7, was similar to that seen in Thal10 cells.
Equal amounts of RNA from each of the 96-h time points in Figure 4A ▶ (indicated with a star) were next analyzed by S1 nuclease protection. The portion of the gel corresponding to the fully protected probe is shown in Figure 4B ▶, with the relative amount of intact β-globin mRNA in each sample listed below. These data compared favorably with those obtained by Northern blot in Figure 4A ▶. There was ~10-fold less intact β-globin mRNA in Thal10 cells than in Norm2 cells, and the level of intact β-globin mRNA in three PMR1-transfected cell lines compared with Norm2 cells ranged from a minimum of 7% in line #7, to a maximum of 44% in line #5. For reasons that we do not currently understand, the relative amount of β-globin mRNA remaining does show an inverse correlation with the relative amount of PMR60 expressed in the different cell lines.
A longer exposure of the gel was required to visualize β-globin mRNA decay intermediates (Fig. 4C ▶). In all cases, the only decay intermediates that were observed correspond to those produced from nonsense-containing β-globin mRNA in Thal10 cells. If myc-PMR60 cleaved at different sites within β-globin mRNA than the endogenous β-globin mRNA endonuclease, we would have expected to observe additional decay products. Scanning densitometry was used to quantify the relative amount of decay intermediates compared with full-length mRNA in each lane. In Norm2 cells, full-length β-globin mRNA accounted for 92% of the total signal in the lane. In contrast, only 36% of the signal from Thal10 cells corresponded to intact mRNA. Full-length-protected probe accounted for 81% of the signal for cell lines #5 and #11, and slightly more for cell line #7. Although not dramatic, these data demonstrate a consistent decrease in the relative quantity of full-length β-globin mRNA versus decay products in cells expressing exogenous PMR1. We conclude that the endonucleolytic cleavage of human β-globin mRNA in cells transfected with Xenopus PMR1 is indistinguishable from cleavage by the endogenous β-globin mRNA endonuclease. Furthermore, the expression of the exogenous endonuclease both destabilized β-globin mRNA and increased the quantity of cytoplasmic β-globin mRNA decay intermediates.
Cycloheximide stabilizes nonsense-containing β-globin mRNA
Inhibiting translation with cycloheximide can stabilize nonsense-containing mRNAs (Maquat and Carmichael 2001; Noensie and Dietz 2001). The experiment in Figure 5 ▶ examined the impact of inhibiting translation on steady-state levels of human β-globin mRNA in Norm2 cells, Norm2 cells transfected with plasmid vector (Norm2+pcDNA3), Norm2 cells transfected with vector expressing PMR1 (Norm2+PMR60#11), and Thal10 cells. Cells were induced for 48 h with DMSO, followed by addition of 1/5 volume of fresh medium without (Fig. 5A ▶) or with cycloheximide (Fig. 5B ▶) and incubation for an additional 8–12 h. RNA extracted at the indicated times was separated by Northern blot and probed for human β-globin mRNA and for GAPDH mRNA as an internal control.
FIGURE 5.
Impact of inhibiting translation on steady-state levels of human β-globin mRNA in transfected MEL cells. The MEL cell transfectants described in the legend to Figure 4 ▶ were induced with DMSO for 48 h. At time 0, the medium the cells either received fresh medium (A) or medium containing cycloheximide (B). Cytoplasmic RNA was isolated at the indicated time points and analyzed by Northern blot for human β-globin mRNA (top, each set) and GAPDH (bottom, each set).
Full-length β-globin mRNA was evident at all time points in Norm2 cells and Norm2 transfectants treated with medium alone (Fig. 5A ▶). Only modest amounts of β-globin mRNA were seen in Thal10 cells by Northern blot, and both full-length and degraded β-globin mRNA were evident. In Norm2 cells and the Norm2 transfectants, inhibiting translation with cycloheximide resulted in a slight (20%) increase in the amount of β-globin mRNA over an 8-h period (Fig. 5B ▶). Within 1 h of cycloheximide addition to Thal10 cells, the amount of β-globin mRNA increased twofold, and this continued to increase, ultimately increasing β-globin mRNA fivefold by 8 h. Cycloheximide treatment of Thal10 cells also caused a marked alteration in the electrophoretic mobility of nonsense-containing β-globin mRNA, from a diffuse smear to a band that was more discrete than any observed in Norm2 cells or the Norm2 transfectants. The mobility of this band increased over the 8-h interval examined here, indicating either a shortening of the poly(A) tail or the accumulation of products resulting from endonucleolytic cleavage within the 5′ portion of β-globin mRNA. In all four cell lines, cycloheximide treatment had little effect on GAPDH mRNA expression.
An actinomycin D time course was used to confirm whether the accumulation of nonsense-containing β-globin mRNA following cycloheximide addition resulted from an increase in mRNA half-life. In the experiment in Figure 6 ▶ Norm2 cells, the Norm2 transfectants and Thal10 cells were treated for 4 h with cycloheximide prior to addition of actinomycin D. Again, cycloheximide treatment slightly increased (by 10%–20%) the amount of β-globin mRNA in Norm2, Norm2+pcDNA3, and Norm2+PMR60#11 cells. These mRNAs were stabilized by treatment with both inhibitors to the extent that they effectively did not decay over the 8-h interval following addition of actinomycin D. A similar result was also observed in Thal10 cells, indicating that the increase in steady-state levels of human β-globin mRNA following translation inhibition resulted from the dramatic stabilization of the nonsense-containing allele. These results are consistent with a role for NMD in the cytoplasmic degradation of nonsense-containing β-globin mRNA in erythroid cells.
FIGURE 6.
Impact of inhibition translation mRNA on turnover in Norm2 cells, Thal10 cells, or Norm2 cells transfected with vector or PMR1. The MEL cell transfectants described in Figure 4 ▶ were induced with DMSO for 48 h. Cycloheximide was added at time 0 (0) and RNA was isolated 4 h later to monitor the impact on inhibiting translation on mRNA steady state levels (+CHX). Actinomycin D was added at this point and cytoplasmic RNA was isolated at the indicated times over the next 8 h. The isolated RNAs were analyzed by Northern blot for human β-globin mRNA (top, each set) and GAPDH mRNA (bottom, each set).
DISCUSSION
Previous work showed that endonuclease-mediated mRNA decay constitutes one step in the degradation of nonsense-containing human β-globin mRNA in erythroid cells (Stevens et al. 2002). That study also showed that a crude in vitro mRNA decay system derived from polysomes of DMSO-induced MEL cells contains an endonuclease activity that cleaves human β-globin mRNA at many of the same sites that are cleaved in vivo. An endonuclease activity selective for β-globin mRNA was identified previously on polysomes of murine S-180 sarcoma cells (Bandyopadhyay et al. 1990). Both that activity and the one described in our previous study have biochemical properties that are strikingly similar to Xenopus PMR1, a polysome-associated mRNA endonuclease that catalyzes the estrogen-regulated destabilization of albumin and other serum protein mRNAs (Pastori et al. 1991b; Dompenciel et al. 1995; Cunningham et al. 2001b). The experiment in Figure 1 ▶ shows that the polysome-associated endonuclease from MEL cells and PMR1 also have similar cleavage properties. PMR1 purified from Xenopus liver polysomes cleaved human β-globin mRNA into products that were similar to those generated by either crude or partially purified activity extracted from MEL cell polysomes, and the polysome-associated endonuclease extracted from MEL cells cleaved albumin mRNA in much the same way as purified Xenopus PMR1.
If the β-globin mRNA endonuclease is a PMR1-like enzyme, we reasoned that the half-life of human β-globin mRNA should be selectively reduced in Norm2 MEL cells stably transfected with a vector expressing a catalytically active form of Xenopus PMR1. Norm2 cells were transfected with a plasmid expressing the catalytically active 60-kD form of Xenopus PMR1 (Chernokalskaya et al. 1998) bearing a myc epitope tag, and 12 cell lines were recovered for further analysis. Expression of Xenopus PMR1 was confirmed by Western blots using both a monoclonal antibody to the myc epitope and a polyclonal antibody to Xenopus PMR1 (Fig. 2 ▶). Interestingly, the latter antibody weakly cross-reacted with a single, slightly larger 67-kD protein in MEL cells. This protein also cross-reacted with a peptide-specific antibody prepared to the sequence of Xenopus PMR1 (Chernokalskaya et al. 1998; data not shown). PMR1 is a member of the peroxidase gene family (Chernokalskaya et al. 1998), and database searches identified an uncharacterized murine peroxidase-like gene that is predicted to encode a 67-kD protein with 45.5% sequence identity to Xenopus PMR1. The predicted protein also retains the epitope of the peptide-specific antibody, making it a good candidate for mammalian PMR1.
Wang and Kiledjian (2000) described an endonuclease with selectivity for α-globin mRNA that is enriched in erythroid cells. The cleavage properties of this enzyme are distinct from PMR1, and unlike the restricted distribution of the erythroid-enriched endonuclease, PMR1 is present in most stages of Xenopus embryos, all adult Xenopus tissues, and A6 kidney cells (M.N. Hanson, J. Peng, and D.R. Schoenberg, unpubl.). Although we have not extensively screened mammalian cell lines for polysome-associated endonuclease activities, we observed an activity similar to that described here and a 67-kD protein that is bound by the PMR1 antibody on polysomes from MCF7 mammary tumor cells (K.A. Bremer and D.R. Schoenberg, unpubl.). This raises the interesting possibility that PMR1 may target different mRNAs in a cell-specific manner.
The expression of Xenopus PMR1 had no effect on the expression of endogenous GAPDH mRNA, but reduced the half-life of human β-globin mRNA in Norm2 cells from 12 to 1 h, approaching that of nonsense-containing mRNA in Thal10 cells (Fig. 3 ▶). Nonsense-containing β-globin mRNA in Thal10 cells decayed in a biphasic manner, in which ~50% of input mRNA was lost rapidly after addition of Actinomycin D, followed by a slower rate of decay. Unexpectedly, a similar biphasic pattern was seen for wild-type β-globin mRNA in Norm2 cells expressing catalytically active PMR1. The reason for this unusual behavior is not known.
In addition to the similar biphasic decay curves seen in Figure 3 ▶, nonsense-containing β-globin mRNA in Thal10 cells and normal β-globin mRNA in cells expressing catalytically active PMR1 generated the same pattern of degradation intermediates (Fig. 4 ▶). The only way this can occur is if both enzymes cleaved β-globin mRNA at the same sites in vivo (Hanson and Schoenberg 2001). Although the relative percentage of decay intermediates increased in PMR1-expressing cells compared with control cells, these do not accumulate to the extent seen in cells expressing the nonsense-containing allele. We speculate that this may be due to stabilization of decay products of the nonsense-containing mRNA by a cap-like structure on their 5′ ends (Lim and Maquat 1992).
It is generally accepted that NMD in mammalian cells acts on newly synthesized mRNA-bearing proteins of the exon-junction complex. Ribosomes scan this mRNP for premature termination codons in a pioneer round of translation, and the nonsense-containing mRNA is degraded in association with the nucleus (Maquat and Carmichael 2001). The mechanism of mRNA degradation in nucleus-associated NMD has not been described. In yeast, the major decay pathway for nonsense-containing mRNA entails decapping followed by 5′–3′ degradation of the mRNA body (Tucker and Parker 2000). Mammalian Upf1 can be recovered with Dcp2 (Lykke-Anderson 2002; van Dijk et al. 2002), raising the possibility of a similar pathway in mammals. However, in yeast there is a secondary 3′–5′ pathway for degradation of nonsense-containing mRNA that is catalyzed by the exosome (Mitchell and Tollervey 2003). Therefore, it is quite reasonable to expect that additional pathways exist in mammals to degrade nonsense-containing mRNA.
Several features distinguish the degradation of nonsense-containing β-globin mRNA in erythroid cells. Decay clearly occurs in the cytoplasm, both in erythroid cells from transgenic mice (Lim et al. 1992) and in MEL cells expressing the same allele (Stevens et al. 2002). The degradation of nonsense-containing β-globin mRNA results in the accumulation of 5′-truncated intermediates that are generated by endonuclease cleavage (Stevens et al. 2002). No decay intermediates have been identified for nucleus-associated mRNA decay (Maquat and Carmichael 2001). Cycloheximide treatment dramatically stabilized nonsense-containing β-globin mRNA (Fig. 6 ▶), resulting in a fivefold increase in its steady-state level (Fig. 5 ▶). The rapid cytoplasmic stabilization of nonsense-containing β-globin mRNA following addition of cycloheximide indicates that ongoing translation is required for mRNA degradation, and that steady-state mRNA must be monitored continuously by some form of surveillance. A similar dependence on ongoing translation for surveillance was observed for exosome-mediated 3′–5′ degradation of nonsense-containing mRNA by Mitchell and Tollervey (2003).
Cycloheximide treatment also changed the appearance of β-globin mRNA on Northern blots, from a diffuse smear to a discrete band. This was not seen in any of the cells expressing wild-type human β-globin mRNA, even when steady-state levels were reduced by expression of exogenous PMR1. Following both cycloheximide and actinomycin D, nonsense-containing human β-globin mRNA also underwent a time-dependent increase in electrophoretic mobility. This could reflect either a shortening of the poly(A) tail or the accumulation of 5′-truncated β-globin mRNA decay intermediates. The latter would be consistent with the observation that only 36% of S1 nuclease-protected products from Thal10 cells in Figure 4 ▶ correspond to the intact 5′ end of human β-globin mRNA. In general, endonuclease-mediated mRNA decay does not involve prior deadenylation (Schoenberg and Chernokalskaya 1997), and experiments are in progress to address the basis for this change in β-globin mRNA. On the basis of the results described above, we propose that the degradation of nonsense-containing human β-globin mRNA in erythroid cells is a specialized form of NMD, in which surveillance occurs on steady-state mRNA in the cytoplasm, and decay is initiated by endonucleolytic cleavage by an enzyme related to PMR1.
MATERIALS AND METHODS
Preparation of 5′ end-labeled transcript
The human β-globin cDNA plasmid pSPkβC was linearized with BamHI, and the Xenopus albumin cDNA plasmid pXa470 was linearized with SauI. Transcription and 5′-end labeling was performed with SP6 or T3 RNA polymerase using the Ambion MAXlscript transcription kit with the following modifications. In addition to the buffer, 5 mM DTT and 10 U of RNase Inhibitor (GIBCO-BRL) were added. The transcription reactions contained 750 mM each of ATP, CTP, UTP, 150 mM of GTP, and 60 μCi of [γ 32P]GTP (6000 Ci/mmole). The reactions were incubated for 1 h with T3 polymerase at room temperature for pXa470 and for 1 h with SP6 polymerase at 37°C for pSPkβC. The transcripts were treated with 2 U of DNase I for 15 min at 37°C and then gel purified.
Endonuclease activity assays
Purified PMR1 and MEL cell polysome extracts were prepared as described previously (Dompenciel et al. 1995; Schoenberg and Cunningham 1999; Stevens et al. 2002). Activity assays were performed in a 20-μL volume containing 50 mM Tris (pH 7.2), 50 mM MgCl2, and 10 mM dithiothreitol using 5 μg of polysome extract, 2.5 μg of Mono S fractions of polysome extract, or 20 U of purified PMR1 plus 105 cpm of 5′ end-labeled transcript. Reactions containing MEL cell protein samples were performed at 37°C for indicated times, whereas reactions with purified PMR1 were performed at 25°C for 20 min. The reactions were terminated by addition of SDS to a final concentration of 1% plus 20 μg of proteinase K, followed by incubation for 15 min at 65°C. The samples were then extracted with phenol:chloroform (50:50) and ethanol precipitated. The recovered RNAs were electrophoresed on denaturing 6% polyacrylamide urea gels and visualized by autoradiography.
Plasmids
The portion of Xenopus PMR1 that corresponds to the processed, catalytically active 60-kD protein extracted from liver was amplified from a full-length cDNA clone using primers JP3 (5′TGACAAGCTTCCGCCATTACAGGACAGTGC-3′) and KC42 (5′-GATCGCGGCCGCTTAAGCCACTTTCCAAGGAT-3′). This was digested with HindIII and NotI and ligated into the corresponding sites in pTRE Myc (Clontech). To generate pcDNA3-myc-PMR60, the insert from pTRE Myc was PCR amplified with JP5 (5′-ACTG GAATTCACCATGGCATCAATGC-3′) and KC42, and the products digested with EcoRI and NotI were ligated into the corresponding sites in pcDNA3.
Cell culture
The development of C88 murine erythroleukemia cells stably transfected with either a normal human β-globin gene (Norm2) or one bearing a premature termination codon (PTC) between codons 60 and 61 (Thal10) was described previously (Stevens et al. 2002). Each cell line carries the human β-globin transgenes under control of a dimethylsulfoxide (DMSO)-inducible locus control element (Antoniou and Grosveld 1990; Collis et al. 1990; Antoniou 1991). Cells were cultured in α-minimal essential medium (α-MEM) plus 100 μg/mL G418, and the transcription of the transfected β-globin genes was induced by addition of 1.5% DMSO to the medium for 48 h. To develop stable cell lines expressing myc-PMR60 1 × 106, Norm2 cells were transfected with 1.2 μg of pcDNA3-myc-PMR60 plus 0.4 μg of ptkHyg and selected in medium containing both 100 μg/mL G418 and 800 μg/mL of hygromicin. Twelve colonies were selected and amplified for further analysis.
RNA isolation and analysis
Methods for the isolation of cytoplasmic RNA and S1 nuclease protection assays were described previously (Stevens et al. 2002). The relative amounts of intact and degraded β-globin mRNA in the S1 nuclease protection assays were determined by scanning densitometry of several exposures using a Protein Databases 420oe scanning densitometer.
Acknowledgments
We thank Jing Peng and Feng Yang for preparing the pcDNA3-myc-PMR60 plasmid, Rob Hoepfner and Emily Welch for technical assistance with this project, and Lynne Maquat for her helpful discussions. This work was supported by PHS grant GM38277 (D.R.S.) from the National Institute of General Medical Sciences, and the Oak Ridge National Laboratory Director’s R&D Fund 3211-002Q (A.S.). Support for core facilities was provided by PHS grant CA16058 from the National Cancer Institute to the Ohio State University Comprehensive Cancer Center.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
REFERENCES
- Antoniou, M. 1991. Induction of erythroid-specific expression in murine erythroleukemia (MEL) cell line. In Methods in molecular biology. Gene transfer and expression protocols (ed. E.J. Murray), pp. 421–434. Humana Press, Clifton, NJ. [DOI] [PubMed]
- Antoniou, M. and Grosveld, F. 1990. β-globin dominant control region interacts differently with distal and proximal promoter elements. Genes & Dev. 4: 1007–1013. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay, R., Coutts, M., Krowczynska, A., and Brawerman, G. 1990. Nuclease activity associated with mammalian mRNA in its native state: Possible basis for selectivity in mRNA decay. Mol. Cell. Biol. 10: 2060–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov, V.I., Scherthan, H., Solinger, J.A., Buerstedde, J.M., and Heyer, W.D. 1997. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 136: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder, R., Horowitz, J.A., Basilion, J.P., Koeller, D.M., Klausner, R.D., and Harford, J.B. 1994. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 13: 1969–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y., Gherzi, R., Ong, S.E., Chan, E.L., Raijmakers, R., Pruijn, G.J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464. [DOI] [PubMed] [Google Scholar]
- Chernokalskaya, E., Dompenciel, R.E., and Schoenberg, D.R. 1997. Cleavage properties of a polysomal ribonuclease involved in the estrogen-regulated destabilization of albumin mRNA. Nucleic Acids Res. 25: 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernokalskaya, E., DuBell, A.N., Cunningham, K.S., Hanson, M.N., Dompenciel, R.E., and Schoenberg, D.R. 1998. A polysomal ribonuclease involved in the destabilization of albumin mRNA is a novel member of the peroxidase gene family. RNA 4: 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collis, P., Antoniou, M., and Grosveld, F. 1990. Definition of the minimal requirements within the human β-globin gene and the dominant control region for high level expression. EMBO J. 9: 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, K.S., Dodson, R.E., Nagel, M.A., Shapiro, D.J., and Schoenberg, D.R. 2000. Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′-UTR by the mRNA endonuclease PMR-1. Proc. Natl. Acad. Sci. 97: 12498–12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham, K.S., Hanson, M.N., and Schoenberg, D.R. 2001a. Polysomal ribonuclease 1. Methods Enzymol. 342: 28–44. [DOI] [PubMed] [Google Scholar]
- ———. Polysomal ribonuclease 1 exists in a latent form on polysomes prior to estrogen activation of mRNA decay. Nucleic Acids Res. 29: 1156–1162. [DOI] [PMC free article] [PubMed]
- Dompenciel, R.E., Garnepudi, V.R., and Schoenberg, D.R. 1995. Purification and characterization of an estrogen-regulated Xenopus liver polysomal nuclease involved in the selective destabilization of albumin mRNA. J. Biol. Chem. 270: 6108–6118. [DOI] [PubMed] [Google Scholar]
- Gallouzi, I., Parker, F., Chebli, K., Maurier, F., Labourier, E., Barlat, I., Capony, J.P., Tocque, B., and Tazi, J. 1998. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: A potential link between signal transduction and RNA stability. Mol. Cell. Biol. 18: 3956–3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, M.N. and Schoenberg, D.R. 2001. Identification of in vivo mRNA decay intermediates corresponding to sites of in vitro cleavage by polysomal ribonuclease 1. J. Biol. Chem. 276: 12331–12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick, D.J. and Ross, J. 1994. The half-life of c-myc mRNA in growing and serum-stimulated cells—influence of the coding and 3′ untranslated regions and role of ribosome translocation. Mol. Cell. Biol. 14: 2119–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinova, I.M., Kulichkova, V.A., Evteeva, I.N., Mittenberg, A.G., Volkova, I.V., Ermolaeva, J.B., and Gause, L.N. 1999. The specific endoribonuclease activity of small nuclear and cytoplasmic α-RNPs. FEBS Lett. 462: 407–410. [DOI] [PubMed] [Google Scholar]
- Lee, C.H., Leeds, P., and Ross, J. 1998. Purification and characterization of a polysome-associated endoribonuclease that degrades c-myc mRNA in vitro. J. Biol. Chem. 273: 25261–25271. [DOI] [PubMed] [Google Scholar]
- Lim, S.K. and Maquat, L.E. 1992. Human β-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates lacking regions of exon I or exons I and II that have a cap-like structure at the 5′ termini. EMBO J. 11: 3271–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S., Mullins, J.J., Chen, C.M., Gross, K.W., and Maquat, L.E. 1989. Novel metabolism of several β°-thalassemic β-globin mRNAs in the erythroid tissues of transgenic mice. EMBO J. 8: 2613–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, S.K., Sigmund, C.D., Gross, K.W., and Maquat, L.E. 1992. Nonsense codons in human β-globin mRNA result in the production of mRNA degradation products. Mol. Cell. Biol. 12: 1149–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22: 8114–8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat, L.E. 2000. Nonsense-mediated RNA decay in mammalian cells: A splicing-dependent means to down-regulate the levels of mRNAs that prematurely terminate translation. In Translational control of gene expression (eds. N. Sonenberg et al.), pp. 849–868. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Maquat, L.E. and Carmichael, G.G. 2001. Quality control of mRNA function. Cell 104: 173–176. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E., Kinniburgh, A.J., Rachmilewitz, E.A., and Ross, J. 1981. Unstable β-globin mRNA in mRNA-deficient β° thalassemia. Cell 27: 543–553. [DOI] [PubMed] [Google Scholar]
- Mitchell, P. and Tollervey, D. 2003. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell 11: 1405–1413. [DOI] [PubMed] [Google Scholar]
- Mukherjee, D., Gao, M., O’Connor, J.P., Raijmakers, R., Pruijn, G., Lutz, C.S., and Wilusz, J. 2002. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 21: 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noensie, E.N. and Dietz, H.C. 2001. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat. Biotechnol. 19: 434–439. [DOI] [PubMed] [Google Scholar]
- Pastori, R.L., Moskaitis, J.E., Buzek, S.W., and Schoenberg, D.R. 1991a. Coordinate estrogen-regulated instability of serum protein-coding messenger RNAs in Xenopus laevis. Mol. Endocrinol. 5: 461–468. [DOI] [PubMed] [Google Scholar]
- Pastori, R.L., Moskaitis, J.E., and Schoenberg, D.R. 1991b. Estrogen-induced ribonuclease activity in Xenopus liver. Biochemistry 30: 10490–10498. [DOI] [PubMed] [Google Scholar]
- Schoenberg, D.R. and Chernokalskaya, E. 1997. Ribonucleases involved in eukaryotic mRNA turnover. In mRNA metabolism and post-transcriptional gene regulation (eds. J. Harford and D.R. Morris), pp. 217–240. J. Wiley, New York.
- Schoenberg, D.R. and Cunningham, K.S. 1999. Characterization of mRNA endonucleases. Methods 17: 60–73. [DOI] [PubMed] [Google Scholar]
- Stevens, A., Wang, Y., Bremer, K., Zhang, J., Hoepfner, R., Antoniou, M., Schoenberg, D.R., and Maquat, L.E. 2002. β-globin mRNA decay in erythroid cells: UG site-preferred endonucleolytic cleavage that is augmented by a premature termination codon. Proc. Natl. Acad. Sci. 99: 12741–12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere, H., Chebli, K., and Tazi, J. 2002. mRNA degradation machines in eukaryotic cells. Biochimie 84: 821–837. [DOI] [PubMed] [Google Scholar]
- Tucker, M. and Parker, R. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69: 571–595. [DOI] [PubMed] [Google Scholar]
- van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., and Seraphin, B. 2002. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21: 6915–6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk, E.L., Sussenbach, J.S., and Holthuizen, P.E. 2000. Distinct RNA structural domains cooperate to maintain a specific cleavage site in the 3′-UTR of IGF-II mRNAs. J. Mol. Biol. 300: 449–467. [DOI] [PubMed] [Google Scholar]
- Wang, Z. and Kiledjian, M. 2000. Identification of an erythroid-enriched endoribonuclease activity involved in specific mRNA cleavage. EMBO J. 19: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Jiao, X., Carr-Schmid, A., and Kiledjian, M. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. 99: 12663–12668. [DOI] [PMC free article] [PubMed] [Google Scholar]