Abstract
The ribosome is universally responsible for synthesizing proteins by translating the genetic code transcribed in mRNA into an amino acid sequence. Ribosomes use cellular accessory proteins, soluble transfer RNAs, and metabolic energy to accomplish the initiation, elongation, and termination of peptide synthesis. In translocating processively along the mRNA template during the elongation cycle, ribosomes act as supramolecular motors. Here we demonstrate that ribosomes adsorbed on a surface, as for mechanical or spectroscopic studies, are capable of polypeptide synthesis and that tethered particle analysis of fluorescent beads connected to ribosomes via polyuridylic acid can be used to estimate the rate of polyphenylalanine synthesis by individual ribosomes. This work opens the way for application of biophysical techniques, originally developed for the classical motor proteins, to the understanding of protein biosynthesis.
Keywords: Ribosome, single molecule, tethered particle, poly(U), poly(Phe)
INTRODUCTION
Astonishing progress on the structure of the ribosome and its accessory factors has been obtained from cryo-electron microscopy (Stark et al. 2000; Frank 2001) and X-ray diffraction studies (Ban et al. 2000; Yusupov et al. 2001; Ramakrishnan 2002; Yonath 2002). The kinetics of the elongation cycle have been elucidated by steady-state and transient kinetics studies (Rodnina et al. 2000). Although the energy required for formation of the peptide bond comes from the ester bond of tRNA charged with its cognate amino acid, the remarkable rate of translation (10–20 peptide bonds/sec; Kjeldgaard and Gausing 1974; Kennell and Riezman 1977), fidelity of amino acid selection, and maintenance of the reading frame during translocation (~10−4 error rate; Loftfield and Vanderjagt 1972; Kurland 1992) require the GTPase activities of two G-protein elongation factors, EF-Tu and EF-G. The mechanisms of action of these factors are not fully understood because the structural changes they undergo and the mechanical events they facilitate have not been elucidated on functioning ribosomes.
Single-molecule techniques have been used to measure directly the elementary events of production of force and displacement by molecular motors (Kinosita 1999; Mehta et al. 1999; Ishijima and Yanagida 2001) and nucleic acid processing enzymes (Rich 1998; Wang et al. 1998; Strick et al. 2000; Wuite et al. 2000), helping to elucidate their properties and mechanisms. Because the sliding of the ribosome along the mRNA template is a mechanical output, analogous single-molecule measurements should provide information relevant toward the understanding of the molecular mechanism of protein synthesis. For instance, applying an external force to the mRNA and measuring the relationship between the rate of translocation and the force applied could help to distinguish between proposed mechanisms of translocation (Czworkowski and Moore 1997; Keller and Bustamante 2000; Wintermeyer and Rodnina 2000).
It has been shown previously that ribosomes adsorbed on a mica surface are competent to bind aminoacyl-tRNA in the peptidyl transfer site (P-site) and to form a peptide bond with added puromycin (Sytnik et al. 1999). Here we extend this work by measuring bulk poly(Phe) synthesis on poly(U)-programmed mica-bound ribosomes and detecting translocation of individual ribosome-bound poly(U) templates using optical microscopy to examine mRNA-tethered beads.
RESULTS AND DISCUSSION
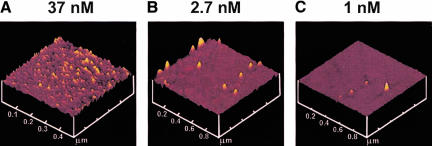
We investigated whether poly(U)-programmed ribosomes, adsorbed nonspecifically to mica surfaces, could synthesize TCA-precipitable poly([3H]-Phe) and translocate along poly(U) mRNA. Ribosomes were adsorbed to mica sample chambers from solutions containing 500 nM or 10 nM concentrations of 70S particles. These concentrations approximate the conditions used in measuring poly(Phe) synthesis in bulk solution and ribosome activity in the microscopic experiments or poly(Phe) synthesis on mica, respectively. Quantification of adsorbed material was achieved using 70S particles labeled with [32P]-phosphate. At both loading concentrations, a layer of stably bound ribosomes remained on the surface after extensive washing, although removal of non-stably bound ribosomes required more washes at the lower applied concentration. We obtained 1.5 ± 0.2 (n = 4) and 0.020 ± 0.002 (n = 21; data not shown) pmole of bound ribosomes/cm2 for the higher and lower concentrations, respectively. These measurements indicate that at 500 nM, a multilayer of ribosomes (4–5 deep) is obtained, whereas at 10 nM, a sparse distribution of ribosomes is achieved, with ~8% of the surface occupied. For the experiments described below, the number of ribosomes present in solution due to detachment from the surface was negligible. Surface images of ribosomes adsorbed to mica obtained by tapping-mode atomic force microscopy (Fig. 1 ▶) also showed that the density of tightly bound ribosomes is related to the initial concentration applied to the surface. At 37 nM loading concentration, the surface was covered with densely packed ribosomes, often in clear multilayers. At ≤2.7 nM loading, a more sparse coverage was achieved, with single ribosomes generally isolated from one another.
FIGURE 1.
AFM images of ribosomes tightly adsorbed to freshly cleaved mica. The concentrations of ribosomes applied in binding buffer are shown.
Mica-bound ribosomes synthesize poly(Phe) in an elongation-factor-dependent manner (Fig. 2A ▶). On average, peptide synthesis proceeded at 0.062 ± 0.012 (n = 4) peptide bonds/min per adsorbed ribosome, measured at room temperature (21° ± 2°C) over 60 min. Increasing [Phe-tRNAPhe] threefold, from 0.16 μM to 0.5 μM, had little effect on this rate. Under comparable conditions, but at 37°C, ribosomes in solution synthesized poly(Phe) at ~1.0 peptide bond/min per ribosome for the first 3 min, followed by a slower rate of ~0.2 peptide bond/min per ribosome over the next 30 min (Fig. 2B ▶). As raising the temperature from 21°C to 37°C increases the rate 1.8 ± 0.1-fold (data not shown), the specific enzymatic activity of surface-bound ribosomes is ~50% of that for ribosomes in solution, which may reflect a surface inactivation phenomenon. Approximately 10% of the ribosomes in solution were active, as determined by the amount of 14C present in the precipitated poly(Phe) as a result of initiation by [14C]-N-AcPhe-tRNAPhe (Wagner et al. 1982), so that the peptide synthesis rate per active ribosome was ~10 peptide bonds/min over the initial 3-min period.
FIGURE 2.
Poly(Phe) synthesis as measured by incorporation of [3H]Phe in a TCA-precipitable peptide. (A) Synthesis by ribosomes adsorbed on mica (squares: in the presence of elongation factors Tu, G, and Ts; diamonds: in the absence of elongation factors). (B) Synthesis by ribosomes in solution. The values shown are the mean and standard deviation (n = 4).
Microscopic measurements of changes in the range of motion of the 3′ end of mRNA were carried out using the tethered particle method (TPM; Schafer et al. 1991), the principle of which is illustrated in Figure 3 ▶. Long poly(U) molecules (8000–10,000 bases) were selectively biotinylated at the 3′ end, incubated with a stable multilayer of mica-bound ribosomes, and tagged with fluorescent neutravidin-coated 0.2-μm diameter beads. After removal of excess beads by washing, three different groups of beads could be distinguished through visualization by epi-fluorescence with an inverted microscope: beads displaying free Brownian diffusion, immobilized beads, and a small fraction of beads (~5%) displaying diffusive motion within a restricted volume (i.e., root-mean-squared horizontal displacement from the average position of the bead, Drms > 100 nm; Fig. 4A ▶). This latter group, denoted “tethered beads,” was only observed in the presence of biotinylated long-chain poly(U). Control experiments in the absence of 70S ribosomes showed that a few beads were localized at the surface whether or not short poly(U) was added. These observations indicate that, in the TPM experiments, most of the tethered beads are attached to surface-bound ribosomes through a long-chain poly(U), with their ranges of restricted Brownian motion determined by the lengths of the poly(U) tethers connecting the ribosome and bead. The distributions of positions of the tethered beads about a central nodal point were sometimes circularly symmetrical, but in other cases were oblong or exhibited a gap (upper right quadrant in Fig. 4A ▶), possibly caused by obstacles on the surface or an insufficient sampling interval (8 sec) for the bead to explore the entire set of accessible positions. In most cases, the distribution of radial distances was approximately uniform within the restricted range.
FIGURE 3.
Principle of the tethered particle method. (A) A microsphere (shown in pink) is tethered at the 3′ end of an mRNA molecule, which is bound to a ribosome immobilized on a microscope slide. The mRNA molecule is shown as a curvy black line, the end-to-end length of the tether is shown as a black dotted line, and the peptide synthesized by the ribosome is shown in orange. The microsphere can diffuse only within the range limited by the surface of the slide and the length of the tether, as shown by the green dashed line and shading. (B) As peptide synthesis proceeds, the ribosome pulls the 3′ end of the mRNA toward itself, reducing the range of restricted diffusion (schematic not drawn to scale).
FIGURE 4.
Restricted diffusion. (A) Centroid distributions of a bead displaying restricted diffusion. The flow chamber contained all the constituents necessary to support poly(U) programmed poly(Phe) synthesis by the ribosomes immobilized on the surface. The distribution was measured at 17, 54, and 95 min after sealing the chamber, as indicated by the magenta, orange, and black symbols, respectively. (B) For the same bead, Drms is plotted as a function of time (green circles); Drms of an immobile bead present in the same microscopic field is also plotted (red squares), as well as the Drms of a bead in a different field, displaying a large range of diffusion throughout the time of the experiment (blue triangles). (C) TPM data pooled from six beads displaying a reduction of Drms with time. The recordings from different beads were aligned by shifting each set of data to the origin at the first measurement. Each set of data is represented by a different symbol. The line is a linear regression through the pooled data, yielding a slope of −2.8 ± 0.3 (standard error) nm/min; r2 = 0.73.
The Drms was used to quantify the length of the tether when all the ligands, substrates, and factors necessary to support poly(Phe) synthesis were added to the flow chamber (see Materials and Methods). Some of the tethered beads initially displaying a large range of restricted diffusion (Drms > 250 nm) exhibited a time-dependent reduction of Drms (Fig. 4A ▶, magenta, orange, and black symbols; Fig. 4B ▶, green symbols). Such a decrease is expected as the tether length between the bead and ribosome decreases, owing to translocation of the poly(U) during poly(Phe) synthesis (as shown in Fig. 3 ▶). In contrast, the Drms of immobilized beads did not change with time (Fig. 4B ▶, red squares). Some of the beads displaying a large Drms at the beginning of the experiment did not undergo a reduction of the range of diffusion with time (as shown, for one bead, by the blue triangles in Fig. 4B ▶). The lack of time dependence in the signal from all immobilized beads and some tethered beads supports the idea that the decline in Drms observed for some beads is not caused by instrumental artifacts, as these should affect all beads in the same manner.
Figure 4C ▶ shows the pooled data from six beads, demonstrating that the measured rate of decrease of Drms is fairly uniform among all measured samples, further indicating that this observation arises from an active process. The slope measured from the pooled data is −2.8 ± 0.3 (standard error) nm/min.
Calculation of the peptide synthesis rate from these measurements requires knowledge of the physical properties of poly(U). From classic polymer mechanics, the dependence of the average squared end-to-end length of the polymer can be written as ≤r2> = 2NLpd, where N is the number of nucleotides in the tether, d is distance between adjacent nucleotides along the polymer backbone, and Lp is the persistence length, a measure of the polymer bending stiffness (Howard 2001). From this expression, Drms can be calculated using the projection of the end-to-end length onto the microscope image plane, obtaining the following relation:
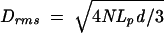 |
Using an explicit Gaussian model for the distribution of end-to-end distances, Qian and Elson (1999) described quantitatively the range of restricted diffusion of beads tethered to double-stranded DNA molecules of known lengths (Yin et al. 1994); the two-dimensional projection of the Gaussian distribution onto the microscope image plane yields the same expression for Drms as given above. In the range of values of N measured by gel electrophoresis for the poly(U) molecules used in these experiments, and using values of 0.8 nm for Lp (Smith et al. 1996; F. Vanzi, Y. Takagi, H. Shuman, B. Cooperman, and Y. Goldman, in prep.), and 0.65 nm for d (Inners and Felsenfeld 1970), the quadratic expression for Drms is well approximated by a line with slope 0.0072 nm/base. However, this approximation, based purely on the physical properties of a flexible polymer, takes no account of the self-avoidance of the mRNA, the volume excluded by the mica surface or the bead, or the mechanics of the biotin–neutravidin linkage, and each of these factors could have significant effects. To resolve some of these uncertainties, we have initiated studies to calibrate the range of bead diffusion with known-length homopolymer RNA molecules. Preliminary results, obtained using an optical trap to measure N and Lp and video microscopy to measure Drms for a series of ribosome tethered beads (Vanzi et al. 2003), indicate that the dependence of Drms on N is, in fact, characterized by a larger slope (0.023 nm/base) than that of the theoretical approximation used above. Applying the two slope values (0.0072 and 0.023 nm/base) to the present TPM data gives apparent rates of peptide synthesis of 2.2 ± 0.2 and 0.66 ± 0.07 peptide bonds/sec, respectively. The correct value is likely to fall within this range, indicating that some ribosomes adsorbed to the mica surface are capable of synthesizing protein at close to the physiological rate.
The apparent discrepancy between the synthesis rate estimated for individual ribosomes using the tethered particle method and the rates measured for bulk poly(Phe) synthesis by mica-bound ribosomes may be plausibly attributed to selection in the single-molecule experiments of tethered beads showing the largest decreases in restricted diffusion with time (i.e., the most catalytically active ribosomes). As well, the fraction of mica-bound ribosomes working at near-physiological rates may be very small. At present, the rate of polypeptide synthesis achieved in bulk measurements is low, and the calibration of the single-particle observations is uncertain. Nevertheless, the demonstration, in this work, of polypeptide synthesis by single ribosomes immobilized on a surface heralds the use of single-molecule techniques for detailed mechanistic study of such questions as the accuracy of tRNA selection and the mechanisms of peptide bond formation and translocation during the elongation cycle.
MATERIALS AND METHODS
Materials were obtained from Sigma unless otherwise indicated.
Preparations
Escherichia coli Q13 70S ribosomes and 30S and 50S subunits were prepared and activated as described earlier (Alexander et al. 1994). 32P-labeled ribosomes were prepared by incubating 30S ribosomal subunits with [32P]pCp (NEN Life Sciences Products) and T4 RNA ligase (Promega), combining the [32P]-30S subunits with a 1.5 M excess of 50S subunits and purifying the labeled 70S ribosomes by fractionation following ultracentrifugation through a linear sucrose concentration gradient (15%–30%). His6-tag EF-Tu and His6-tag EF-G were isolated by Ni-column purification (Ni-NTA agarose, QIAGEN). EF-Ts fused to an intein-chitin-binding domain was purified by binding to a chitin column (New England Biolabs). Yeast Phe-tRNAPhe synthetase was purified as described (Von der Haar 1979). Labeled Phe-tRNAPhe (900 pmole/A260) was formed by incubation of 40 μM yeast tRNAPhe with yeast Phe-tRNAPhe synthetase, 7 mM ATP, 0.2 mM CTP, 100 mM Tris-HCl (pH 7.5), 50 mM NH4Cl, 1 mM dithiothreitol, and 200 μM Phe, using either 14C- or 3H-labeled Phe (NEN Life Sciences Products). Acetylation of Phe-tRNAPhe was performed as described (Rappoport and Lapidot 1974).
Mica cells
Mica sheets (Electron Microscopy Sciences) were cleaved into thin sheets just prior to use. Adsorption experiments were carried out either within a minicell formed by building a closed rectangular border of epoxy glue on the mica surface (2.1 cm2), or using a flow cell assembled by attaching a 1.5 cm × 1.5 cm mica coverslip onto a larger mica or standard glass microscope slide. Spacers gave a chamber height of 0.1–0.25 mm.
Adsorption of ribosomes to the flow cell
[32P] 70S ribosomes (500 nM) and Sigma poly(U) (2.5 μg/μL) in 50 μL of binding buffer were applied for 5 min to an open (2.1 cm2) minicell. The solution was removed, and the surface was washed three times with polymix (solution constituents given below), once with blocking solution, and three further times with polymix, followed by incubation with factor mix 2, and incubation with polymix containing 1 mM puromycin. Each wash or incubation was performed for 5 min or 1 h, respectively, with a 60-μL volume. The number of ribosomes remaining on the surface was measured as the sum of the number of 32P-labeled ribosomes collected from the mica following puromycin incubation and washing with a solution containing 0.2% SDS, 10 mM EDTA, and 10 mM Phe (55%–60% of the total) plus the number remaining on the mica (40%–45% of the total).
Tapping mode atomic force microscopy
Ribosomes were applied to a mica slide in a petri dish for 5 min, the solution was removed, and the surface was washed six times with 20 μL of binding buffer (50 mM Tris-HCl at pH 7.4, 50 mM KCl, 10 mM MgCl2). The petri dish was then filled with 5 mL of binding buffer to cover the mica and prevent sample drying. Observations were made with a Digital Instruments Extended BioScope with Nanoscope IIIa controller, equipped with a silicon nitride cantilever (tip diameter, 20 nm; spring constant, 0.32 N/m).
Poly(Phe) synthesis
By ribosomes adsorbed to mica
Adsorbed ribosomes were prepared by application of 10 nM ribosomes in 15 μL of polymix (5 mM KH2PO4 at pH 7.3, 95 mM KCl, 5 mM NH4Cl, 5 mM Mg acetate, 0.5 mM CaCl2, 8 mM putrescine, 1 mM spermidine, 1 mM dithiothreitol) to open minicells (2.1 cm2) for 5 min, followed by three washes with binding buffer, one with blocking solution (Block Aid [Molecular Probes, Inc.] diluted 1:20 in polymix containing 1.2 U/μL RNA guard RNase inhibitor [Amersham Pharmacia Biotech, Inc.] and 0.5 mM Phe), and three further washes with polymix containing 0.5 mM Phe. Each minicell was then preincubated (1 h) with a mixture of 20 μg of Sigma poly(U), 0.2 pmole yeast N-AcPhe-tRNAPhe, and 5 nmole Phe in 10 μL of polymix. Synthesis at room temperature (21° ± 1°C) was commenced by addition of 10 μL of factor mix 1 (3.14 pmole of yeast [3H]-Phe-tRNAPhe [92 Ci/mmole]), 200 pmole of EF-G, 40 pmole of EF-Ts, 380 pmole of EF-Tu, 33 nmole of GTP, 20 nmole of ATP, 200 nmole of phosphoenol pyruvate, 5 nmole of Phe, 1 μg of pyruvate kinase, and 0.06 μg of myokinase dissolved in polymix and incubated at 30°C for 15 min before use) and terminated by addition of 2 μL of 10 mM puromycin (1 h incubation). Following addition of 2 μL of 0.5 M EDTA (pH 8.0, 5 min incubation), the solution was removed from the mica slide and joined with three 20 μL consecutive washes of the surface (25 mM Tris-HCl at pH 7.6, 10 mM EDTA, 0.5 mM Phe; each wash had a 5-min incubation). BSA (5 μg) was added to the pooled washes, and an aliquot was transferred into 10% trichloroacetic acid (TCA). The sample was placed at 85°C (30 min) to hydrolyze the remaining [3H]-Phe-tRNAPhe, cooled on ice (10 min), and filtered through a 0.45-μm nitrocellulose filter. The filter was washed three times with cold 10% TCA, dried, and dissolved in 1 mL of ethyl acetate. Incorporation of [3H]-Phe in TCA-precipitable peptides was quantified by liquid scintillation counting.
By ribosomes in solution
Ribosomes (100 pmole) were preincubated at 30°C for 15 min with Sigma poly(U) (260 μg) and N-Ac[14C]-Phe-tRNAPhe (100 pmole) in 100 μL of polymix. Synthesis at 37°C was commenced by addition of 100 μL of factor mix 1, made up exactly as above but containing a higher concentration of yeast [3H]-Phe-tRNAPhe (400 pmole/100 μL). Aliquots withdrawn at different times after the addition of factor mix were treated as above to determine TCA-precipitable poly(Phe).
Synthesis and biotinylation of high-molecular-weight poly(U)
Long-chain poly(U) was synthesized by polymerization of UDP catalyzed by polynucleotide phosphorylase (from Micrococcus lysodeikticus or E. coli). The reaction was carried out in a solution containing 100 mM Tris-HCl (pH 9.0), 5 mM MgCl2, 0.4 mM EDTA, 20 mM UDP, 1.5 mg/mL short chain poly(U) (Sigma), and polynucleotide phosphorylase to a final concentration of 50 μg/mL. After an incubation at 37°C for 2 h, additional UDP was added to a final concentration of 40 mM, and the mixture was incubated at 37°C for another 2 h. The sample was then loaded on a Sephacryl S400HR column (Pharmacia), pre-equilibrated with PBS buffer (1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl at pH 7.4), and eluted in PBS buffer, yielding an early fraction with chains of 5000–12,000 nucleotides, as determined by agarose gel electrophoresis and methylene blue staining (Sambrook et al. 1989). The high-molecular-weight fractions were concentrated by ethanol precipitation and biotinylated as described (Odom et al. 1980), except that elution through an NAP-5 column (Pharmacia) was used in place of phenol extraction to eliminate unreacted biotin. Successful biotinylation was confirmed by dot blot on nitrocellulose and detection of bound biotin with horseradish peroxidase-conjugated neutravidin (Pierce) and ECL detection reagents (Pharmacia).
Restricted diffusion
Ribosomes (1 μM), N-Ac[14C]-Phe-tRNAPhe (1 μM), and biotinylated long-chain poly(U) (2.6 μg/μL) were preincubated in polymix at 30°C for 15 min and applied to a mica flow chamber (10–15 μL) for 5 min. The chamber was then washed three times with 15 μL of polymix, and blocking solution (15 μL) was flowed in and incubated for 5 min. Next, a 0.02% suspension of fluorescent beads (0.2 μm diameter, neutravidin-labeled Fluospheres; Molecular Probes, Inc.) in blocking solution was flowed into the chamber and incubated for 30–60 min. The chamber was then washed 5–10 times with 15 μL of polymix. Finally, 15 μL of factor mix 2 (150 pmole of yeast [3H]Phe-tRNAPhe, 150 pmole of EF-G, 30 pmole of EF-Ts, 150 pmole of EF-Tu, 1.5 nmole of GTP, 15 nmole of ATP, 150 nmole of phosphoenolpyruvate, 0.75 μg of pyruvate kinase, and 0.045 μg of myokinase in polymix; preincubated at 30°C for 15 min) was flowed into the chamber, which was then sealed with clear nail polish and transferred to a Nikon Diaphot inverted microscope for observation. Images of the beads were acquired at room temperature (22° ± 2°C) under epi-illumination. Excitation was from a xenon arc lamp, with a fluorescein filter block (485/520 nm excitation/emission; Omega Optical). Video sequences were recorded using a video camera (VE-1000CCD; Dage-MTI), stored on sVHS magnetic tape, and digitized at 5 Hz by a frame grabber (DT3152; National Instruments). The centroid position of each bead was determined weighting each pixel position by its grayscale level above background. The distribution of centroid positions of the bead over an 8-sec interval was determined at ~10-min intervals after addition of the factor mix.
Acknowledgments
We thank Paul Janmey and Tony Yeung for their help in obtaining the AFM images, Kevin S. Wilson for the EF-G plasmid, Mandana Sassanfar for the EF-Tu plasmid, Henrik Karring for the EF-Ts plasmid, Akira Kaji and Måns Ehrenberg for useful support and discussions, and Nora Zuño for technical assistance. This work was supported by NIH grants GM53146 and GM63205.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
poly(Phe), polyphenylalanine
poly(U), polyuridylic acid
TCA, trichloroacetic acid
TPM, tethered particle method
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5800303.
REFERENCES
- Alexander, R.W., Muralikrishna, P., and Cooperman, B.S. 1994. Ribosomal components neighboring the conserved 518–533 loop of 16S rRNA in 30S subunits. Biochemistry 33: 12109–12118. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Czworkowski, J. and Moore, P.B. 1997. The conformational properties of elongation factor G and the mechanism of translocation. Biochemistry 36: 10327–10334. [DOI] [PubMed] [Google Scholar]
- Frank, J. 2001. Cryo-electron microscopy as an investigative tool: The ribosome as an example. BioEssays 23: 725–732. [DOI] [PubMed] [Google Scholar]
- Howard, J. 2001. Mechanics of motor proteins and the cytoskeleton. Sinauer Associates, Inc., Sunderland, MA.
- Inners, L.D. and Felsenfeld, G. 1970. Conformation of polyribouridylic acid in solution. J. Mol. Biol. 50: 373–389. [DOI] [PubMed] [Google Scholar]
- Ishijima, A. and Yanagida, T. 2001. Single molecule nanobioscience. Trends Biochem. Sci. 26: 438–444. [DOI] [PubMed] [Google Scholar]
- Keller, D. and Bustamante, C. 2000. The mechanochemistry of molecular motors. Biophys. J. 78: 541–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennell, D. and Riezman, H. 1977. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J. Mol. Biol. 114: 1–21. [DOI] [PubMed] [Google Scholar]
- Kinosita Jr., K. 1999. Real time imaging of rotating molecular machines. FASEB J. 13: S201–S208. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard, N.O. and Gausing, K. 1974. Regulation of biosynthesis of ribosomes. In Ribosomes (eds. M. Nomura et al.), pp. 369–392. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Kurland, C.G. 1992. Translational accuracy and the fitness of bacteria. Annu. Rev. Genet. 26: 29–50. [DOI] [PubMed] [Google Scholar]
- Loftfield, R.B. and Vanderjagt, D. 1972. The frequency of errors in protein biosynthesis. Biochem J. 128: 1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, A.D., Rief, M., Spudich, J.A., Smith, D.A., and Simmons, R.M. 1999. Single-molecule biomechanics with optical methods. Science 283: 1689–1695. [DOI] [PubMed] [Google Scholar]
- Odom Jr., O.W., Robbins, D.J., Lynch, J., Dottavio-Martin, D., Kramer, G., and Hardesty, B. 1980. Distances between 3′ ends of ribosomal ribonucleic acids reassembled into Escherichia coli ribosomes. Biochemistry 19: 5947–5954. [DOI] [PubMed] [Google Scholar]
- Qian, H. and Elson, E.L. 1999. Quantitative study of polymer conformation and dynamics by single-particle tracking. Biophys. J. 76: 1598–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan, V. 2002. Ribosome structure and mechanism of translation. Cell 108: 557–572. [DOI] [PubMed] [Google Scholar]
- Rappoport, S. and Lapidot, Y. 1974. The chemical preparation of acetylaminoacyl-tRNA. Methods Enzymol. 29: 685–688. [DOI] [PubMed] [Google Scholar]
- Rich, A. 1998. The rise of single-molecule DNA biochemistry. Proc. Natl. Acad. Sci. 95: 13999–14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina, M.V., Stark, H., Savelsbergh, A., Wieden, H.-J., Mohr, D., Matassova, N.B., Peske, F., Daviter, T., Gualerzi, C.O., and Wintermeyer, W. 2000. GTPase mechanisms and functions of translation factors on the ribosome. Biol. Chem. 381: 377–387. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schafer, D.A., Gelles, J., Sheetz, M.P., and Landick, R. 1991. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature 352: 444–448. [DOI] [PubMed] [Google Scholar]
- Smith, S.B., Cui, Y., and Bustamante, C. 1996. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 271: 795–799. [DOI] [PubMed] [Google Scholar]
- Stark, H., Rodnina, M.V., Wieden, H.-J., van Heel, M., and Wintermeyer, W. 2000. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 100: 301–309. [DOI] [PubMed] [Google Scholar]
- Strick, T., Allemand, J.-F., Croquette, V., and Bensimon, D. 2000. Twisting and stretching single DNA molecules. Prog. Biophys. Mol. Biol. 74: 115–140. [DOI] [PubMed] [Google Scholar]
- Sytnik, A., Vladimirov, S., Jia, Y., Li, L., Cooperman, B.S., and Hochstrasser, R.M. 1999. Peptidyl transferase center activity observed in single ribosomes. J. Mol. Biol. 285: 49–54. [DOI] [PubMed] [Google Scholar]
- Vanzi, F., Takagi, Y., Vladimirov, S., Shuman, H., Cooperman, B.S., and Goldman, Y.E. 2003. Single-molecule mechanics of poly(U): Elastic properties and interactions with ribosomes. Biophys. J. 84: 299a. [Google Scholar]
- Von der Haar, F. 1979. Purification of aminoacyl-tRNA synthetases. Methods Enzymol. 59: 257–267. [DOI] [PubMed] [Google Scholar]
- Wagner, G.H., Jelenc, P.C., Ehrenberg, M., and Kurland, C.G. 1982. Rate of elongation of polyphenylalanine in vitro. Eur. J. Biochem. 122: 193–197. [DOI] [PubMed] [Google Scholar]
- Wang, M.D., Schnitzer, M.J., Yin, H., Landick, R., Gelles, J., and Block, S.M. 1998. Force and velocity measured for single molecules of RNA polymerase. Science 282: 902–907. [DOI] [PubMed] [Google Scholar]
- Wintermeyer, W. and Rodnina, M.V. 2000. Translational elongation factor G: A GTP-driven motor of the ribosome. Essays Biochem. 35: 117–129. [DOI] [PubMed] [Google Scholar]
- Wuite, G.J.L., Smith, S.B., Young, M., Keller, D., and Bustamante, C. 2000. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature 404: 103–106. [DOI] [PubMed] [Google Scholar]
- Yin, H., Landick, R., and Gelles, J. 1994. Tethered particle motion method for studying transcript elongation by a single RNA polymerase molecule. Biophys. J. 67: 2468–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonath, A. 2002. The search and its outcome: High-resolution structures of ribosomal particles from mesophilic, thermophilic, and halophilic bacteria at various functional states. Annu. Rev. Biophys. Biomol. Struct. 31: 257–273. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G. Zh., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H.D., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]