Abstract
We previously isolated from random sequences ribozymes able to form a glycosidic linkage between a ribose sugar and 4-thiouracil in a reaction that mimics protein-catalyzed nucleotide synthesis. Here we report on two serial in vitro selection experiments that defined the core motif of one of the nucleotide synthase ribozymes and provided improved versions of this ribozyme. The first selection experiment started from a degenerate sequence pool based on the previously isolated sequence and used a selection-amplification protocol that allowed the sequence requirements at the 3′ terminus of the ribozyme to be interrogated. Comparing the active sequences identified in this experiment revealed the complicated secondary structure of the nucleotide synthase ribozyme. A second selection was then performed to remove nonessential sequence from the ribozyme. This selection started with a pool with variation introduced in both the sequence and the length of the nonconserved loops and joining regions. This pool was generated using a partial reblocking/deblocking strategy on a DNA synthesizer, allowing the combinatorial synthesis of both point deletions and point substitutions. The consensus ribozyme motif that emerged was an ~71 nt pseudoknot structure with five stems and two important joining segments. Comparative sequence analysis and a cross-linking experiment point to the probable location of nucleotide synthesis. The prototype isolate from the second selection was nearly 35 times more efficient than the initial isolate and at least 108 times more efficient than an upper limit of an as-yet undetectable uncatalyzed reaction, supporting the idea that RNA-catalyzed nucleotide synthesis might have been important in an RNA world.
Keywords: Catalytic RNA, in vitro selection, pyrimidine nucleotide synthesis
INTRODUCTION
Previously we used in vitro selection to isolate three ribozyme families able to synthesize tethered 4-thiouridine starting from 4-thiouracil (4SUra) and an activated ribose presented as phosphoribosyl 1-pyrophosphate (pRpp) attached to the ribozyme 3′ terminus (Fig. 1 ▶; Unrau and Bartel 1998). The ability of RNA to promote the synthesis of a pyrimidine nucleotide is notable because uncatalyzed synthesis of pyrimidine nucleotides has not been detected even under a variety of extreme prebiotic conditions (Orgel 1998). Members of one of the ribozyme families (family A) promoted the pyrimidine nucleotide synthesis reaction at least 106 times faster than the upper limit of the analogous uncatalyzed reaction (Unrau and Bartel 1998). Moreover, nucleotide synthesis and hydrolysis reactions, when promoted by protein enzymes, involve intermediates with considerable oxocarbenium character (Tao et al. 1996; Chen et al. 2000). This type of dissociative mechanism differs from the concerted reaction mechanisms that RNA is known to promote (Bartel and Unrau 1999), making the structure and mechanism of this ribozyme family of considerable interest.
FIGURE 1.
Reaction of 4-thiouridine synthase ribozyme. Ribozyme tethered to pRpp at its 3′ end is incubated with 4SUra. After the displacement of pyrophosphate (PPi), a glycosidic linkage is formed between the ribose sugar and the pyrimidine base to form 4-thiouridine.
We selected for further study the a15 isolate, a member of the family A nucleotide synthase ribozymes. This ribozyme has an apparent kcat/Km of 4.3 M−1min−1 for its 4SUra substrate and reacts approximately 10,000 times slower when uracil is used as a substrate (Unrau and Bartel 1998). The ribozyme was originally isolated from a large pool of RNA sequences (approximately 1.5 × 1015 different sequences), each possessing three stretches of random sequence totaling 228 random nucleotides (Fig. 2A ▶).
FIGURE 2.
Designs of RNA pools used to isolate the original nucleotide synthase ribozymes and generate improved variants of the a15 isolate. The RNA pools were transcribed from double-stranded DNA pools. Regions of random or degenerate sequence are shown as boxes; regions of fixed sequence are shown as lines. (A) The initial random-sequence pool (Unrau and Bartel 1998). This pool had three 76 nt random-sequence regions (N76), joined by StyI and BanI restriction sites. A population of ~1.5 × 1015 different sequences was used in the initial selection and yielded three nucleotide synthase families. (B) The degenerate-sequence pool based on a 5′ truncated variant of a15, the prototype isolate of the family A nucleotide synthase ribozymes. This pool was constructed to generate ribozyme variants with sequence diversity suitable for identifying critical residues and inferring secondary structure by comparative analysis. It was made from two pieces linked by a BanI site. Most residues of the a15 ribozyme were within the 73 and 93 nt degenerate regions (M73 and M93), which were mutagenized such that on average 80% of the residues would be the same as the starting sequence and 20% would be substituted by one of the other three possibilities. A 10 nt random-sequence region (N10) was inserted into the terminal loop of stem I. Note that in its RNA form this pool has variation extending to its 3′ terminus. (C) The pool with variation in both sequence and length. This pool was based on the a.6.10 ribozyme isolated from the degenerate-sequence pool (panel B). Two pools were synthesized, each containing five regions (R1–R5) in which a modified coupling cycle was used to introduce both point deletions and substitutions. The number of positions in each mutagenized region varied from 7 to 25, as indicted by the numbers prefixed by a D (D15, D25, D8, D10, D11, and D7; note that R4 spans both the D10 and D11 boxes). Each position subjected to the modified coupling cycle incurred a 20% substitution frequency and a deletion frequency of either 17% or 25%, depending on the pool. The remaining sequence was kept fixed to form the ribozyme helices (stems I to VI), the arms of which are indicated by Roman numerals. Equimolar amounts of the two pools were combined to generate a starting pool with ~5 × 1013 unique deletion mutants.
Long random-sequence pools, such as the one used in the isolation of a15, are expected to dramatically increase the probability of isolating complicated ribozyme motifs, due to a combinatorial increase in the number of ways core elements can be arranged within the longer RNA (Sabeti et al. 1997). Although the use of long random-sequence pools is advantageous, and sometimes necessary, to isolate rare motifs that efficiently promote challenging reactions, ribozymes isolated from such pools are more difficult to define, because the core ribozyme motifs are imbedded within stretches of sequence not absolutely required for function. Such nonessential regions can be identified either by a comparative sequence analysis of active variants (Ellington and Szostak 1990; Ekland and Bartel 1995) or through use of chemical- or modification-interference experiments (Strobel 1998). In the present analysis of the nucleotide synthase ribozyme, we opted to use the comparative sequence analysis of active variants, because this approach has the added benefit of identifying variants with improved reaction efficiency. Investigating the sequence requirements near the 3′ terminus of the ribozyme, which is the site of pRpp attachment, required developing a method to amplify the ribozyme without using the 3′-terminal portion of the ribozyme as a primer-binding site.
Once nonessential regions have been identified, it is useful to delete them to create compact versions of the ribozyme more suitable for structural and kinetic characterization. Typically, extraneous sequences found in loops or beyond the 5′ or 3′ boundaries of the ribozyme are easily recognized and removed, whereas interhelical joining regions are more difficult to shorten in a manner that retains full activity. In the present study, we also found joining regions that did not appear to be critical for function, based on their low sequence conservation among active ribozyme variants, yet were difficult to shorten without diminishing nucleotide synthase activity. We therefore explored methods to generate diverse pools of sequence variants that included a high number of deletions in addition to nucleotide substitutions, with the idea that selection of active variants from such pools would identify deletions that perhaps in combination with altered local sequence contexts could support full ribozyme activity.
Several strategies have been used previously to introduce point deletions into DNA pools. One strategy is the split-and-pool approach (Giver et al. 1993) wherein the solid support used in DNA synthesis is temporarily removed before particular couplings. Although useful for pools with low diversity, this method is limited by the small number of beads making up the solid support in a DNA synthesis column. Another strategy for introducing point deletions into DNA pools is to omit the capping step during DNA synthesis (Bartel et al. 1991). This method is limited because the phosphoramidite coupling efficiency must also be reduced in a controlled manner in order to reliably achieve a deletion frequency greater than 1%–5% per coupling cycle. Yet another approach is the “partial deblock” strategy, where the dimethoxytrityl (DMT) blocking group is only partially removed, so that some, but not all, of the molecules can participate in the next phosphoramidite coupling cycle (Treiber and Williamson 1995). However, dichloroacetic acid-catalyzed removal of DMT is strongly sequence-dependent, which results in a strong deletion sequence bias. To overcome these difficulties, we modified the procedure of Treiber and Williamson in order to allow the construction of large libraries (1014–1015 different sequences) of deletion and mutation variants with little sequence bias. In vitro selection was then used to isolate the shortest and most active sequences, thus avoiding the tedious production and screening of individually designed deletion constructs. Here we outline the selection and characterization of improved, internally truncated nucleotide synthase ribozymes using this methodology.
RESULTS
Ribozyme secondary structure and tertiary constraints
The prototype member of the family A nucleotide synthase ribozyme is isolate a15, which is 272 nt long. Truncation experiments indicated that 98 nucleotides could be removed from the 5′ end of a15 while still retaining full activity. A large pool of RNA molecules related to this truncation construct were synthesized containing regions of sequence that were fixed, random, or mutagenized to a level of 20% (Fig. 2B ▶). The RNA of this pool was then ligated to its pRpp substrate and incubated with 4SUra. Reactive sequences able to form tethered 4-thiouridine were purified away from unreactive sequences using N-acryloylaminophenylmercuric acetate (APM) polyacrylamide gels (Igloi 1988; Unrau and Bartel 1998).
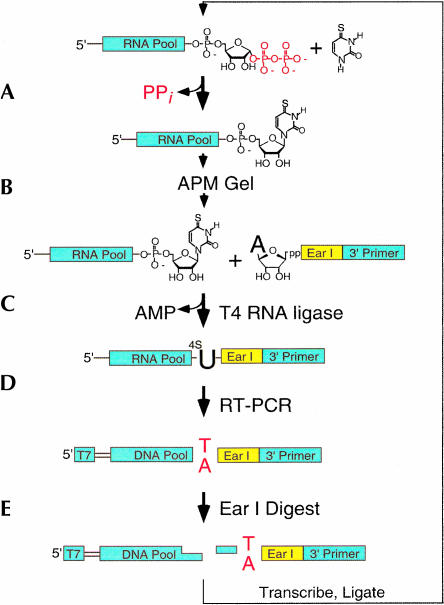
A notable feature of this selection was that we were able to select ribozymes with sequence variability at their very 3′ ends. Conventional in vitro selection experiments require an invariant sequence at the 3′ end of an RNA strand in order to allow RT-PCR-based amplification. This was a potentially serious limitation, because the 3′ end of the nucleotide synthase ribozyme is tethered to the pRpp substrate and might therefore be proximal to the active site of the ribozyme. We circumvented this problem by ligating an invariant DNA sequence to the 3′ end of active RNA sequences purified out of an APM gel. Ligation of this DNA oligo created an RT primer-binding site, making it possible to amplify the pool by RT-PCR. Before the next round of selection, digestion with EarI endonuclease removed the RT primer, allowing transcription of RNA sequences with variable 3′ ends (Fig. 3 ▶).
FIGURE 3.
Selection scheme designed to retain sequence variation all the way to the 3′ end of RNA transcripts. (A) Pool RNA tethered to pRpp was incubated with 4SUra. Active sequences react to form tethered 4-thiouridine. (B) Active sequences were purified away from unreactive sequences, using an APM gel. (C) Using T4 RNA ligase, purified ribozymes were then ligated to an adenylated DNA oligo containing an EarI restriction site and 3′ RT-PCR primer binding region. (D) RT-PCR was performed to amplify ribozyme DNA sequence and add a T7 promoter (T7) to the 5′ end of the sequences. (E) Digestion with EarI-generated DNA sequence lacking a fixed 3′ primer-binding site, which was then transcribed to generate RNA for another round of selection.
After six cycles of selection, we found that the pool activity was significantly faster than that of the parent sequence. Individual ribozyme sequences, obtained by cloning from the pool, were screened for activity. Their observed reaction rates ranged from 0.025 to 0.10 min−1 when assayed at 1 mM 4SUra substrate concentration. The fastest ribozymes were approximately 25 times more efficient than the a15 parent.
An alignment of 30 unique family A ribozyme DNA sequences from this selection revealed a diverse set of ribozymes, each with the potential to assume the same complicated secondary structure consisting of at least five helical regions, three loop regions, and seven interhelical joining regions. A secondary structure model for family A was determined by a statistical analysis of the aligned sequence data. Nineteen of the 166 residues mutagenized to a level of 20% (Fig. 2B ▶) in the degenerate pool were found to be absolutely conserved. This level of sequence conservation is exceedingly unlikely to be due to chance and implies that the majority if not all of these conserved residues play an important role in ribozyme function. Moreover, these highly conserved residues were found within larger blocks of highly, but not absolutely, conserved sequence. All but one of these conserved blocks could be base-paired with complementary sequence found within the degenerate regions of the pool. The remaining conserved block was complementary to a region of sequence found in the invariant 5′ primer binding region of the pool. Importantly, low levels of variability within these regions could often be attributed to covariations consistent with the proposed Watson-Crick helixes. Such covariation was taken as strong evidence for base-pairing, because the probability of a double mutation leaving two residues base-paired by chance [P = (4/9) × (0.2)2 = 1.8%, including G:U wobbles] is much smaller than the probability of finding single or double mutations that violate pairing [P = 1 − (0.82 + ((4/9) × (0.2)2)) = 34%].
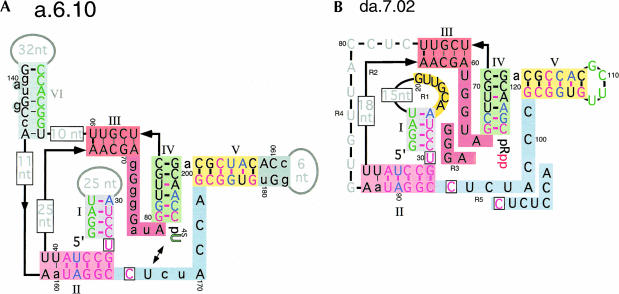
Figure 4A ▶ summarizes the statistical data and a model of the ribozyme. Stems I and II are linked by an absolutely conserved uridine residue. Opposite the conserved uridine is an absolutely conserved cytidine in the core of the ribozyme. Immediately 3′ to the cytidine residue is a well conserved stretch of 6–8 nucleotides connecting stem II to stem V. Two other stems, III and IV, stack coaxially and are linked topologically by a short guanosine-rich strand to form a pseudoknot. One end of stem IV forms the 3′ terminus of the ribozyme and is covalently attached to the tethered pRpp substrate, and the other end is linked to stem V by a single-nucleotide spacer. Stem V is closed by a hairpin loop containing highly variable sequence. The ends of stems II and III are linked together by two long, poorly conserved regions of RNA. Weak statistical evidence suggested the presence of a sixth stem in the larger of these regions.
FIGURE 4.
Inferred secondary structure of family A 4-thiouridine synthase ribozyme. (A) The structure of isolate a.6.10 obtained from the secondary structure determination selection is shown after having reacted with 4SUra to produce 4-thiouridine 5′ phosphate (p4SU). The ribozyme consists of six stems (I–VI), three terminal loops, and seven interhelical regions. U34 and C166 were absolutely conserved in every isolate sequenced (black boxed residues). Cross-linking the 4-thiouridine product of the ribozyme using UV light resulted in a efficient covalent linkage to U167 (arrows). Stems and conserved regions of sequences are denoted by colored boxes. Nucleotide colors: green, sequence fixed due to pool construction; magenta, no sequence variation observed (probability due to chance, P = 0.0012); blue, always covaries with partner (includes G-U wobble pairs). Uppercase black, ≤ 4/30 variations from consensus; lowercase black, > 4/30 variants from consensus. Residues showing poor sequence conservation are indicated in gray. Conserved potential for pairing is indicted as follows: thick purple bar, always base-paired (P = 3.6 × 10−5 if both residues can vary); thick black bar, 29/30 bp (P = 4.8 × 10−4); thin black bar, 26/30 or more bp (P<0.04). (B) The selection designed to minimize the ribozyme sequence in regions 1 to 5 (R1–R5) resulted in considerably shorter isolates, as represented by da.7.02. The consensus sequence for isolates resulting from round 7 of the selection differs from da.7.02 in R3 and R5 and is highlighted adjacent to the da.7.02 sequence. Conservation of sequence found in loop I is highlighted in yellow. The uppercase gray and boxed gray residues indicate variable sequence that could not be uniquely aligned, preventing the statistical analysis of mutational frequencies. The base-pairing information inferred from comparative analysis of the previous isolates (panel A) is shown within the helical regions of ribozyme da.7.02.
Large amounts of covariation and stabilization in helix IV made this an area of particular interest. Helix IV is tethered to the pRpp substrate and must therefore point into the active site. In all of the round-six isolates, helix IV had evolved from the a15 parent sequence (GCAACpRpp, 3′ terminus of a15) to sequences thermodynamically stabilized by the addition of at least one C:G pair: GCAACCpRpp (15/30 sequences, a.6.10 residues 199–204), RCAGCpRpp (8/30, R any purine), GCAAGCpRpp (3/30), GCGAC CpRpp (2/30), GCAGCCpRpp (1/30), and GCGCCpRpp (1/30). The underlined nucleotides indicate the location of an insertion or mutation with respect to the parental a15 sequence. In all cases, covariation was observed in the complementary helical arm (a.6.10 residues 80–85). These data indicated a strong preference for a stable stem terminating in a conserved C(204):G(80) base pair immediately adjacent to the pRpp substrate.
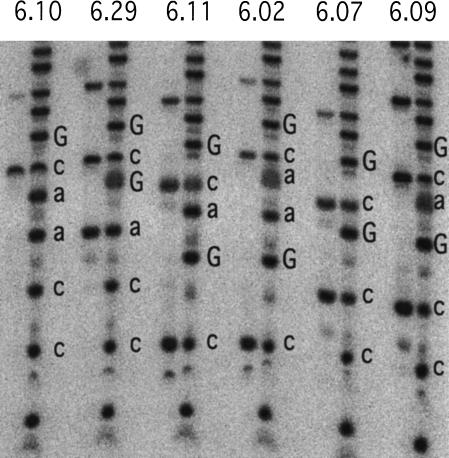
As T7 RNA polymerase often produces RNA transcripts with nontemplated terminal residues (Milligan et al. 1987), functional ribozyme sequences might not directly correspond with our DNA sequence data. We therefore characterized the terminal sequence of the parental a15 ribozyme along with six new ribozyme isolates, each possessing one of the unique stem IV sequence modifications discussed in the previous paragraph. After reacting these ribozymes with 4SUra, the ribozymes were 3′ end-labeled, purified using an APM gel, and either lightly hydrolyzed or digested with RNase T1 (Fig. 5 ▶, a15 not shown). In all cases, the observed RNA digestion pattern corresponded precisely to the sequence predicted from DNA sequence.
FIGURE 5.
Characterization of 3′-terminal RNA sequence of ribozymes from round 6 of the selection from the degenerate pool (Fig. 2B ▶). The locations of G residues in ribozymes having different terminal sequences were mapped to confirm that the sequence of the active RNA species corresponded to the DNA sequence in this region. Ribozymes were ligated to pRpp and reacted with 4SUra. After 3′ end-labeling with [32P]-α-CoTP and poly(A) polymerase, RNA was either lightly digested with RNase T1 to generate a G ladder (left lane of each pair) or partially hydrolyzed with sodium bicarbonate to produce a hydrolysis ladder (right lane of each pair). Uppercase letters, location of G residues; lowercase, the residues inferred from DNA sequence (the cleavage of RNA 3′ to G residues combined with the 3′ end-labeling strategy displaces the G ladder upwards by 1 nt in the gel). The lowest spot on the gel is 4SUpCo, as confirmed by APM gel analysis. No discrepancies between DNA and RNA sequence were observed.
Another key element of the ribozyme was stem II, which maintained perfect Watson-Crick pairing for the five base pairs of the helix closest to the absolutely conserved U and C residues found at its terminus (boxed residues in Fig. 4A ▶). A single covariation was observed in the entire helix, and it was located 4 bp away from the absolutely conserved unpaired nucleotides located at the end of the helix. This high level of sequence conservation implied that helix II plays an important role in ribozyme function.
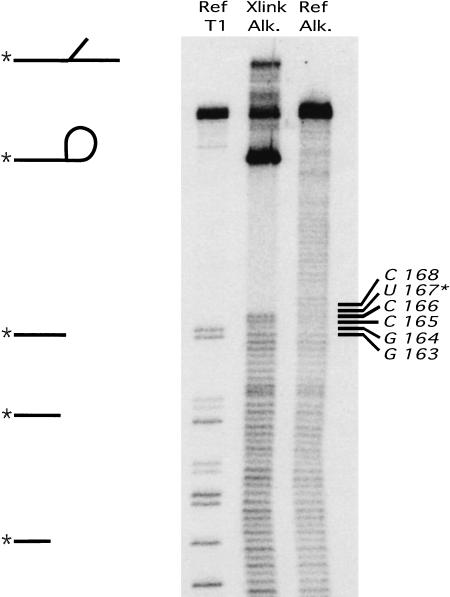
The importance of this region was reinforced by UV cross-linking experiments. Because 4-thiouridine is known to be a good UV cross-linking reagent (Favre et al. 1998) we performed cross-linking experiments on reacted ribozymes. The UV-induced cross-link was nearly 100% efficient and mapped to residue U167 in isolate a.6.10 (Fig. 6 ▶). This residue was immediately adjacent to the absolutely conserved cytidine (C166 of a.6.10) and across from the conserved uridine residue linking stem I and stem II (U34 of a.6.10). At the location of the cross-link, 21/30 isolates contained the sequence CUC (bold indicates absolutely conserved C166 of a.6.10). Of these, nine had a longer consensus sequence CUCUACCA found also in a.6.10 (underline indicates location of cross-link in a.6.10). Notably, three of the nine sequences differing from the CUC-type sequence had mutations at the location of the cross-link observed in a.6.10. Isolate a.6.03 (CCUCACAA) also cross-linked immediately downstream of the invariant C, whereas isolate a.6.13 (CACUGCCA) failed to cross-link. Considering that 4-thiouridine cross-links with adenosine in solution (Favre et al. 1998), the absence of a cross-link in this case might be explained by the orientation of the 4-thiouridine product relative to the adenine base of this ribozyme.
FIGURE 6.
Mapping of UV cross-link between 4-thiouridine product and residue U167 of the a.6.10 ribozyme. A partial hydrolysis ladder of 5′-radiolabeled cross-linked RNA (Xlink, Alk) was compared to that of 5′-radiolabeled linear ribozyme partially digested with RNase T1 (Ref, T1) or partially hydrolyzed (Ref, Alk.) The gap in the ladder indicates the location of the lariat branchpoint in the cross-linked RNA.
Combinatorial minimization of nonessential sequence
An interesting feature of family A is that it contains significant tracts of sequence whose only apparent purpose is to link helical elements together (Fig. 4A ▶). For example, the two strands linking stems II and III are poorly conserved and yet proved resistant to truncation by manual design and screening of constructs. Nearly a dozen deletion constructs (including variants eliminating stem VI) were constructed, and all were inactive. In contrast, the terminal loops of stems V and VI, also poorly conserved, could easily be replaced by UUCG tetraloops without noticeable loss of activity. The same replacement for the loop closing stem I greatly diminished activity, indicating the functional importance of sequence within this loop. In order to more thoroughly investigate the length and sequence requirements of the family A nucleotide synthase ribozyme, we first developed a method to synthesize a very diverse population of sequences with numerous deletions and substitutions in the extended joining regions. Subsequent analysis of active sequences selected from this population revealed sequence contexts tolerant of deletions.
The “partial deblock” strategy produces point deletions by using dichloracetic acid to partially remove protective DMT groups during the phosphoramidite coupling cycle (Treiber and Williamson 1995). However, dichloroacetic acid-catalyzed removal of DMT is strongly sequence-dependent. Our measurements with trichloroacetic acid (TCA) confirmed that protected deoxyguanosine (dG) detritylates approximately six times faster than protected deoxyadenosine, cytidine, or thymidine (dA, dC, or T, respectively). As a consequence, deletion libraries produced by partial deblocking will preferentially fail to delete bases 5′ to dG. Given this limitation, we explored an approach that uses DMT-Cl to partially reblock a completely deblocked oligonucleotide. We measured the reblocking rates of 5′ hydroxyls during an oligonucleotide synthesis and found again that the 5′ hydroxyl of dG was much more reactive than those of the other three nucleotides. As with partial deblocking, unbalanced levels of deletion were produced, but this time sequences 5′ to dG were overdeleted. Therefore we tried partial reblocking with DMT-Cl followed by partial deblocking with TCA. This approach generated more balanced levels of deletion (see supplemental data at http://www.sfu.ca/mbb/mbb/faculty/unrau/Chapple_RNA_Supp.pdf) and was still compatible with automated oligonucleotide synthesis techniques commonly used to generate point substitutions.
We synthesized pools requiring a total of 162 couplings that consisted of alternating regions of fixed and variable sequence (Fig. 2C ▶). The variable-sequence regions, requiring 76 couplings, had both point deletions and point substitutions. Two pools were constructed and then mixed together, one with a deletion frequency of 17% and a nucleotide substitution frequency of ~20% in the variable regions, and a second with a deletion frequency of 25% and a nucleotide substitution frequency of ~20%. Both pools were based on the sequence of isolate a.6.10, with the following changes. The loops closing stems V and VI were replaced with tetraloops, and helix V was shortened (eliminating segments 180–183 and 189–191). Two helical modifications were also made based on the consensus structure of the previous isolates. The first swapped the second-to-last base pair of stem IV from 203C:81G to 203G:81C. The second repaired the G-U wobble found in stem V. The loop closing stem I was made variable in sequence length and base composition to allow the selection of faster ribozyme variants optimized by deletion and mutation in the only terminal loop region of demonstrated importance in ribozyme function. Figure 2C ▶ shows the seven fixed-length and six variable-length regions of the deletion/substitution pool and specifies the maximum length possible for each variable-length section. The regions of fixed sequence include all of the proposed helical regions, including the poorly conserved stem VI (Fig. 4A ▶).
A total of eight rounds of selection were performed using APM gels to select short functional ribozymes from an equimolar mix of these two pools. After activity was observed (round 4), the faster and slower migrating 4-thiouridine-containing products were separately excised, reverse transcribed, and amplified. To apply selective pressure for shorter sequences without risking a loss in activity, the activity of RNA derived from these longer and shorter molecules was compared before carrying forward the shorter RNA to the next round of selection. We reasoned that ribozymes from round 7 were a reasonable compromise between length and activity, because in the next round the shorter sequences reacted more slowly than the longer sequences. The mean pool length decreased to 132 nt from the starting length of 163 nt (31 bases deleted on average or a net deletion frequency within the variable-length regions of 41%, n = 20, Fig. 7 ▶). This is to be compared with the mean starting deletion frequencies of 17% and 25% for the initial deletion pools (13 nt and 19 nt average deletion, respectively). The difference is presumably a consequence of the selection of rare sequences found at the shorter tail of the deletion distribution, perhaps combined with additional deletions occurring during amplification.
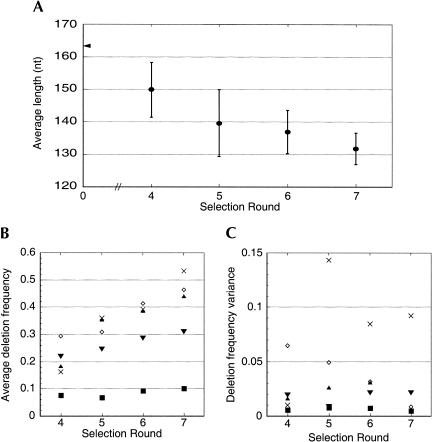
FIGURE 7.
(A) Length of pool molecules with successive rounds of selection from the variable-length pool. Error bars indicate the average and standard deviation of the lengths of cloned ribozymes (n = 17, 19, 20, 20 for rounds 4, 5, 6, and 7 respectively). The length of the RNA sequence template containing no deletions (163 nt) is indicated by the arrow on the vertical axis. (B) Deletion frequencies for each of the five variable regions of isolates shown in Fig. 8 ▶ as a function of selection round. (▾) R1; (▴) R2; (⋄) R3; (×) R4; (▪) R5. (C) Deletion frequency variance for each variable region during the deletion selection. R4 has a large variance in part due to the occasional elimination of stem VI during the selection. R3 variance decreases dramatically during the selection, appearing to converge on an optimal solution by round 7. The length of R5 appears to have been nearly optimal since the start of the selection. Symbols as in panel B.
An alignment of sequenced clones from rounds 4, 5, 6, and 7 showed the repeated elimination of nucleotides within regions not critical for catalysis (Fig. 8 ▶). The deletion frequency and variance for each of the regions R1–R5 was calculated as a function of selection round (Fig. 7B,C ▶), and show how each region accommodated deletions as the selection progressed. Some regions, such as R3 (linking stems III and IV) and R5 (linking stems II and V) favored particular sequence motifs and exhibited low final variance in deletion and mutation frequency. Other regions, such as R2 and R4, showed high deletion and mutation frequency variance that was consistent with the poor sequence conservation observed in these regions during the earlier selection. R1 was a blend of both extremes, apparently containing both conserved and variable sequence segments.
FIGURE 8.
Alignment of DNA sequences of clones from rounds 4, 5, 6, and 7 of the selection, relative to the undeleted parent sequence, 163.1. Seven constant sequence regions form stems I–VI and are color-coded to correspond with Fig. 4 ▶. The five variable regions (R1–R5) show higher frequencies of deletion each successive round (Fig. 7 ▶). R3 and R5 appear to converge to particular sequences, whereas R2 and R4 show little sequence preference. A conserved motif at the 3′ end of R1, as well as interactions between the 3′ end of R3 and the 5′ end of R5, are evident in the round 7 sequence data. All isolates from this selection experiment are named as da.x.y, where x is the round, and y is the isolate.
The single-stranded R5 sequence, which was inferred to be proximal to the active site of the ribozyme by cross-linking data, exhibited little sequence variation, even though seven bases were subjected to deletion and substitution in this region. The R5 consensus sequence observed among the ribozyme isolates of the earlier selection and manifested in sequence a.6.10, CUCUACCAGC... (bold indicating the absolutely conserved C166 in a.6.10, italics indicating the variable region of the pool used in this experiment) shifted to the sequence CUCUCCAGC, through the elimination of a single adenosine. The uniformly low deletion-frequency variance of R5 throughout the selection indicated the importance of the initial sequence in this region. In contrast, R3, which is part of the pseudoknot element, started with eight variable residues and was dominated by the 4 nt long sequence GGGA by round 7 (Figs. 7 ▶, 8 ▶).
Other regions, in particular R2 and R4, tolerated a wider range of sequence mutations and lengths. R4 (linking stems III and II and containing stem VI) was notable in that mutants which completely eliminate stem VI were found in rounds 5, 6, and 7. Our original pool had been designed to generate a maximum of 21 deletions in this region, and we observed in one case the removal of 27 nucleotides (isolate da.5.03). During the process of reverse transcription, the RT enzyme might occasionally hop over the 14 nucleotides involved in the helix-loop structure of stem VI, resulting in a larger than anticipated net deletion. It is therefore not surprising that this region has a large variance in deletion frequency (Fig. 7C ▶). Likewise, R2 tolerated the removal of large amounts of sequence (Fig. 7B ▶) but with a considerably smaller deletion frequency variance. In contrast to R3 and R5, no apparent sequence preference was observed in these regions.
R1, like R2 and R4, showed no tendency to converge on a particular sequence and yet, like R3 and R5, it contained a short region of preferred sequence: ...GUUGCA immediately 5′ of the AUCC 3′ arm of stem I. Since a.6.10 also contained this preferred sequence and was shown to be active when the entire loop closing stem I was replaced by a UUCG tetraloop (albeit with greatly diminished activity), this sequence cannot be essential for catalysis but possibly enhances the folding or chemistry of the ribozyme.
Family A noncanonical interactions
The cross-link between the 4-thiouridine product and U167 in isolate a.6.10 (Figs. 4A ▶, 6 ▶) implies that sequence found in regions R3 and R5 are in close proximity. To explore this possibility further, we examined the aligned sequence data from round 7 of the combinatorial minimization selection (Fig. 8 ▶) for evidence of noncanonical interactions. A65, the terminal 3′ nucleotide of R3, which lies opposite the pRpp substrate (Fig. 4B ▶), changed from an A to another nucleotide in seven out of 20 isolates. In all seven of these isolates, the change in A65 was accompanied by changes in the otherwise highly conserved CUCU at the 5′ terminus of R5 (segment 94–97 in Fig. 4B ▶; the absolutely conserved cytidine is underlined). In four of the seven isolates, the A:CUCU starting sequence changed to U:CUGU; in two it changed to C:CUGA. Conversely, in 11 of the 13 isolates in which A65 did not change, the CUCU segment also remained constant (in the other 2 of the 13, there were wholesale changes in both R3 and R5). Similar, though less striking, patterns were seen in the round 4, 5, and 6 sequence data, perhaps indicating that these covariations confer an advantage in response to selective pressure for shorter sequence elements.
Kinetic properties of deletion construct, da.7.02
We screened the shortest isolates from rounds 5, 6, and 7 for activity. We selected sequences that had deletions in different parts of the ribozyme sequence, as indicated in Table 1 ▶. The variants reported had first-order rate constants in the range 0.04–0.13 min−1 when assayed using 4SUra concentrations near 1 mM. Isolate da.7.02 (Figs. 4B ▶, 8 ▶) was the only sequence isolated twice, and because it appeared to be marginally faster than the other isolates, was chosen for further study.
TABLE 1.
Ribozyme activities and deletions for isolates from round 5, 6, and 7 of the selection from the deletion/substitution pool
| Number of deleted nucleotides | ||||||||
| Ribozyme | Activity (min−1) | Length (nt) | R1 | R2 | R3 | R4 | R5 | Total |
| Init. pool | n/a | 163 | (15) | (25) | (8) | (10 + 11) | (7) | (76) |
| da.5.03 | 0.04 | 119 | 3 | 9 | 4 | 27 | 1 | 44 |
| da.5.11 | 0.06 | 124 | 3 | 6 | 4 | 24 | 2 | 39 |
| da.6.05 | 0.07 | 120 | 4 | 9 | 4 | 26 | 0 | 43 |
| da.6.06 | 0.06 | 131 | 10 | 12 | 2 | 6 | 2 | 32 |
| da.6.13 | 0.05 | 137 | 2 | 22 | 0 | 2 | 0 | 26 |
| da.6.17 | 0.04 | 136 | 4 | 14 | 4 | 5 | 0 | 27 |
| da.7.02 | 0.08–0.13 | 124 | 4 | 7 | 3 | 25 | 0 | 39 |
| da.7.08 | 0.05–0.09 | 131 | 9 | 10 | 4 | 8 | 1 | 32 |
| da.7.13 | 0.07–0.10 | 123 | 2 | 12 | 4 | 21 | 1 | 40 |
| da.7.18 | 0.05–0.06 | 127 | 10 | 11 | 4 | 10 | 1 | 36 |
In parentheses are the numbers of deletion/substitution couplings used to create a given variable region (Fig. 2C ▶). Bold numbers highlight deletions significantly larger than expected by chance (assuming a deletion frequency of 0.25, P < 0.01).
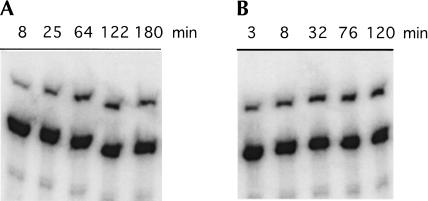
The kinetic behavior of this ribozyme was explored keeping the da.7.02 ribozyme concentration constant and varying the concentration of 4SUra from 0.1 mM to 12 mM. The resulting rate profile did not display standard Michaelis-Menten-like behavior but rather increased almost linearly from 0.1 mM to 4 mM 4SUra, reaching a maximum apparent first-order rate constant of 0.21 min−1 at 4 mM 4SUra (Fig. 9 ▶), before smoothly dropping to 0.08 min−1 at 12 mM 4SUra. A similar apparent inhibition by high concentrations of 4SUra was observed for isolates from the earlier selections as well. The apparent efficiency of the da.7.02 ribozyme measured at 0.1 mM 4SUra concentration was 150 M−1min−1. This was ~35-fold faster than the initial a15 isolate (4.3 M−1min−1) and slightly faster than isolate a.6.10 (120 M−1min−1) from the earlier selection. This gain in efficiency over the initial a15 isolate was accomplished while decreasing the ribozyme length from 272 to 124 nt.
FIGURE 9.
Ribozyme da.7.02 kinetics at two different 4sUra substrate concentrations. Timepoints were stopped in formamide loading dye and loaded on an APM polyacrylamide gel. (A) At 0.1 mM 4SUra, the apparent first-order rate constant was 0.015 min−1 (β = 0.10). (B) At 4 mM 4SUra, the apparent first-order rate constant had risen to 0.21 min−1 (β = 0.17). The fraction able to react, β, is dependent on ligation and transcription conditions together with the rate of pRpp hydrolysis.
Secondary structure confirmation and minimization
Of the five stems shown in Figure 4B ▶, only stem I contained a helical arm that was not part of the degenerate sequence region of the pool of Figure 2B ▶. This sequence was not mutated because it formed part of the 5′ PCR primer-binding site required for the RT-PCR amplification of the ribozyme. Thus stem I, while highly conserved, was not supported by covariational data.
Replacing residues 1–24 of da.7.02 with GG completely disrupted ribozyme function, demonstrating the general importance of sequence predicted to form stem I and its associated loop. Stem I was explored further by introducing the point mutations U27G and A3C. Individually these two mutations, predicted to destabilize the stem, led to 20-fold and 3.5-fold decreased activity, respectively. When combined, the predicted pairing and the ribozyme activity were restored, supporting the hypothesis that these nucleotides participate in the formation of stem I. Ribozyme activity was also sensitive to the sequence loop I. Replacing the loop found in da.7.02 with the sequence found in da.7.18 (6 nt shorter) lowered ribozyme activity by twofold. The parental sequence a.6.10 suffered a much larger activity drop when a UUCG tetraloop was inserted into stem–loop I.
The importance of stem length in helix V was explored and found not to significantly affect ribozyme activity. Isolate da.7.02 was found to have the same activity when the length of stem V was truncated from 6 to 4 bp. Together, truncating stem V and using the shorter stem–loop I from da.7.18 allowed us to construct a ribozyme variant 114 nt long that was only twofold slower than da.7.02.
DISCUSSION
UV cross-linking and phylogenetic data both indicate that the active site of the ribozyme is nestled in a structure formed from three conserved sequence elements that are in turn surrounded by the core helices I, II, and IV. The first of these sequence elements is the absolutely conserved uridine linking helices I and II. Proximal to this nucleotide is the absolutely conserved cytidine that emerges from the opposite strand of helix II starting the second sequence element. This element appears to interact in a noncanonical fashion with the nucleotide emerging from helix IV in R3. This residue is opposite the pRpp substrate, which itself is tethered to the 3′ end of helix IV. The interaction between R3 and R5 sequence would naturally place the pRpp substrate close to the absolutely conserved uridine and cytidine nucleotides. The roles of helices II and IV and loop I have not been established in this context, but they likely help orient the sequence found in regions R3 and R5. Further evidence for the proposed geometry is provided by the high-yield cross-link that forms between the 4-thiouridine ribozyme product and the nucleotide adjacent to the absolutely conserved cytidine in R5.
Like the Class I ligase ribozyme (Ekland and Bartel 1995), the family A ribozyme contains loop regions that were easily truncated and replaced with tetraloops. Unlike the Class I ligase, the family A nucleotide synthase has significant stretches of sequence that link the five helices of the ribozyme together. The size of the core motif can be estimated by counting the nucleotides either involved in base-pairing or in well conserved sequence regions. The ribozyme has 27 well conserved base pairs, of which at least two can be removed by truncating helix V. This implies that ~50 nucleotides are required to form helical elements. Important nucleotides found in conserved sequence regions can be estimated by examining the consensus sequence information. Two nucleotides were absolutely conserved in every selection performed (boxed residues in Fig. 4 ▶). Selection from the variable-length pool indicated that the minimum sequence length in R3 was 4 nt. Seven nucleotides were required in R5, of which four nucleotides were well conserved (in addition to the absolutely conserved cytidine residue). Another four nucleotides are required to form stem loop V, and it is likely that at least six nucleotides are important within stem–loop I. If the single-nucleotide spacer between stems III and IV is added to this count, a total of ~71 nucleotides are required to form the core motif of the ribozyme. The remaining sequence found in R2 and R4 does not appear to be essential and might be required only to connect the elements of the core motif together.
Of the nucleotides required to build the core motif, the equivalent of ~40 nucleotides are required to specify the information encoded by both the conserved sequence elements and helical structures. The probability of finding a ribozyme motif with this informational complexity can be roughly estimated assuming that the ribozyme can be built from modular elements able to tolerate the insertion of variable intervening sequence. Probable insertion sites can be found in R1, R2, R4, and the loop closing stem V, implying that the ribozyme has as many as five modular subcomponents. Large modularities can dramatically enhance the probability of isolating complicated motifs in long sequence length pools (Sabeti et al. 1997). Indeed, the likelihood of isolating the 4-thiouridine synthase core motif from a pool containing 228 random positions grows from about 1 in 1022 for a modularity of one to about 1 in 1015 if a modularity of five is assumed.
The large size and complexity of the core motif of the family A 4-thiouridine nucleotide synthase imply that pyrimidine nucleotide synthesis may not be easily catalyzed by RNA. Not only must the ribozyme recognize a small pyrimidine substrate, but the synthesis of pyrimidine nucleotides is known to be thermodynamically less favorable (by 25–30 kJ/mole) than purine nucleotide synthesis (Dewolf et al. 1986; Bhatia et al. 1990). In addition, the very slow hydrolysis rate of pyrimidine nucleotides relative to purine nucleotides means that the uncatalyzed rate of pyrimidine nucleotide synthesis might be dramatically slower than for purine nucleotide synthesis. Consistent with this, prebiotic experiments have failed to demonstrate pyrimidine nucleotide synthesis (Orgel 1998).
Kinetic isotope experiments indicate that the mechanism of the nucleotide synthase ribozyme involves an oxocarbenium intermediate (P. Unrau and D. Bartel, in prep.). This is an unusual reaction mechanism, even for protein-catalyzed reactions, and so exploring the transition state of the family A nucleotide synthase is of considerable interest. The proposed secondary structure of the family A nucleotide synthase might allow the engineering of a ribozyme able to perform multiple turnover reactions using a short RNA-pRpp substrate. This ribozyme construct could be used to measure the binding affinities of transition-state analogs commonly used to inhibit protein-catalyzed nucleotide synthesis (Shramm 1998; Li et al. 1999), thus providing a deeper understanding of the ribozyme’s catalytic mechanism.
The selection from oligonucleotides that varied in both length and sequence proved to be useful for the efficient characterization of the nucleotide synthase core motif. Although on statistical grounds no one isolate would be expected to be maximally deleted, a significant number of ribozymes should contain maximal deletions over short intervals of sequence. For example, at a deletion frequency of 0.2 per nucleotide, a particular 10 nt segment would be completely eliminated by successive point deletions in about one in 107 pool molecules [P = (0.2)10]. Additional deletions are probably generated during the reverse transcription and PCR steps of each round. An inspection of Table 1 ▶ reveals that in addition to R3 and R5, large randomly localized deletions are found scattered throughout R1, R2, and R4. Because only two out of 20 round 7 isolates sequenced were identical, there was still a high diversity of deletion mutants in the pool at round 7. Systematically recombining these sequences using homologous recombination might allow further minimization of the ribozyme sequences, as well as further optimization of ribozyme function and exploration of the evolutionary potential of RNA as a catalyst.
MATERIALS AND METHODS
Degenerate pool synthesis and selection protocol
Two oligonucleotides based on the parent a15 sequence (Unrau and Bartel 1998) were made using 1000 Å 1 μmole columns: a1-5′-ggataatgcgcacaagtNNNNNNNNNNAATCCTGCCTATTTAAACACTAAATCTGTCGCAATGGAAACGCGGTGGCTAGGTTGCGCGTTACTACCATATTggcacctgtccacgctc; a2:-5′-ctatcccgactggcaccAGATACGCAGTAGAGTAATCGGGAAGTACTGGATGCCATAGTGTAGACAATAGGCCTCTACCAGCGGTGTGTTGTAACGCACATCGCAGCAACtgaagagcctacgac (lowercase, fixed sequence; N, random coupling; uppercase, 20% phosphoramidite mutagenesis). PCR reactions (30 mL, 10 mM Tris pH 8.3, 50 mM KCl, 4 mM MgCl2, 0.2 mM dNTPs, 0.01% gelatin, Taq enzyme, and 1 μM primers) were performed using the following primers: a1-5′-ttctaatacgactcactataggataatgcgcacaagt and 5′-gagcgtggacag gtgcc; a2-5′-ctatcccgactggcacc and 5′-tcgtcgtaggctcttca. Strands a1 and a2 were digested with BanI and ligated together using T4 DNA ligase, following previously published methods (Unrau and Bartel 1998).
In each round, the DNA population was digested with EarI in order to generate DNA with the correct 3′ end. DNA was transcribed into RNA (with or without [32P]-α-UTP, 40 mM Tris pH 8.6, 26 mM MgCl2, 5 mM DTT, 2.5 mM Spermidine, 0.01% TritonX-100, 8 mM GTP, 4 mM ATP, 4 mM CTP, 2 mM UTP, and 5 U/μL T7 RNA polymerase, incubated for 1 h at 37°C), and the RNA was gel-purified on 5% polyacrylamide gels. RNA was precipitated and then ligated at the 3′ end to pRpp using adenylated pRpp (AppRpp) and T4 RNA ligase (Amersham Pharmacia). Ligation conditions were 50 mM HEPES pH 8.3, 10 mM MgCl2, 3.3 mM dithiothreitol (DTT), 10 μg/mL BSA, 8.3% v/v glycerol, 65 μM AppRpp, 1 U/μL enzyme, for 4 h at 23°C. RNA ligated to pRpp was prepared for incubation with 4SUra by heating at 80°C for 1 min, and cooling to 23°C. RNA was then supplemented with a reaction buffer (~1 μM RNA, 50 mM Tris-HCl pH 7.6, 150 mM KCl, 25 mM MgCl2) and incubated at 23°C in the presence of 4SUra. Incubation times and 4SUra concentrations for each round (R) were: R1, 1200 min, 4 mM, 1 gel; R2, 120 min, 1mM, 1 gel; R3, 60 min, 1mM, 2 gels; R4, 6 min, 0.1 mM, 1 gel; R5, 6 min, 0.1 mM, 2 gels; R6, 6 min, 0.1 mM, 2 gels (gel number refers to the number of 5% polyacrylamide 80 μM APM purification gels used each round). A marker RNA the same length as the pool and terminated with 4-thiouridine was used to identify the location of catalytic RNAs within the selection gels (Unrau and Bartel 1998). RNA with the mobility of the 4-thiouridine marker was excised and eluted from the gel and ethanol-precipitated. Isolated ribozymes were then ligated to an oligo called App16.5 that created an RT-PCR primer binding site containing an EarI DNA restriction site (Fig. 3B ▶). Ligation conditions were the same as for AppRpp ligation, except that 50 μM App16.5 and a 2-h incubation were used. RT-PCR was performed as described (Unrau and Bartel 1998). The next round of selection was initiated by digesting the pool with EarI and transcribing. App16.5 was synthesized by first kinasing and then incubating the oligo 5′-gaagagcctacgacga (underline EarI site) with imidazole-activated AMP in the presence of magnesium (Lohrmann and Orgel 1978; Unrau and Bartel 1998). The construct was purified on a 20% polyacrylamide gel before use.
Deletion/substitution pool and selection protocol
Two pools were synthesized having different deletion frequencies and using sequence a.6.10 (Fig. 4A ▶) as a template: 5′-ggataatgcgcaca AGTAAGCGTGTTGCAatcctgcctattGAAACACGAAATCAGTCGTA ATGGAaacgaGGGGGATAgcttg c t c g t t A C T C C C A T T T t g g c c t t c g g g c c a TAGTGGCACAAaataggccTCTACCAgcggtgttcgcaccgcagcaagc (lowercase nucleotides, fixed sequence; uppercase, deletion/substitution protocol, 20% mutagenesis using either 17% or 25% deletion frequency). After deprotection (in saturated aqueous ammonia at 55° C for 18 h), each synthesis was gel-purified on a 6% polyacrylamide gel, excising DNA of about 90–180 nt. DNA was PCR-amplified (36 mL, 1 μM of primers 5′-ttctaatacgactcactataggataat gcgcac and 5′-gcttgctgcggtgcga, 75 nM gel-purified pool) for eight cycles of PCR as described for the previous selection. PCR-amplified DNA from each pool was resolved on a 4% agarose gel. DNA that migrated between the 125 and 200 bp markers was excised from the gel, cloned, and sequenced. Sequences were aligned using the constant sequence elements found in each pool, and deletion frequency was calculated. Equimolar amounts of the 17% and 25% deletion frequency pools were mixed, generating a starting pool containing ~5 × 1013 different sequences.
RNA was transcribed and purified on a denaturing 6% polyacrylamide gel and ligated to AppRpp. RNA was reacted with 4SUra using conditions identical to those described in the previous section, except that selection times and 4SUra concentrations were as follows: R1, 120 min, 1 mM; R2, 12 min, 0.2 mM; R3, 0.5 min, 0.2 mM; R4, 0.1 min, 0.1 mM; R5, 0.1 min, 0.1 mM; R6, 2 min, 1 mM; R7, 0.25 min, 1mM; R8, 1 min, 1mM. Incubation was terminated by the addition of 1 volume of denaturing gel loading buffer (50 mM EDTA, 90% formamide, and 10 μM tRNA). The incubated sample was purified on a denaturing 6% polyacrylamide gel containing ~40 μM APM. Times and 4SUra concentrations were increased after round 5 in order to allow 1%–5% of the pool population to react, making it possible to locate and excise faster migrating material for the next round of selection. A marker sequence 86 nt long ligated to 4-thiouridine (using App4SU) was used to delimit the shortest likely ribozymes. In later rounds (R6, R7, and R8), reacted RNA was divided into upper and lower halves (the lower half consisting of material above the marker sequence and below the point in the gel containing half the reacted RNA). Reactive RNA was eluted from the gel in the presence of DTT, precipitated, and converted to DNA using the 16 nt RT primer (5′-gcttgctgcggtgcga) and reverse transcriptase (Superscript II, Invitrogen). After heating to 90° for 10 minutes in 100 mM KOH followed by neutralization with Tris-HCl, cDNA was PCR-amplified (primers listed above). The reaction rates of the RNA transcribed from each RT-PCR was compared, and the shorter RNA was carried forward for the next round of selection.
Deletion/substitution coupling protocol and analysis
Oligonucleotides were made on 2000 Å 1 μmole columns (Glen Research) using an 8909 Expedite DNA synthesizer (Applied Biosystems). For the deletion/substitution couplings, the column was deblocked, using the standard protocol (10 rapid pulses of 3% TCA in dichloromethane followed by 50 pulses of TCA over 49 sec, followed by 40 rapid pulses of acetonitrile). Next, 100 mM DMT-Cl (Fluka) in 95% dichloromethane and 5% pyridine was delivered to the deblocked column to achieve a partial reblock (10 rapid pulses from the spare reagent port, followed by 16 pulses spaced over a coupling time of either 2 or 4 min). After washing the column with acetonitrile (40 rapid pulses), 3% TCA was delivered to achieve a partial deblock (7 rapid pulses). The column was washed (40 rapid pulses of acetonitrile), and standard coupling, capping, and oxidation steps followed. Each reagent pulse delivered 17.6 μL to the column, and consecutive rapid pulses were separated by 0.36 sec on our DNA synthesizer.
The 2-min reblocking produced sequence which, when cloned and sequenced, had on average 12.8 nt deletions (n = 14, s.d. = 4.4) within the 76 nucleotides subject to the deletion/substitution protocol for an overall deletion frequency of 17%. The 4-min reblocking yielded an average of 19.4 nt deleted (n = 14, s.d. = 6.7) resulting in a deletion frequency of 25%. These averages agreed well with the observed agarose gel mobilities of the deletion pools (previous section). Alignment of the DNA cloned from each of two initial pools showed that the deleted nucleotides were in areas synthesized using the deletion/substitution protocol (see supplemental material at http://www.sfu.ca/mbb/mbb/faculty/unrau/Chapple_RNA_Supp.pdf). The deleted residues also appear to be evenly dispersed throughout the deletion/substitution variable regions, with the consensus sequence matching almost identically that of the parent sequence. Deletion frequencies were calculated from the aligned sequence data and were well balanced, showing less than a twofold bias with respect to the identity of the 3′-proximal nucleotide.
When developing this procedure, two assays were used to monitor deblocking and the deletion rates. To monitor the extent of deblocking, the DMT was collected in deblock solution and the optical density was measured at 504 nm. Deletion rates were measured by synthesizing the four pentanucleotide homopolymeric DNA sequences, with the third coupling cycle using the deletion protocol under investigation. Deprotected oligonucleotides were kinased with an excess of [32P]-γ-ATP using polynucleotide kinase. Products were resolved on a 20% sequencing gel and quantified by phosphorimaging.
Phosphoramidite mixtures
Each mutagenic coupling used phosphoramidite mixtures designed to generate a 20% mutation rate. Individual 0.5-g batches of phosphoramidites (Perkin Elmer) were diluted to concentrations resulting in equal coupling activity (0.26:0.25:0.29:0.20 dA:dC:dG:T molar ratios phosphoramidite). Equal volumes of these solutions were then combined to give an “N mix”. Twenty-percent doping was achieved by mixing 3.6 mL of this N mix with 10.0 mL of each equi-active phosphoramidite stock.
Ribozyme assays
Unless otherwise stated, ribozyme pools and isolates were assayed with 1 mM 4SUra and the reaction conditions used during the selections. Typically, five timepoints were taken to determine rates. Aliquots were stopped with a sevenfold excess of denaturing loading dye containing 10 μM tRNA. Samples were loaded onto analytical 6% denaturing polyacrylamide APM gels and quantified by phosphorimager analysis. Rates were determined by fitting k and β to the equation F = β(1−e−kt) where F is fraction reacted, k is the apparent first-order reaction rate, and β is the fraction able to react. Typical values of β ranged from 0.1 to 0.3, and appear to be most influenced by the efficiency of pRpp ligation and 3′ heterogeneity of RNA transcripts.
UV cross-linking
Ribozyme transcribed in the presence of γ-[32P]-GTP was ligated to pRpp using AppRpp and then reacted to completion with 4SUra. Unreacted 4SUra was removed by dialysis (Centricon) with ribozyme reaction buffer. RNA was placed on ice and exposed to a handheld 302-nm UV light (UVP, LMW-20, 10 cm from sample). A 5% denaturing APM gel revealed the nearly quantitative conversion of the ribozyme-4SU to a faster migrating species (~97% in 40 min of UV exposure). This faster migration can be explained by the loss of the thio-keto group and the formation of a lariat structure. The cross-linked species was excised from the gel, phosphatased, and end-labeled using T4 polynucleotide kinase and γ-[33P]-ATP, to generate 5′-labeled material with greater specific activity. The RNA was then either partially digested with RNase T1 or lightly hydrolyzed with sodium bicarbonate, and products were resolved on a 6% sequencing gel.
Characterization of ribozyme 3′ terminus
The reaction products were end-labeled with [32P]-α-cordycepin triphosphate (CoTP) and poly(A) polymerase (Unrau and Bartel 1998). Active ribozymes sequences possessing both a 4-thiouridine and CoTP label were purified using an APM polyacrylamide gel. The purified RNAs were then partially digested with RNase T1 or lightly hydrolyzed. A 20% denaturing polyacrylamide gel confirmed that active RNA sequences contained G residues at exactly the locations predicted by the DNA sequence data.
Acknowledgments
We are grateful for the careful reading of this manuscript by members of the Unrau laboratory, M.S. Lawrence and M.L. Unrau. This work was supported in part by funds from the Michael Smith Foundation for Health Research (P.J.U.), National Sciences and Engineering Research Council of Canada (P.J.U.), and National Aeronautics and Space Administration (D.P.B.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5500603.
REFERENCES
- Bartel, D.P. and Unrau, P.J. 1999. Constructing an RNA world. Trends Biochem. Sci., Trends Genet., Trends Cell Biol. 24,15,9: M9–M13. [PubMed] [Google Scholar]
- Bartel, D.P., Zapp, M.L., Green, M.R., and Szostak, J.W. 1991. Hiv-1 Rev regulation involves recognition of non-Watson-Crick base-pairs in viral-RNA. Cell 67: 529–536. [DOI] [PubMed] [Google Scholar]
- Bhatia, M.B., Vinitsky, A., and Grubmeyer, C. 1990. Kinetic mechanism of orotate phosphoribosyltransferase from Salmonella typhimurium. Biochemistry 29: 10480–10487. [DOI] [PubMed] [Google Scholar]
- Chen, X.Y., Berti, P.J., and Schramm, V.L. 2000. Ricin A-chain: Kinetic isotope effects and transition state structure with stem–loop RNA. J. Am. Chem. Soc. 122: 1609–1617. [Google Scholar]
- Dewolf, W.E., Emig, F.A., and Schramm, V.L. 1986. AMP nucleosidase—Kinetic mechanism and thermodynamics. Biochemistry 25: 4132–4140. [DOI] [PubMed] [Google Scholar]
- Ekland, E.H. and Bartel, D.P. 1995. The secondary structure and sequence optimization of an RNA ligase ribozyme. Nucleic Acids Res. 23: 3231–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington, A.D. and Szostak, J.W. 1990. In-vitro selection of RNA molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Favre, A., Saintome, C., Fourrey, J., Clivio, P., and Laugaa, P. 1998. Thionucleobases as intrinsic photoaffinity probes of nucleic acid strucuture and nucleic acid-protein interactions. Photochem. Photobiol. B, Biol. 42: 109–124. [DOI] [PubMed] [Google Scholar]
- Giver, L., Bartel, D., Zapp, M., Pawul, A., Green, M., and Ellington, A.D. 1993. Selective optimization of the Rev-binding element of Hiv-1. Nucleic Acids Res. 21: 5509–5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi, G.L. 1988. Interaction of transfer-RNAs and of phosphorothioate-substituted nucleic-acids with an organomercurial—Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 27: 3842–3849. [DOI] [PubMed] [Google Scholar]
- Li, C.M., Tyler, P.C., Furneaux, R.H., Kicska, G., Xu, Y.M., Grubmeyer, C., Girvin, M.E., and Schramm, V.L. 1999. Transition-state analogs as inhibitors of human and malarial hypoxanthine-guanine phosphoribosyltransferases. Nat. Struct. Biol. 6: 582–587. [DOI] [PubMed] [Google Scholar]
- Lohrmann, R. and Orgel, L.E. 1978. Preferential formation of (2′–5′)-linked internucleotide bonds in nonenzymatic reactions. Tetrahedron 34: 853–855. [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., and Uhlenbeck, O.C. 1987. Oligoribonucleotide synthesis using T7 RNA-polymerase and synthetic DNA templates. Nucleic Acids Res. 15: 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel, L.E. 1998. The origin of life—A review of facts and speculations. Trends Biochem. Sci. 23: 491–495. [DOI] [PubMed] [Google Scholar]
- Sabeti, P.C., Unrau, P.J., and Bartel, D.P. 1997. Accessing rare activities from random RNA sequences: The importance of the length of molecules in the starting pool. Chem. Biol. 4: 767–774. [DOI] [PubMed] [Google Scholar]
- Schramm, V.L. 1998. Enzymatic transition states and transition state analog design. Annu. Rev. Biochem. 67: 693–720. [DOI] [PubMed] [Google Scholar]
- Strobel, S.A. 1998. Ribozyme chemogenetics. Biopolymers 48: 65–81. [DOI] [PubMed] [Google Scholar]
- Tao, W., Grubmeyer, C., and Blanchard, J.S. 1996. Transition state structure of Salmonella typhimurium orotate phosphoribosyltransferase. Biochemistry 35: 14–21. [DOI] [PubMed] [Google Scholar]
- Treiber, D.K. and Williamson, J.R. 1995. A simple method for preparing pools of synthetic oligonucleotides with random point deletions. Nucleic Acids Res. 23: 3603–3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unrau, P.J. and Bartel, D.P. 1998. RNA-catalysed nucleotide synthesis. Nature 395: 260–263. [DOI] [PubMed] [Google Scholar]