Abstract
The synthesis of the Gag-Pol polyprotein, the precursor of the enzymes of the human immunodeficiency virus type 1 (HIV-1), requires a programmed −1 ribosomal frameshift. This frameshift has been investigated so far only for subtype B of HIV-1 group M. In this subtype, the frameshift stimulatory signal was found to be a two-stem helix, in which a three-purine bulge interrupts the two stems. In this study, using a luciferase reporter system, we compare, for the first time, the frameshift efficiency of all the subtypes of group M. Mutants of subtype B, including a natural variant were also investigated. Our results with mutants of subtype B confirm that the bulge and the lower stem of the frameshift stimulatory signal contribute to the frameshift in addition to the upper stem–loop considered previously as the sole participant. Our results also show that the frameshift stimulatory signal of all of the other subtypes of group M can be folded into the same structure as in subtype B, despite sequence variations. Moreover, the frameshift efficiency of these subtypes, when assessed in cultured cells, falls within a narrow window (the maximal deviation from the mean value calculated from the experimental values of all the subtypes being ~35%), although the predicted thermodynamic stability of the frameshift stimulatory signal differs between the subtypes (from −17.2 kcal/mole to −26.2 kcal/mole). The fact that the frameshift efficiencies fall within a narrow range for all of the subtypes of HIV-1 group M stresses the potential of the frameshift event as an antiviral target.
Keywords: Ribosomal frameshift, HIV-1, group M
INTRODUCTION
The nucleotide sequence of the human immunodeficiency virus type 1 (HIV-1) is highly variable, due in part to the lack of proofreading activity of the reverse transcriptase that copies the viral RNA into DNA (Roberts et al. 1988). Analysis of this genetic variation resulted in a phylogenetic tree with three main branches for HIV-1, M (main), O (outlier), and N (non-M, non-O or new). Group M accounts for >99% of the viral isolates responsible for the worldwide pandemic and is further subdivided into nine subtypes (A, B, C, D, F, G, H, J, and K), on the basis of their env, gag, or pol gene sequences. Subtypes A and C (in Africa) are prevalent worldwide, followed by subtype B (in North America and Western Europe), AE recombinants (in Southeast Asia), and AG recombinants (in Western Africa; McCutchan 2000; Robertson et al. 2000; Moore et al. 2001).
Like all retroviruses, HIV-1 contains three major genes that are expressed as precursors of structural proteins (Gag), enzymatic proteins (Pol), and envelope proteins (Env). The gag and pol products are translated from the full-length viral RNA, but pol lacks its own initiation codon and is in a −1 reading frame relative to gag. Thus, HIV-1, like many other retroviruses, uses a programmed −1 ribosomal frameshift to produce the precursor of its enzymes, the Gag-Pol polyprotein, whereas conventional translation of the same RNA produces Gag. The programmed recoding event allows a minority of ribosomes to shift the reading frame at a specific sequence and translate the pol coding region (Jacks et al. 1988). A programmed −1 ribosomal frameshift requires two cis-acting elements in the RNA, a heptameric slippery sequence, in which the shift occurs, with an X XXY YYZ consensus (the zero frame is indicated by spaces), which is U UUU UUA in all HIV-1 groups, and a downstream stimulator that strongly increases the frameshift efficiency. The slippery sequence is such that when ribosomes bearing the peptidyl-tRNA and the aminoacyl-tRNA shift the reading frame by one base in the 5′ direction, the anticodons of both tRNAs can re-pair with the codons of the mRNA after the shift (Jacks and Varmus 1985). The downstream stimulatory signal, which is a pseudoknot in most cases, can also be a stem–loop (Brierley 1995; Farabaugh 2000; Brierley and Pennel 2001, and references therein). In HIV-1, this signal was assumed previously to be a simple 11-bp stem–loop (Jacks et al. 1988; Le et al. 1991; Chamorro et al. 1992; Kang 1998), and it was suggested that the frameshifting efficiency was directly related to the stability of this stem–loop (Bidou et al. 1997). However, it was found recently by Dinman et al. (2002) and by our group (Dulude et al. 2002) that the frameshift region of HIV-1 is longer than commonly assumed. Using structure probing and mutagenesis studies, we showed that the frameshift stimulatory signal of HIV-1 is a larger helix containing two stems separated by a three-purine bulge. The upper stem–loop corresponds to the classic stem–loop, whereas the lower stem is formed by pairing the purine-rich spacer between the slippery site and the classic stem–loop to a pyrimidine-rich sequence downstream of the classic stem–loop (Dulude et al. 2002). However, Dinman et al. (2002) proposed an alternative structure for the frameshift stimulatory signal, which consists in a pseudoknot, in which the pyrimidine-rich downstream segment forms a 3-bp stem with bases in the loop of the classic stem–loop. This short stem is stabilized by the formation of a triple helix between the classic stem and a portion of the pyrimidine-rich segment, but as discussed in Dulude et al. (2002), such triplex structure is sterically unfavorable.
The Gag-Pol to Gag ratio in HIV-1 is critical for the RNA dimerization, particle assembly, replication, and viral infectivity (Park and Morrow 1991; Karacostas et al. 1993; Shehu-Xhilaga et al. 2001), which suggests that altering the frameshift efficiency could affect viral replication. So far, the frameshift efficiency has only been studied for subtype B of HIV-1 group M. However, the development of an antiviral strategy directed against frameshifting requires the characterization of the frameshift efficiency of the other subtypes. All of the other subtypes of group M (A, C, D, F, G, H, J, K) possess the same slippery sequence as subtype B (U UUU UUA), but exhibit variations in the sequence of the stimulatory region (Los Alamos National Laboratory 2002; http://hiv-web.lanl.gov). In this study, we assessed the frameshift efficiency of all subtypes of HIV-1 group M. The sequence variations in the frameshift stimulatory signal of these subtypes compared with subtype B affect the predicted stability of the stimulator (see below), and consequently, could alter the frameshift efficiency. Using a luciferase reporter system, the frameshift efficiency of the different subtypes was assessed in vitro and in cultured cells. Whereas the in vitro system is not sensitive enough to analyze the efficiency of the frameshift stimulatory signal of HIV-1 group M, we found that the frameshift efficiency in cultured cells did not significantly differ between the different subtypes, despite the predicted variations in the stability of the stimulator.
RESULTS
Frameshift efficiency of different mutants of subtype B of HIV-1 group M
We first examined the frameshift efficiency in 293T cultured cells and in vitro in a rabbit reticulocyte lysate (RRL) of a construct containing the complete frameshift region of subtype B of HIV-1 group M, the so-called 3′ extended frameshift region (Fig. 1A ▶). The frameshift efficiency of subtype B was 1.9% in 293T cells and 7.0% in vitro in a RRL (Table 1 ▶). These values are comparable with those obtained previously for the frameshift efficiency of subtype B, using an appropriate reporter gene, which were found to vary from 2.1% to 5.5% in cultured cells and 5.6% to 11.6% in vitro (Reil and Hauser 1990; Cassan et al. 1994; Kim et al. 2001; Dulude et al. 2002). It can be noted that the values that we obtained are inferior to those obtained by Dulude et al. (2002) with a similar construct. This difference can be ascribed to a difference in the sequence upstream the slippery site, as it is known that this sequence affects the frameshift efficiency by influencing the rate with which the ribosomes encounter the slippery site (Kim et al. 2001). The frameshift efficiency of a construct containing only the slippery sequence was 0.2% and 2.8% in 293T cells and in vitro, respectively, showing that the corresponding stimulation of the frameshift efficiency with the 3′ extended region was about 10-fold and 3-fold (data not shown).
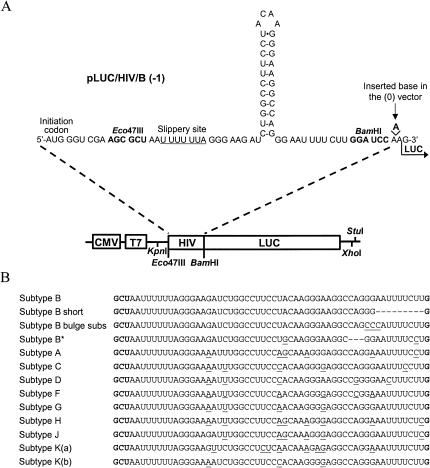
FIGURE 1.
Vectors expressing luciferase used to study the programmed −1 ribosomal frameshift of HIV-1 group M in vitro and in cultured cells. (A) The frameshift region of subtype B was inserted upstream of the coding sequence of the luciferase reporter gene, which is under control of a CMV and a T7 promoter, generating pLUC/HIV/B (−1). The classic structure of the frameshift stimulatory signal is represented here. For the study of all subtypes of group M and mutants of subtype B, the corresponding vectors were constructed by exchanging the Eco47III–BamHI fragment of pLUC/HIV/B (-1) with an appropriate oligonucleotide cassette. For the (-1) constructs, the luciferase sequence is in the −1 reading frame relative to the AUG initiation codon, so that a −1 frameshift is required to produce luciferase. An adenine nucleotide was added between the frameshift region and luciferase sequence for the (0) constructs, so that luciferase is expressed by ribosomes that do not shift the reading frame. (B) Sequences of the frameshift region of all of the constructs used in this study. Nucleotides modified or deleted relative to subtype B are underlined or represented by broken lines, respectively.
TABLE 1.
Frameshift efficiency of different mutants of subtype B and of all subtypes of HIV-1 group M
| Frameshift efficiency (%) | |||
| Construct | Description | In vitro | In cultured cells |
| Frameshift efficiency of mutants of subtype B | |||
| pLUC/HIV/B | Frameshift region of subtype B | 7.0 | 1.9 |
| pLUC/HIV/B short | Classic HIV-1 frameshift region | 7.3 | 0.9 |
| pLUC/HIV/B bulge subs | Substitution in the bulge | 8.5 | 1.4 |
| pLUC/HIV/B* | Natural variant of subtype B | 5.4 | 0.8 |
| Frameshift efficiency of subtypes A, B, C, D, F, G, H, J, K(a), and K(b) | |||
| pLUC/HIV/A | Frameshift region of subtype A | 5.9 | 1.8 |
| pLUC/HIV/B | Frameshift region of subtype B | 7.0 | 1.9 |
| pLUC/HIV/C | Frameshift region of subtype C | 5.3 | 2.0 |
| pLUC/HIV/D | Frameshift region of subtype D | 6.4 | 2.0 |
| pLUC/HIV/F | Frameshift region of subtype F | 4.6 | 1.8 |
| pLUC/HIV/G | Frameshift region of subtype G | 5.4 | 1.6 |
| pLUC/HIV/H | Frameshift region of subtype H | 5.6 | 2.4 |
| pLUC/HIV/J | Frameshift region of subtype J | 5.9 | 1.8 |
| pLUC/HIV/K(a) | Frameshift region of subtype K(a) | 6.4 | 1.2 |
| pLUC/HIV/K(b) | Frameshift region of subtype K(b) | 8.4 | 2.0 |
The pLUC/HIV constructs contain the HIV-1 gag/pol frameshift region. The frameshift region of each subtype is identified by the corresponding letter [A, B, C, D, F, G, H, J, K(a), and K(b)] and corresponds to the 3′ extended HIV-1 frameshift region. Mutants of the subtype B frameshift region are further identified by either a short description (B short, B bulge subs) or by an asterisk for the natural variant (see details in Fig. 1B ▶). Results are the means of five independent experiments. The standard error on the mean was inferior or equal to 10%.
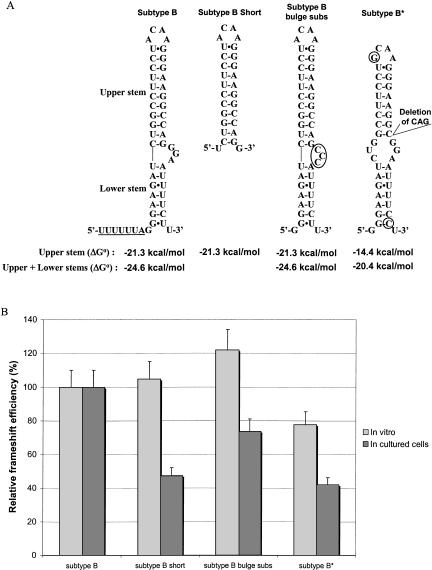
Prior to the analysis of the frameshift efficiency in the different subtypes of HIV-1 group M, we examined two mutants of subtype B as follows: subtype B short, in which the frameshift stimulatory signal contains only the classic stem–loop, and subtype B bulge subs, in which the purine bulge is substituted with pyrimidines (Fig. 1B ▶). The structure of the stimulator of each mutant and its thermodynamic stability, calculated using mfold (Mathews et al. 1999; http://bioinfo.math.rpi.edu/~mfold/rna/form1.cgi), are presented in Figure 2A ▶. This stability was calculated either for the extended bulged stem–loop structure (upper + lower stems) found by Dulude et al. (2002) or for the classic structure of the stimulator (only the upper stem–loop). In cultured cells, the frameshift efficiency compared with subtype B decreased by ~60% and 40% for subtype B short and subtype B bulge subs, respectively (Fig. 2B ▶). Dulude et al. (2002) had also investigated the effect on the frameshift efficiency of mutations preventing the formation of the lower stem and substituting the purine bulge with pyrimidines. Although the experimental values we obtained for the frameshift efficiency of subtype B short and subtype B bulge subs are inferior to the values they obtained for the corresponding mutations, the decrease relative to the subtype B is the same, confirming that the lower stem and the bulge contribute to the frameshift efficiency. No decrease in the frameshift efficiency was seen in vitro, which suggests that the frameshift efficiency under these conditions depends only upon the classic stem–loop, and that the in vitro system is not sensitive enough to characterize the structure of the frameshift region of HIV-1, as discussed previously (Dulude et al. 2002).
FIGURE 2.
Relative frameshift efficiency of mutants of subtype B of HIV-1 group M. (A) Secondary structure of the frameshift stimulatory signal for subtype B, subtype B short, subtype B bulge subs, and subtype B*, a natural variant that has a deletion of three nucleotides in the frameshift stimulatory signal. The stimulatory signal is represented with the extended bulged stem–loop structure found for HIV-1 subtype B (Dulude et al. 2002). Circles indicate nucleotides that differ from subtype B. The thermodynamic stability, ΔG° (kcal/mole), of the frameshift stimulatory signal of the different mutants of subtype B is indicated. The predicted ΔG° were calculated with the mfold program of M. Zuker (Mathews et al. 1999) for the classic stem–loop structure (upper stem) and the extended bulged stem–loop structure (upper stem + lower stem). The complete frameshift region with the slippery sequence underlined is represented for subtype B, whereas only the frameshift stimulatory signal is shown for the mutants. (B) Relative frameshift efficiency of mutants of subtype B. In vitro studies in RRL and assays in cultured cells (293T) were performed as described in the text. For assays in cultured cells, the experimental values were normalized for variations in transfection efficiency by cotransfecting the various constructs used for measuring the frameshift efficiency with pcDNA3.1/Hygro(+)/lacZ and monitoring the β-galactosidase activity. The frameshift efficiency of subtype B is arbitrarily set at 100%. The values of the frameshift efficiency are the means of at least five independent experiments, with bars representing the standard error on the means.
The frameshift efficiency of a third mutant called subtype B* was also investigated. The sequence of the frameshift stimulatory signal of this mutant originates from a natural isolate of subtype B and has a 3-nucleotide deletion (Fig. 1B ▶). This deletion changes the structure of the stimulator, which can be predicted to be a variant of the two-stem structure, with a shorter upper stem separated from the lower stem by a symmetric internal loop. The lower stem has the same size as in subtype B, but is slightly more stable due to the replacement of a G-U pair with a G-C pair. The predicted stability for this structure was lower than that of the stimulator of subtype B, whether one considers only the upper stem–loop or the extended bulged stem–loop structure (Fig. 2A ▶). The frameshift efficiency of subtype B* compared with subtype B was decreased by ~25% and 60% in vitro and in cultured cells, respectively (Fig. 2B ▶). The lower in vitro frameshift efficiency can be related directly to the lower stability of the upper stem of this construct. In cultured cells, the frameshift efficiency of subtype B* is the same as that of subtype B short, showing that the presence of the lower stem counteracts the destabilization of the upper stem. The effect of the symmetrical internal loop between the two stems in subtype B* will be analyzed below (see Table 1 ▶ for a summary of the results).
Structure and stability of the frameshift stimulatory signal of all the subtypes of HIV-1 group M
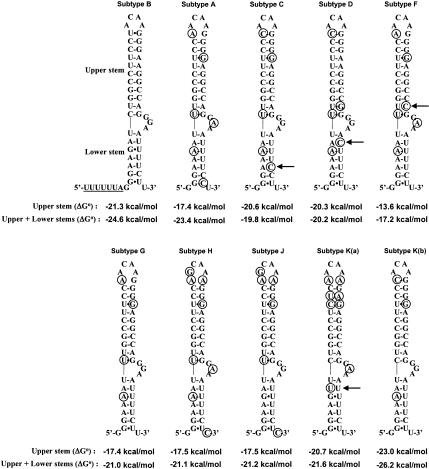
Next, the frameshift efficiency of all the subtypes of group M was investigated. The sequences used are the consensus sequences of each subtype, except for subtype K, and were obtained from the Los Alamos HIV Sequence Database (Los Alamos National Laboratory 2002; http://hiv-web.lanl.gov). For subtype K, only two highly divergent sequences [named arbitrarily K(a) and K(b)] are available, making the identification of a consensus sequence impossible. Figure 3 ▶ shows that although the frameshift stimulatory signal sequence displays variations in the different subtypes, it can be folded into the extended bulged stem–loop structure demonstrated previously for subtype B. The majority of the sequence variations allow the conservation of the base pairing in the two stems. As to the bulge, it contains purines in every subtype (GGA or GAA), which fully agrees with the demonstrated requirement of purines in the bulge to obtain maximal frameshift efficiency in subtype B (see above).
FIGURE 3.
Secondary structure of the frameshift stimulatory signal of all of the subtypes of HIV-1 group M. The frameshift stimulatory region is folded according to the extended bulged stem–loop structure found for HIV-1 subtype B stimulator (Dulude et al. 2002). Circles indicate nucleotides that differ from subtype B. The arrows point to noncanonical base pairs and mismatches. The thermodynamic stability, ΔG° (kcal/mole), of the frameshift stimulatory signal of the different subtypes is indicated. The predicted ΔG° were calculated as described in the legend to Figure 2 ▶.
The thermodynamic stability of the stimulator of every subtype was calculated for the classic structure (upper stem–loop only) and for the extended bulged stem–loop structure (upper + lower stems), as has been done for subtype B and its mutants. The hierarchy, by order of decreasing stability, is as follows: subtype K(b) > B > K(a) ≅ C > D > H = J ≅ G = A > F, when considering only the upper stem–loop (see also Chang et al. 1999). Subtypes A, F, G, H, J, and K(a) have a loop of six nucleotides instead of four in subtypes B, C, D, and K(b), which decreases their upper-stem length by 1 bp. Subtype F is further destabilized by an U-C mismatch in the lower part of the upper stem. When considering the extended bulged stem–loop, the hierarchy of stability changes to K(b) > B > A > K(a) > J ≅ H ≅ G > D > C > F. The predicted stability of subtypes C and D is reduced due to an A-C mismatch in their lower stem. However, we cannot exclude the possibility that the A-C pair forms a non-Watson–Crick base pair geometrically similar to a G-U pair (Nowakowski and Tinoco Jr. 1999), and that the thermodynamic stability calculated with mfold does not correspond to the actual stability of the structure. In subtype K(a), a U-U mismatch destabilizes the lower stem.
Frameshift efficiency of all the subtypes of HIV-1 group M
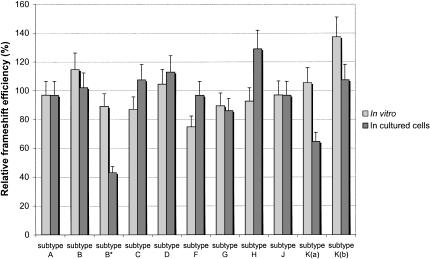
The frameshift efficiency of all of the subtypes of HIV-1 group M was measured with the luciferase reporter system and is presented in Figure 4 ▶ (summarized in Table 1 ▶). It can be seen that the in vitro frameshift efficiency varies between 4.5% (subtype F) and 8.4% [subtypes K(b)], with a mean value of 6.0%. As discussed above for subtype B and its mutants, the stimulation of the frameshift efficiency in vitro appears to depend only upon the upper stem–loop. Our results with the different subtypes of group M confirm this observation, as the frameshift efficiency varies in parallel to the predicted stability of the upper stem in vitro. For example, subtype F has the lowest frameshift efficiency in vitro, and is the only subtype with a mismatch in its upper stem, drastically reducing its thermodynamic stability. In cultured cells, however, the frameshift efficiency of the different subtypes ranges from 1.2% [subtype K(a)] to 2.4% (subtype H), with a mean value of 1.9%, clearly showing that the complete extended bulged stem–loop and not just the upper stem, contributes to the frameshift efficiency. This is best seen with subtypes F and K(a). The upper stem of subtype F has a low stability, but its lower stem is very stable, and its frameshift efficiency relative to the other subtypes is higher in cultured cells than in vitro. In contrast, the upper stem of subtype K(a) has a high stability and its frameshift efficiency in vitro is proximal to the mean value, but a U-U mismatch in its lower stem, which compromises the formation of this stem, likely accounts for the low frameshift efficiency of this subtype in cultured cells, compared with the other subtypes.
FIGURE 4.
Relative frameshift efficiency of all subtypes of HIV-1 group M. In vitro studies in RRL and assays in cultured cells (293T) were performed as indicated in Figure 2 ▶. The mean value of the frameshift efficiency, calculated from the values obtained for all of the subtypes (1.9% in cultured cells and 6.0% in vitro), is arbitrarily set at 100%. The frameshift efficiency of subtype B* has been added to the figure to facilitate comparison with the different subtypes. The values of the frameshift efficiency are the means of at least five independent experiments, with bars representing the standard error on the means.
It can be observed that the frameshift efficiency of the different subtypes varies within a narrow range in cultured cells, the maximal deviation from the mean value calculated from the experimental values of all the subtypes being ~35%. This does not reflect the range of variations of the thermodynamic stability of the frameshift stimulator (from −17.2 kcal/mole to −26.2 kcal/mole). However, as suggested by Kim et al. (2001), it is not the stability of the isolated stimulatory signal alone that must be considered, but its stability when it interacts with the ribosome, an interaction that remains to be defined. It is worth recalling that a stimulator makes the ribosome pause at a slippery sequence, and that the duration of the pause likely correlates with the stability of the signal, but, on the other hand, pausing is not sufficient to promote frameshifting (Tu et al. 1992; Somogyi et al. 1993; Kontos et al. 2001). We found that the thermodynamic stability of subtype B* is higher than that of subtype F, but its frameshift efficiency in cultured cells is twofold less than subtype F. A direct explanation for this observation is that this low frameshift efficiency of subtype B* results from the absence of the bulge, which is replaced by a symmetric internal loop. It is likely that this internal loop does not interact with the ribosome in a way promoting a productive interaction of the frameshift stimulatory signal.
DISCUSSION
Our analysis presents, for the first time, an assessment of the frameshift efficiency of the different subtypes of HIV-1 group M. It shows that the frameshift efficiencies of the different subtypes of HIV-1 group M investigated fall within a narrow window, despite variations in the stimulatory signal sequence and in its predicted thermodynamic stability. This likely reflects the existence of a selective pressure to maintain the Gag-Pol to Gag ratio at a critical value for optimal virus assembly and infectivity. It was observed that a threefold increase of the Gag-Pol to Gag ratio, caused by cotransfecting HIV-1 proviral DNA with an HIV-1 Gag-Pol expression vector in 293T cells, produces a 10-fold decrease in virion infectivity (Shehu-Xhilaga et al. 2001). In this study, we show that the frameshift efficiencies of the different subtypes of group M are found within 35% of the mean frameshift efficiency, calculated from the experimental values obtained for all subtypes. Telenti et al. (2002) recently investigated how mutations destabilizing the classic stem–loop of natural variants and laboratory mutants of HIV-1 affect the frameshift efficiency and the capacity of the mutant virions to replicate in peripheral blood lymphocytes. Their results show that a decrease of the frameshift efficiency >60% compared with subtype B severely impairs viral replication, whereas a decrease of up to 40% has no effect. With the natural subtype B* variant we investigated here, the frameshift efficiency decreased by ~60% compared with either subtype B or to the mean value obtained from all subtypes, but whether subtype B* is infectious has yet to be determined. Our results coupled to those of Telenti et al. (2002) suggest that agents targeted against the frameshift event should affect the frameshift efficiency by >60% to efficiently impair viral replication.
We can also use the natural sequence variations of the different subtypes of group M to address the possible formation of the alternate frameshift stimulatory signal structure proposed by Dinman et al. (2002). This structure is derived from the analysis of sequences from GenBank, which include mostly subtype B sequences. In this structure, which is a pseudoknot, the pyrimidine-rich segment downstream of the classic stem–loop forms an additional 3-bp stem with bases in the loop of the classic stem–loop. This stem is further stabilized by a triple helix, in which a portion of the pyrimidine-rich segment interacts with the classic stem. If the frameshift region of all of the subtypes investigated here was folded according to the model of Dinman et al. (2002), subtypes H and J would have mismatches in two of the three base pairs of the additional stem, whereas subtypes A and K(a) would have one mismatch in this stem. However, we found that the frameshift efficiency of these subtypes was not significantly lower than the mean value calculated for all of the subtypes, except for subtype K(a). In this latter case, as discussed above, the presence of a mismatch in the lower stem of the extended bulged stem–loop structure can readily account for the decrease of the frameshift efficiency of this subtype. These observations raise an additional criticism against the model of Dinman et al. (2002), in addition to the fact that the pyrimidine-rich strand of the triplex is in an antiparallel orientation relative to the purine-rich strand of the duplex with which it interacts, an orientation shown to be sterically unfavorable (Han and Dervan 1993; Frank-Kamenetskii and Mirkin 1995; Lim and Barton 1998).
Agents that decrease the frameshift efficiency in HIV-1 have already been developed (Hung et al. 1998), and although the toxicity of these agents precludes their use for clinical treatments, they illustrate the potential of an approach targeting the frameshift event. The absence of treatments able to eradicate HIV-1 and the emergence of viruses resistant to the protease and reverse transcriptase inhibitors presently in use (Condra 1998; O’Brien 2000) make it imperative to develop alternative strategies against novel viral targets (Dinman et al. 1998; Gallego et al. 2001). Our results identify the frameshift event as such a potential target, as the frameshift efficiency is very similar in the different subtypes of HIV-1 group M. Any agent perturbing the frameshift event, by bringing the Gag-Pol to Gag ratio outside a narrow window, could impair the viral propagation in a broad population of HIV-1 subtypes.
MATERIALS AND METHODS
Construction of plasmids
A plasmid construct was derived from pcDNA3.1/Hygro(+) (Invitrogen), in which a reporter gene coding for the firefly luciferase was inserted under the control of the T7 and CMV promoter, allowing the assessment of its expression in vitro and in cultured cells, respectively. The luc sequence originated from pGEM-luc (Promega) and was bracketed between a BamHI and a XhoI site. The AUG initiation codon of luc was deleted (so as to avoid aberrant initiations), and an oligonucleotide fragment containing an AUG codon followed by a short linker encompassing an Eco47III site and the frameshift region of subtype B of the HIV-1 group M (GenBank K02007) was inserted upstream of the luc sequence between the KpnI site of the vector and the BamHI site preceding the luc sequence. This generated the pLUC/HIV/B (−1) construct (Fig. 1A ▶). The frameshift region in this construct, coined the 3′ extended frameshift region, encompasses the slippery sequence, the spacer, the classic stem–loop plus 10 nucleotides downstream of this stem, that is, the stimulator defined in a previous study (Fig. 2A ▶; Dulude et al. 2002). The slippery site was preceded by an Eco47III site, so that constructs corresponding to mutants or other subtypes of HIV-1 could be generated readily by replacing the Eco47III–BamHI fragment of subtype B with an appropriate oligonucleotide cassette, corresponding to either subtype A (GenBank AF069673), C (GenBank AF110962), D (GenBank U88822), F (GenBank AJ249238), G (GenBank AF061640), H (GenBank AF005496), J (GenBank AF082394), K(a) (GenBank AJ249235), or K(b) (GenBank AJ249239; see Fig. 1B ▶). Different mutants of subtype B, that is, subtype B short, subtype B bulge subs, and subtype B*, a natural variant of subtype B (GenBank M17449), were also derived from the pLUC/HIV/B (−1) construct (Fig. 1B ▶). In all (−1) constructs, a −1 ribosomal frameshift is required for expression of the luc reporter gene, otherwise translation terminates at a stop codon overlapping codon six of the luc sequence. For each (−1) construct, a pLUC/HIV (0) in-frame control was made, in which an additional adenine was introduced before the luc coding sequence (see Fig. 1A ▶), so that luciferase is synthesized by ribosomes that do not make a frameshift. All constructs were verified by sequencing the entire insert.
Transient transfections and luciferase assays
For assays in cultured cells, transient transfections were made in 293T cells, using the standard calcium phosphate precipitation method (Jordan et al. 1996). To this end, precipitates were formed with 3 μg of pLUC/HIV and 1.25 μg of pcDNA3.1/Hygro(+)/lacZ (Invitrogen), which codes for β-galactosidase. Precipitates were overlayed over 40% confluent cells grown into 6-well plates and maintained in Dulbecco’s modification of Eagle’s medium with 10% (v/v) fetal bovine serum (BioMedia). Cells were harvested with 600 μL of the Cell Culture Lysis Reagent (Promega) 48 h post-transfection, and the luciferase activity was measured in cell lysates. Luminescence produced by adding 2 μL of the lysate to the Luciferase Assay Reagent (Promega) was measured in relative light units in a Berthold Lumat LB 9507 luminometer. The β-galactosidase activity of the cell lysates was determined using chlorophenolred-β-D-galactopyranoside substrate (Calbiochem; Eustice et al. 1991) and was used to normalize the transfection efficiency. The frameshift efficiency of the different pLUC/HIV constructs was calculated by dividing the level of luciferase activity produced with the (-1) construct by the sum of the luciferase activity of the (0) and (-1) constructs.
In vitro transcription and translation
For in vitro studies, plasmids were linearized at the StuI site located 3′ of the luc coding sequence. HIV-LUC mRNA was generated by run-off transcription with T7 RNA polymerase, and 0.2 μg of this mRNA was translated at 30°C for 15 min in 25 μL of RRL (Promega). The reaction was stopped by addition of EDTA to a final concentration of 6 mM. Luciferase activity was monitored with 2.5 μL of the translation mixture, as described in the “transient transfections and luciferase assays” section. Frameshift efficiencies were calculated as described above.
Acknowledgments
This study was supported by a grant from the Canadian Institutes for Health Research to L.B.-G. and G.L. We thank Pascal Chartrand and Sergey Steinberg for stimulating discussions and for critical reading of this manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5113603.
REFERENCES
- Bidou, L., Stahl, G., Grima, B., Liu, H., Cassan, M., and Rousset, J.P. 1997. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem–loop stimulatory signal. RNA 3: 1153–1158. [PMC free article] [PubMed] [Google Scholar]
- Brierley, I. 1995. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 76: 1885–1892. [DOI] [PubMed] [Google Scholar]
- Brierley, I. and Pennel, S. 2001. Structure and function of the stimulatory RNAs involved in programmed eukaryotic -1 ribosomal frameshifting. In The ribosome, pp 233–248. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Cassan, M., Delaunay, N., Vaquero, C., and Rousset, J.P. 1994. Translational frameshifting at the gag-pol junction of human immunodeficiency virus type 1 is not increased in infected T-lymphoid cells. J. Virol. 68: 1501–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro, M., Parkin, N., and Varmus, H.E. 1992. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc. Natl. Acad. Sci. 89: 713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, S.Y., Sutthent, R., Auewarakul, P., Apichartpiyakul, C., Essex, M., and Lee, T.H. 1999. Differential stability of the mRNA secondary structures in the frameshift site of various HIV type 1 viruses. AIDS Res. Hum. Retroviruses 15: 1591–1596. [DOI] [PubMed] [Google Scholar]
- Condra, J.H. 1998. Resistance to HIV protease inhibitors. Haemophilia 4: 610–615. [DOI] [PubMed] [Google Scholar]
- Dinman, J.D., Ruiz-Echevarria, M.J., and Peltz, S.W. 1998. Translating old drugs into new treatments: Ribosomal frameshifting as a target for antiviral agents. Trends Biotechnol. 16: 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman, J.D., Richter, S., Plant, E.P., Taylor, R.C., Hammell, A.B., and Rana, T.M. 2002. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl. Acad. Sci. 99: 5331–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulude, D., Baril, M., and Brakier-Gingras, L. 2002. Characterization of the frameshift stimulatory signal controlling a programmed −1 ribosomal frameshift in the human immunodeficiency virus type 1. Nucleic Acids Res. 30: 5094–5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustice, D.C., Feldman, P.A., Colberg-Poley, A.M., Buckery, R.M., and Neubauer, R.H. 1991. A sensitive method for the detection of β-galactosidase in transfected mammalian cells. Biotechniques 11: 739–740, 742–743. [PubMed] [Google Scholar]
- Farabaugh, P.J. 2000. Translational frameshifting: Implications for the mechanism of translational frame maintenance. Prog. Nucleic Acid Res. Mol. Biol. 64: 131–170. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetskii, M.D. and Mirkin, S.M. 1995. Triplex DNA structures. Annu. Rev. Biochem. 64: 65–95. [DOI] [PubMed] [Google Scholar]
- Gallego, O., de Mendoza, C., Perez-Elias, M.J., Guardiola, J.M., Pedreira, J., Dalmau, D., Gonzalez, J., Moreno, A., Arribas, J.R., Rubio, A., et al. 2001. Drug resistance in patients experiencing early virological failure under a triple combination including indinavir. AIDS 15: 1701–1706. [DOI] [PubMed] [Google Scholar]
- Han, H. and Dervan, P.B. 1993. Sequence-specific recognition of double helical RNA and RNA. DNA by triple helix formation. Proc. Natl. Acad. Sci. 90: 3806–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, M., Patel, P., Davis, S., and Green, S.R. 1998. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 72: 4819–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks, T. and Varmus, H.E. 1985. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science 230: 1237–1242. [DOI] [PubMed] [Google Scholar]
- Jacks, T., Power, M.D., Masiarz, F.R., Luciw, P.A., Barr, P.J., and Varmus, H.E. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331: 280–283. [DOI] [PubMed] [Google Scholar]
- Jordan, M., Schallhorn, A., and Wurm, F.M. 1996. Transfecting mammalian cells: Optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H. 1998. Direct structural evidence for formation of a stem–loop structure involved in ribosomal frameshifting in human immunodeficiency virus type 1. Biochim. Biophys. Acta 1397: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karacostas, V., Wolffe, E.J., Nagashima, K., Gonda, M.A., and Moss, B. 1993. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193: 661–671. [DOI] [PubMed] [Google Scholar]
- Kim, Y.G., Maas, S., and Rich, A. 2001. Comparative mutational analysis of cis-acting RNA signals for translational frameshifting in HIV-1 and HTLV-2. Nucleic Acids Res. 29: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontos, H., Napthine, S., and Brierley, I. 2001. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 21: 8657–8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, S.Y., Shapiro, B.A., Chen, J.H., Nussinov, R., and Maizel, J.V. 1991. RNA pseudoknots downstream of the frameshift sites of retroviruses. Genet. Anal. Tech. Appl. 8: 191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, A.C. and Barton, J.K. 1998. Rh(phen)2phi3+ as a shape-selective probe of triple helices. Biochemistry 37: 9138–9146. [DOI] [PubMed] [Google Scholar]
- Los Alamos National Laboratory. 2002. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos National Laboratory, Los Alamos, CA.
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- McCutchan, F.E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14: S31–S44. [PubMed] [Google Scholar]
- Moore, J.P., Parren, P.W., and Burton, D.R. 2001. Genetic subtypes, humoral immunity, and human immunodeficiency virus type 1 vaccine development. J. Virol. 75: 5721–5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski, J. and Tinoco Jr., I. 1999. RNA structure in solution. In Oxford handbook of nucleic acid structure, pp. 567–602. Oxford University Press, New York.
- O’Brien, W.A. 2000. Resistance against reverse transcriptase inhibitors. Clin. Infect. Dis. 30: S185–S192. [DOI] [PubMed] [Google Scholar]
- Park, J. and Morrow, C.D. 1991. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 65: 5111–5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reil, H. and Hauser, H. 1990. Test system for determination of HIV-1 frameshifting efficiency in animal cells. Biochim. Biophys. Acta 1050: 288–292. [DOI] [PubMed] [Google Scholar]
- Roberts, J.D., Bebenek, K., and Kunkel, T.A. 1988. The accuracy of reverse transcriptase from HIV-1. Science 242: 1171–1173. [DOI] [PubMed] [Google Scholar]
- Robertson, D.L., Anderson, J.P., Bradac, J.A., Carr, J.K., Foley, B., Funkhouser, R.K., Gao, F., Hahn, B.H., Kalish, M.L., Kuiken, C., et al. 2000. HIV-1 nomenclature proposal. Science 288: 55–56. [DOI] [PubMed] [Google Scholar]
- Shehu-Xhilaga, M., Crowe, S.M., and Mak, J. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75: 1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi, P., Jenner, A.J., Brierley, I., and Inglis, S.C. 1993. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 13: 6931–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenti, A., Martinez, R., Munoz, M., Bleiber, G., Greub, G., Sanglard, D., and Peters, S. 2002. Analysis of natural variants of the human immunodeficiency virus type 1 gag-pol frameshift stem–loop structure. J. Virol. 76: 7868–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, C., Tzeng, T.H., and Bruenn, J.A. 1992. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl. Acad. Sci. 89: 8636–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]