Abstract
The genome of the hepatitis C virus (HCV) is a plus-strand RNA molecule that carries a single long open reading frame. It is flanked at either end by highly conserved nontranslated regions (NTRs) that mediate crucial steps in the viral life cycle. The 3′ NTR of HCV has a tripartite structure composed of an about 40-nucleotide variable region, a poly(U/UC) tract that has a heterogeneous length, and a highly conserved 98-nucleotide 3′-terminal sequence designated the X tail or 3′X. Conflicting data as to the role the sequences in the 3′ NTR play in RNA replication have been reported. By using the HCV replicon system, which is based on the self-replication of subgenomic HCV RNAs in human hepatoma cell line Huh-7, we mapped in this study the sequences in the 3′ NTR required for RNA replication. We found that a mutant with a complete deletion of the variable region is viable but that replication is reduced significantly. Only replicons in which the poly(U/UC) tract was replaced by a homouridine stretch of at least 26 nucleotides were able to replicate, whereas RNAs with homopolymeric guanine, adenine, or cytosine sequences were inactive. Deletions of individual or all stem-loop structures in 3′X were not tolerated, demonstrating that this region is most crucial for efficient RNA replication. Finally, we found that none of these deletions or substitutions within the 3′ NTR affected RNA stability or translation, demonstrating that the primary effect of the mutations was on RNA replication. These data represent the first detailed mapping of sequences in the 3′ NTR assumed to act as a promoter for initiation of minus-strand RNA synthesis.
Hepatitis C virus (HCV), the etiologic agent of non-A, non-B hepatitis, is an enveloped plus-strand RNA virus that belongs to the family Flaviviridae (46). One of the hallmarks of HCV infection is the high frequency of persistence. About 80% of infected individuals are unable to eliminate the virus, and these patients are at high risk to develop chronic liver disease, including liver cirrhosis and hepatocellular carcinoma (32). In spite of recent improvements in therapy of chronic hepatitis C with ribivirin and alpha interferon conjugated with polyethyleneglycol, only about one-half of the patients develop a sustained response, and nonresponders and relapses are common (40). Therefore, more-effective therapies are certainly required.
Like those of other members of the Flaviviridae family, the HCV genome carries a single long open reading frame (ORF), which encodes a polyprotein that is cleaved co- and posttranslationally into a series of products (for reviews see references 4 and 47). For HCV, the cleavage products are arranged in the following order (from the amino to the carboxy terminus): core, envelope protein 1 (E1), E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B. Structural proteins core, E1, and E2 are the main constituents of the virus particle whereas the NS3, NS4B, NS5A, and NS5B proteins are essential and sufficient for RNA replication (36). NS3 is composed of two domains that carry distinct enzymatic activities. The amino-terminal domain contains a serine-type proteinase, which is required for proper processing of the NS3 to NS5B region (19). The carboxy-terminal domain harbors nucleoside triphosphatase (NTPase) and helicase activities that are essential for replication (28, 30). NS4A is an important cofactor of the NS3 proteinase, and it must form a heterodimeric complex to activate this enzyme (14, 33). The function of NS4B so far is not known. As inferred from the clustering of cell culture-adaptive mutations in the center of NS5A, this protein appears to play an important role in RNA replication (6, 31). Moreover, NS5A seems to be a determinant of the success of alpha interferon therapy, probably by counteracting the antiviral effect exerted by double-stranded RNA-activated interferon-induced protein kinase PKR (13, 17). NS5B is the RNA-dependent RNA polymerase (RdRp), and it is essential for RNA replication in vivo and in cell culture (5, 30, 34, 36).
Translation of the HCV ORF is mediated via the 5′ nontranslated region (NTR), which carries an internal ribosome entry site (IRES) (58, 59). Interestingly, this IRES binds the 40S ribosomal subunit in the absence of translation factors, thereby positioning the initiator AUG codon of the ORF directly at the P site (44). Although several studies suggest that sequences at the 5′ end of the core gene are required for full IRES activity, a recent report convincingly shows that this coding region is not directly involved in IRES function but rather prevents the formation of stable secondary structures at the 5′ end of the cistron (49). In addition to being required for expression of the HCV polyprotein, sequences within the 5′ NTR overlapping with the IRES are essential for efficient RNA replication (16).
Another RNA element involved in replication is the 3′ NTR (Fig. 1). It has a tripartite structure composed of an about 40-nucleotide variable region, which is only poorly conserved among different HCV isolates, a poly(U/UC) tract, which is very heterogeneous in length, and a highly conserved 98-nucleotide sequence, which was designated the X tail or 3′X (29, 55, 56, 61). Computer predictions and structure-probing studies suggested the formation of two stem-loops in the variable region (VSL1 and VSL2) and three stable stem-loops in 3′X, designated SL1, SL2, and SL3 (Fig. 1) (7, 23, 56). SL1, either on its own or together with SL2, is a binding site for polypyrimidine tract-binding protein (PTB) (23, 57). The most stable stem-loop is the 3′-terminal SL3, which has a length of 46 nucleotides. While most of this sequence forms regular Watson-Crick base pairs, the terminal nucleotide forms an irregular uracil:guanine base pair, which is less stable. This property may be required to facilitate the melting of the terminal SL3 during initiation of minus-strand RNA synthesis. Interestingly, the closely related pestiviruses, such as bovine viral diarrhea virus (BVDV), also have a very stable 3′ terminal stem-loop that is required for RNA replication (64). However, unlike what is found for HCV, three to five non-base-paired cytidines are present at the very 3′ end, indicating that the requirements for initiation of minus-strand RNA synthesis for the BVDV and the HCV replicases are different. Recent studies suggest that, in addition to playing a major role in RNA replication, the 3′ NTR, in particular 3′X, stabilizes the RNA and enhances translation both in vitro and in transfected cells (24, 54). These effects require the binding to the 3′ NTR of cellular factors such as PTB, the autoantigen La, or some ribosomal proteins (23, 52, 60).
FIG. 1.
(A) Schematic presentation of the basic replicon constructs used in this study. The 5′ and 3′ NTRs (only shown for the selectable replicon) are given with their secondary structures (solid line, EMCV IRES; open bars, coding regions for neomycin phosphotransferase (neo) and firefly luciferase (Ffl-luc), as well as the HCV nonstructural proteins). ∗, positions of the three cell culture-adaptive mutations. (B) Potential structures of the 3′ NTR from references 7 and 29 with two stem-loops in the variable region (VSL1 and VSL2), the 83-nucleotide poly(U/UC) tract, and the three stem-loops in 3′X. The positions of the engineered recognition sequences of restriction enzymes StuI and BamHI as well as the authentic recognition site for NheI are indicated. To generate runoff transcripts with authentic 3′ ends, the two substitutions in SL3 (∗) were introduced. They result in a change of an A:U to a U:A base pairing required to generate a ScaI restriction site. The stop codon of the polyprotein ORF is boxed. Numbers refer to the nucleotide positions of the HCV Con1 isolate (EMBL database accession no. AJ238799).
Conflicting data on whether or not the 3′ NTR is essential for RNA replication have been published. HCV genomes that lack most of this sequence and that terminate with a homopolymeric adenosine or uridine tract have been reported to replicate in transfected human hepatoma cells (12, 63). On the other hand, results obtained from experimental inoculation of chimpanzees with cloned infectious HCV genomes carrying mutations in the 3′ NTR have shown that both the poly(U/UC) region and 3′X are essential for infectivity whereas a mutant with a deletion of part of the variable region was still viable (30, 62). However, because of obvious limitations inherent to this in vivo test, no detailed studies of the sequences involved in RNA replication have been performed.
A clarification of these controversies was hindered by the lack of an efficient cell culture system. This limitation has now been overcome by the development of the HCV replicon system, which utilizes selectable subgenomic HCV RNAs that are composed of the following elements: the HCV 5′ NTR including the first 16 codons of the core gene to direct translation of a selectable marker (the gene encoding the neomycin phosphotransferase), the IRES of the encephalomyocarditis virus (EMCV) to direct translation of the HCV NS3 to NS5B codzng region, and the 3′ NTR (Fig. 1A) (36). Upon transfection of human hepatoma cell line Huh-7 and selection with Geneticin, cell lines that carry large amounts of self-replicating HCV-RNAs could be established. Subsequent studies led to the identification of cell culture-adaptive mutations that enhance RNA replication to a level that can be monitored in transient-replication assays (6, 20, 31). The most efficient adaptation described thus far is achieved with three mutations, two in NS3 and one in NS5A, that increase RNA replication synergistically (31). This discovery allowed the development of luciferase replicons in which the gene encoding the selectable marker was replaced by a convenient reporter gene (the luciferase gene from the firefly Photinus pyralis). Replication of this RNA could be monitored in transient-replication assays by measuring the activity of the luciferase reporter (31).
In this study we characterized the sequences in the 3′ NTR required for RNA replication. We found that both 3′X and a short homouridine tract are indispensable, whereas a deletion of the complete variable region is tolerated but significantly reduces replication efficiency. The minimal length and composition of the homopolymeric tract required for RNA replication were determined, and the effect of mutations at the very 3′ end of the replicon was examined.
MATERIALS AND METHODS
Cell cultures.
Huh-7 human hepatoma cells were grown in Dulbecco's modified minimal essential medium (Life Technologies GmbH, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin, 100 μg of streptomycin, and 10% fetal calf serum (complete DMEM).
Plasmid constructions and sequence analysis.
Unless otherwise stated, standard recombinant DNA technologies were used for all cloning procedures (2). The basic replicon constructs pFK-I389neo/NS3-3′/5.1, pFK-I389luc/NS3-3′/5.1, and pFK-I389luc/NS3-3′/GND have been described recently (31). These constructs carry an engineered recognition sequence for ScaI at the 3′ end upstream of a SpeI site that was used for cloning procedures described below. To facilitate manipulations at the 3′ NTR, construct pBSK8499-9605/Stu-Bam was made in the following way. Two overlapping PCR fragments were generated with primer pair S9392Stu and A9508Bam or S9508Bam and A9605Sca/Spe, with pFK-I389neo/NS3-3′/5.1 as the template (Table 1). Fragments were combined by PCR using the outer primers (S9392Stu and A9605Sca/Spe) and, after restriction with StuI and SpeI, were inserted into pBluescript vector SKI (Stratagene, La Jolla, Calif.) together with a PCR fragment that was generated with primers S8492Xho/Sfi and A9392Stu and restricted with XhoI and StuI. To remove the BamHI restriction site in this construct, a PCR fragment was amplified with primers S9392Stu and A9605Sca/Spe by using pFK-I389neo/NS3-3′/5.1 as the template and, after restriction with StuI and SpeI, was inserted into pBSK8499-9605/Stu-Bam. All further mutations in the 3′ NTR were introduced into the resulting plasmid, pBSK8499-9605/Stu, and SfiI/SpeI fragments (nucleotide positions 8499 to 9605 of our HCV isolate) were transferred into the parental replicon constructs. Replicons carrying homopolymeric uridine stretches of 46, 26, 6, or 1 nucleotide in length were generated by hybridization of the appropriate sense primers listed in Table 1 with A9535NheI, incubation with the Klenow fragment of DNA polymerase (Roche Molecular Biochemicals), restriction with StuI and NheI, and insertion into pBSK8499-9605/Stu. Partial deletion of the variable region was achieved by generating a PCR fragment with primers S8492Xho/Sfi and Adel9375-9401 and insertion of the SfiI- and StuI-restricted fragment into pBSK8499-9605/Stu. The complete variable region was removed by insertion into pBSK8499-9605/Stu of an SfiI- and NheI-restricted DNA fragment generated by PCR with primers S8492Xho/Sfi and Adel-var. To delete stem-loop 1 in the 3′ NTR, oligonucleotides S-SL1 and A-SL1 were hybridized, incubated with the Klenow fragment of DNA polymerase, restricted with StuI and NheI, and inserted into pBSK8499-9605/Stu. The complete deletion of 3′X was achieved in the same way with oligonucleotides SdelX and AdelX (Table 1). Replicon constructs lacking stem-loops 2 and 3 were generated by PCR using oligonucleotide pairs S-SL2/3 and A-SL2 and S-SL2/3 and A-SL3, respectively, and, after restriction with StuI and SpeI, fragments were inserted into pBSK8499-9605/Stu. To obtain constructs carrying 26-nucleotide homopolymeric cytidine, guanine, or adenosine tracts, complementary oligonucleotides listed in Table 1 were hybridized and inserted into pBSK8499-9605/Stu after restriction with StuI and NheI. Replicons carrying five additional cytidine residues at the 3′ end of SL3 were generated by insertion of complementary oligonucleotides 3′C-SbfI-up and 3′C-SbfI-down (Table 1) into the ScaI site at the 3′ end of the HCV sequence. In this way a unique recognition sequence for SbfI was fused to the 3′ end of SL3 via the following sequence: UCA AGU CCC CCT GCA GG (sequences of SL3 and SbfI are underlined). During this cloning procedure, a construct with only three cytidine residues between SL3 and the SbfI site was obtained fortuitously. Nucleotide sequences of all constructs were verified by DNA sequence analysis as described recently (35).
TABLE 1.
Oligonucleotides used for construction of replicons with mutations in the 3′ NTR
| Oligonucleotidea | Sequence | Construct |
|---|---|---|
| S8492Xho/Sfi | GTT ACC TCG AGG CCG CTG CGG CCT GTC GAG CTG CG | pBSK8499-9605/Stu |
| A9392Stu | AAA ACA GGA AGG CCT ATT GGC CAG GAG | pBSK8499-9605/Stu |
| S9392Stu | CAC TCC TGG CCA ATA GGC CTT CCT G | pBSK8499-9605/Stu |
| A9508BamHI | GAC TAG GGA TAA GAT GGA TCC ACC AAA GG | pBSK8499-9605/Stu |
| S9508BamHI | CCT TTG GTG GAT CCA TCT TAT CCC TAG | pBSK8499-9605/Stu |
| A9605Sca/Spe | TGC ACT AGT AGT ACT TGA TCT GCA GAG AGG CCA GTA | pBSK8499-9605/Stu |
| S9140 | GGG GGG GAG GGC TGC CAC TTG TGG C | |
| S9402Stul-50U-Nhel | CCA ATA GGC CTT CCT GTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT GGT GGC TCC ATC TTA GCC CTA GTC ACG GCT AGC TGT GAG | pBSK8499-9605/U46 |
| S9402Stul-30U-Nhel | CCA ATA GGC CTT CCT GTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TGG TGG CTC CAT CTT AGC CCT AGT CAC GGC TAG CTG TGA G | pBSK8499-9605/U26 |
| S9402 Stul-10U-Nhel | CCA ATA GGC CTT CCT GTT TTT TTT TTG GTG GCT CCA TCT TAG CCC TAG TCA CGG CTG CTG TGA G | pBSK8499-9605/U6 |
| S9402 Stul-5U-Nhel | CCA ATA GGC CTT CCT GTT TTT GGT GGC TCC ATC TTA GCC CTA GTC ACG GCT GCT GTG AG | pBSK8499-9605/U1 |
| A9535 Nhel | CTC ACA GCT AGC CGT GAC TAG GGC TAA G | pBSK8499-9605/U46, U26, U6, U1 |
| Adel9375-9401 | CAG GAA GGC CTT CAT CGG TTG GGG AGT AGA TAG ATG CCT ACC CC | pBSKdel9375-9401 |
| Adel-Var | AAA AGC TAG CCG TGA CTA GGG CTA AGA TGG AGC CAC CAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA TCA TCG GTT GGG GAG TAG ATA GAT GCC | pBSKdel9375-9415 |
| S-SL1 | CCT TCC TGT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TGT CAC GGC TAG CTG TG | pBSK8499-9605/ΔSL1 |
| A-SL1 | CAC AGC TAG CCG TGA C | pBSK8499-9605/ΔSL1 |
| S-SL2/3 | ACT CCT GGC CAA TAG GCC TTC C | pBSK8499-9605/ΔSL2 |
| A-SL2 | AAA AAC TAG TAG TAC TTG ATC TGC AGA GAG GCC AGT ATC AGC ACT CTC TGC AGT CAA GCG GCC TAG GGC TAA GAT GGA GCC ACC | pBSK8499-9605/ΔSL2 |
| A-SL3 | AAA AAC TAG TAG TAC TCA CGG ACC TTT CAC AGC TAG CCG TGA CTA GGG CTA AGA TGG | pBSK8499-9605/ΔSL3 |
| SdelX | CCT TCC TGT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TAG TAC TAC TAG TCC CC | pBSK8499-9605/ΔSL3 |
| AdelX | GGG GAC TAG TAG TAC | pBSK8499-9605/Δ3′X |
| S26-A | CAA TAG GCC TTC CTG TTT AAA AAA AAA AAA AAA AAA AAA AAA AAT GGT GGC TCC ATC TTA GCC CTA GTC ACG GCT AGC TGT G | pBSK8499-9605/A26 |
| A26-A | CAC AGC TAG CCG TGA CTA GGG CTA AGA TGG AGC CAC CAT TTT TTT TTT TTT TTT TTT TTT TTT TAA ACA GGA AGG CCT ATT G | pBSK8499-9605/A26 |
| S26-G | CAA TAG GCC TTC CTG TTT GGG GGG GGG GGG GGG GGG GGG GGG GGT GGT GGC TCC ATC TTA GCC CTA GTC ACG GCT AGC TGT G | pBSK8499-9605/G26 |
| A26-G | CAC AGC TAG CCG TGA CTA GGG CTA AGA TGG AGC CAC CAC CCC CCC CCC CCC CCC CCC CCC CCC CAA ACA GGA AGG CCT ATT G | pBSK8499-9605/G26 |
| S26-C | CAA TAG GCC TTC CTG TTT CCC CCC CCC CCC CCC CCC CCC CCC CCT GGT GGC TCC ATC TTA GCC CTA GTC ACG GCT AGC TGT G | pBSK8499-9605/C26 |
| A26-C | CAC AGC TAG CCG TGA CTA GGG CTA AGA TGG AGC CAC CAG GGG GGG GGG GGG GGG GGG GGG GGG GAA ACA GGA AGG CCT ATT G | pBSK8499-9605/C26 |
| 3′C-Sbfl-up | CCC CCT GCA GGA | |
| 3′C-Sbfl-down | CTA GTC CTG CAG GGG G |
Numbers refer to the nucleotide position of HCV isolate Con1 (EMBL database accession no. AJ238799).
In vitro transcription, electroporation, and selection of G-418-resistant cell lines.
These methods have been described in detail elsewhere (35), and they were used with the following modifications. Plasmid DNA was restricted with AseI and ScaI and used for in vitro transcription under standard conditions. For the replicons with three or five additional cytidine residues at the 3′ end, constructs were restricted with SbfI and the four unpaired nucleotides (TGCA) were removed by treatment with DNA polymerase in the presence of high concentrations of nucleotides. In vitro transcripts were treated with DNase, and RNA was extracted with phenol, precipitated with ethanol, and dissolved in RNase-free water. RNA concentrations were determined by measuring the optical density at 260 nm, and the integrity of the transcripts was analyzed by denaturing formaldehyde agarose gel electrophoresis. Various amounts of selectable replicons (0.3 to 100 ng) were adjusted with total RNA from naive Huh-7 cells to a total amount of 10 μg and mixed with 400 μl of a suspension containing 107 Huh-7 cells per ml in Cytomix (35) without dimethyl sulfoxide (DMSO). After electroporation with a Gene Pulser system (Bio-Rad) at 960 μF and 270 V, cells were immediately transferred to 8 ml of complete DMEM without DMSO and seeded into a 100-mm-diameter culture dish. After 24 h, medium was replaced by complete DMEM supplemented with 500 μg of Geneticin (Life Technologies)/ml. Three to 4 weeks later, cells were fixed and stained with Coomassie brilliant blue (0.6 g/liter in 50% methanol-10% acetic acid).
Amplification of HCV replicons from cell lines, cloning, and sequence analysis.
Total RNA was prepared from cell lines by a single-step isolation method (10). About 1 μg of RNA was mixed with 50 pmol of primer A9605Sca/Spe in a total volume of 10 μl and heated for 10 min at 65°C. Reverse transcription was performed with 200 U of murine leukemia virus reverse transcriptase (Gibco Life Sciences) in a total volume of 20 μl as recommended by the manufacturer. After 1 h at 42°C, 1 to 5 μl was withdrawn and used for PCR with the Expand long-template PCR system (Roche) in accordance with the instructions of the manufacturer and primers A9605Sca/Spe and S9140. Amplified fragments were purified by preparative agarose gel electrophoresis and, after elution, were restricted with StuI and SpeI and inserted into vector pBSK-Stu. Nucleotide sequences were determined with a Thermo Sequenase fluorescence-labeled primer cycle sequencing kit with 7-deaza-dGTP (Amersham-Pharmacia Biotech, Freiburg, Germany) and IRD-41-labeled primers (MWG-Biotech, Ebersberg, Germany) in accordance with the instructions of the manufacturers. Reactions were analyzed on a Licor DNA sequencer 4000 (MWG-Biotech).
Determination of RNA half-life by Northern blotting.
Five to 10 μg of total RNA was denatured by treatment with glyoxal and analyzed by denaturing agarose gel electrophoresis and Northern blotting as described recently (31). Plus-strand replicon RNA was detected by hybridization with a 32P-labeled negative-sense riboprobe complementary to the HCV IRES and neo.
Transient replication assays with luciferase replicons.
Huh-7 cells were mixed with 5 to 7.5 μg of a luciferase replicon and used for electroporation as described above. After addition of 9 ml of complete DMEM, aliquots of the cell suspension were seeded in 3.5-cm-diameter culture dishes and harvested at 4, 24, 48, and 72 h. Prior to harvest, cells were washed three times with phosphate-buffered saline and then scraped off the plate into 350 μl of ice-cold lysis buffer (1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM dithiothreitol [DTT]). A 100-μl aliquot of the lysate was mixed with 360 μl of assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, 15 mM K2PO4, pH 7.8) and, after addition of 200 μl of a 200 μM luciferin stock solution, was measured in a luminometer (Lumat LB9507; Berthold, Freiburg, Germany) for 20 s. Values obtained with cells harvested 4 h after electroporation were used to normalize for the transfection efficiency.
In vitro translation and translation studies with transfected cells.
In vitro translations in rabbit reticulocyte lysates (Promega, Mannheim, Germany) were performed in mixture containing 8.75 μl of lysate, 0.25 μl of RNasin, 0.25 μl of an amino acid mixture without methionine, 1.5 μl of 35S-protein-labeling mixture, and 1.75 μl of RNA (corresponding to 0.5 μg), and after 1 h at 30°C proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. To determine the translation efficiency in cells, 7.5 μg of the bicistronic RNA was used for transfection of Huh-7 cells as described above. Three hours later, cells were lysed and luciferase activities were measured by using the dual-luciferase reporter assay system (Promega) according to the instructions of the manufacturer.
RESULTS
The characterization of sequences in the 3′ NTR required for RNA replication was performed with two classes of HCV replicons (Fig. 1A): first, replicons carrying the selectable marker neo, and second, replicons that allow the expression of the firefly luciferase reporter gene (luc). For selectable replicons, RNA replication was measured by determining the number of Geneticin-resistant colonies obtained after transfection of serial dilutions of in vitro transcripts into Huh-7 cells and subsequent selection. Replication efficiency was expressed as CFU per microgram of transfected RNA. For luc replicons, transfected cells were harvested at 4, 24, 48, and 72 h and luciferase activities in cell lysates were measured and normalized for transfection efficiency by using the 4-h value. Since, at this time point, luciferase activities are only determined by translation of the input RNA and are not influenced by replication, they can be used to determine transfection efficiency (31). Thus, for a given time point posttransfection, replication efficiency of the luciferase replicons is expressed as the percentage of reporter activity measured 4 h after transfection. The advantage of the selectable replicons was the high sensitivity and the possibility to select for revertants. In spite of some interassay variability, the luciferase replicons allowed the rapid analysis of a large number of mutants. Moreover, owing to the properties of this transient test, measurements of RNA replication were not affected by compensatory mutations or reversions.
The variable region in the 3′ NTR is not essential for RNA replication.
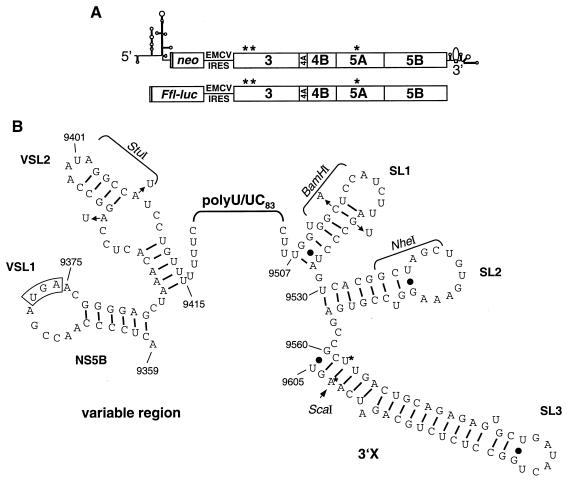
To facilitate genetic manipulations of the 3′ NTR, we first attempted to introduce convenient restriction sites in the immediate vicinity of the poly(U/UC) tract. Therefore, a recognition sequence for the restriction enzyme StuI was engineered into VSL2 of the variable region by introducing a uridine substitution at nucleotide position 9407 and a compensatory mutation at position 9394 in order to avoid a potential base pairing that might lead to a stabilization of the upper stem (Fig. 1B). Downstream of the poly(U/UC) tract we substituted adenosine for cytidine at nucleotide position 9513 to generate a BamHI recognition sequence and a compensatory guanosine-to-uridine transversion to preserve the stem structure. These mutations were inserted into parental replicon rep5.1 in the context of both the selectable marker (neo) and the luciferase reporter gene. Replicon 5.1 harbors three cell culture-adaptive mutations, which are located in NS3 (E1202G and T1280I) and NS5A (S2197P) and which increase RNA replication synergistically (31). As shown in Fig. 2A and C, the introduction of the StuI site into the variable region affected neither replication in the transient-replication assay nor the number of Geneticin-resistant colonies. Surprisingly, in spite of preserving the stem structure, the mutations introduced into SL1 that were required to generate the BamHI recognition sequence abolished replication in both assays, indicating that SL1 plays an important role in RNA replication. Thus, all subsequent manipulations of the 3′ NTR were performed with the replicon carrying only the StuI site. A naturally occurring recognition sequence for NheI present in SL2 was used for all further cloning steps (Fig. 1B).
FIG. 2.
The variable region in the 3′ NTR enhances RNA replication. (A) Representative result of a transient-replication assay with luciferase replicons carrying given engineered restriction sites in the variable region of the 3′ NTR. Cells were lysed at 4, 24, 48, and 72 h posttransfection, and luciferase activities were measured and corrected for transfection efficiency as determined from the 4-h value (set at 100%). Bars, means of quadruplicate determinations and error ranges. (B) Transient replication of luciferase replicons lacking part or the complete variable region (Δvar-9401 and Δvar-9415, respectively). Transfected cells were analyzed as for panel A. (C) Effects of mutations in the variable region of selectable replicons on the number of Geneticin-resistant colonies. Huh-7 cells were transfected with neo replicons carrying the given mutations in the 3′ NTR, subjected to Geneticin selection, and, after about 3 weeks, fixed and stained. The CFU per microgram of RNA (mean ± error range) as determined by transfection of serial dilutions of a given in vitro transcript are given below each culture dish. Representative results obtained after transfection of 100 ng of each RNA are shown. Wt, wild-type replicon (rep5.1); GND and Δ5B, inactive replicons with a single amino acid substitution in motif C of the NS5B RdRp (for luciferase replicons) or a 10-amino-acid deletion in the same motif (for neo replicons), respectively.
It has recently been shown that a mutant with a deletion of the 5′-proximal part of the variable region introduced into an infectious genome is viable in vivo (62). However, this approach did not allow firm conclusions about the efficiency of RNA replication and whether the full complement of the variable region is dispensable for RNA replication, too. Therefore, we generated two different replicons lacking either a part of the variable region (nucleotides 9375 to 9401) or the complete variable region (nucleotides 9375 to 9415) and tested them for RNA replication in the transient-replication assay. As shown in Fig. 2B, both mutants still replicated in transfected Huh-7 cells but at significantly reduced efficiency in comparison to that at which the parental replicon replicates. The analogous result was found when these mutations were introduced into selectable replicons (Fig. 2C). The value of CFU per microgram of RNA was reduced about 10-fold for the partial deletion, and up to 100-fold for complete deletion, of the variable region. It should be noted that, due to technical reasons applicable to the latter replicon, the poly(U/UC) tract was replaced by a 52-nucleotide homopolymeric uridine sequence. However, as described below, this mutation does not affect RNA replication. In summary, these results suggest that the variable region in the 3′ NTR is not essential for but enhances RNA replication.
Importance of the poly(U/UC) tract for RNA replication.
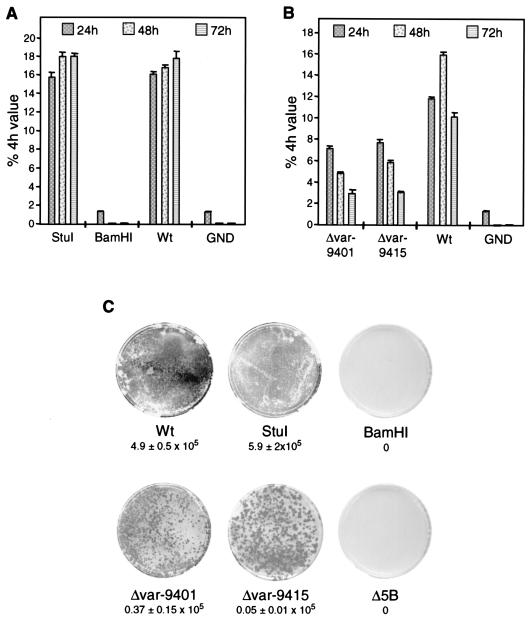
In spite of a significant heterogeneity of the length and composition of the poly(U/UC) tract, its strict conservation in all HCV genomes indicates that it plays an important role in the viral life cycle, in particular for RNA replication. To address this assumption experimentally, we analyzed whether the poly(U/UC) tract is essential and determined the minimal length required for efficient RNA replication. Therefore, we constructed a series of replicons in which the sequence between VSL2 and SL1 (nucleotides 9415 to 9507; Fig. 1B) was replaced by homopolymeric stretches of 46, 26, 6, or 1 uridine residue. As shown in Fig. 3A and B, within the variations of our assays, RNAs with a polyuridine tract of 46 or 26 residues replicated as efficiently as the parental replicon with a poly(U/UC) tract of 83 nucleotides. In contrast, the replicon with a hexauridine tract or only one uridine residue did not replicate, as determined in the transient-replication assay. However, when replication was analyzed in the context of a selectable neo replicon, a low number of Geneticin-resistant colonies were obtained with the replicon carrying the hexauridine sequence (<50 CFU/μg of RNA) whereas no colonies were obtained with the U1 replicon. This low efficiency of colony formation observed with the U6 replicon suggested that the RNAs replicating in these cells corresponded to pseudorevertants. To confirm this assumption, six independent colonies were expanded, replicon RNAs were amplified by reverse transcription-PCR (RT-PCR) and cloned, and the 3′ NTRs of two clones from each colony were sequenced. Interestingly, the replicons recovered from these colonies all carried a poly(U/UC) tract of 40 or more nucleotides (Fig. 3C). Four clones with homopolymeric sequences of between 44 and 52 uridine residues instead of the original 6 uridines were recovered, whereas an interspersed cytidine residue was found with two other clones in the context of homouridine sequences of 52 and 48 nucleotides (clones MP4I and MP2II, respectively). It should be noted that, with the exception of a single mutation at nucleotide position 9570, the sequences of the variable region and of 3′X were fully conserved with all replicons analyzed. These data show that the HCV RNAs recovered from cell lines after transfection with the U6 replicon all corresponded to pseudorevertants.
FIG. 3.
Determination of the minimal length of the poly(U/UC) tract required for RNA replication. (A) Result of a transient-replication assay using luciferase replicons that carry a homopolymeric uridine tract of 1, 6, 26, or 46 nucleotides. Wt and GND are as defined for Fig. 2. (B) Number of Geneticin-resistant colonies obtained after transfection of selectable replicons with the same modifications in the 3′ NTR as in panel A. Representative results obtained after transfection of 100 ng of in vitro transcript are shown. For further details see the legend to Fig. 2. (C) Sequence analysis of the poly(U/UC) tracts of replicons isolated from Huh-7 cells that had been transfected with U6 replicons and subjected to selection with Geneticin. From six independent colonies replicon RNA was amplified by RT-PCR and cloned, and two clones from each colony were sequenced. The sequence of only one clone of each colony is shown below the poly(U/UC) tracts of the wild-type replicon and the U6 mutant. Dashes, deletions; boldface, sequences of VSL2 and SL1.
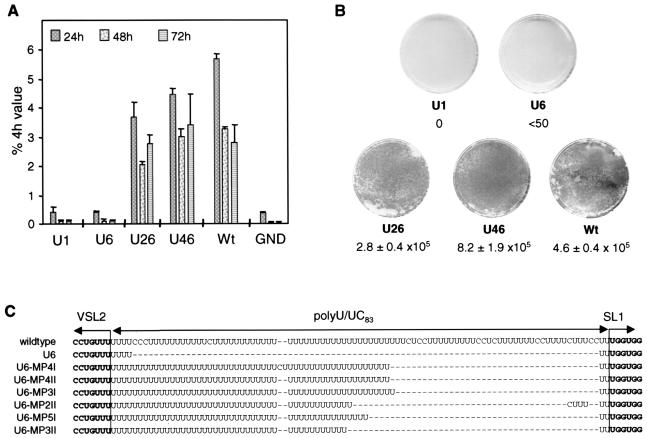
Next we wanted to analyze whether homopolymeric sequences other than uridine could functionally replace the poly(U/UC) tract. Luciferase replicons carrying homopolymers of 26 guanosine or adenosine or cytosine residues were constructed and tested in the transient-replication assay. The results in Fig. 4A show that only the U26 RNA replicated efficiently whereas all other RNAs were not amplified. Given the higher sensitivity achieved by testing mutants with selectable RNAs, neo replicons that carried the various homopolymeric sequences in the 3′ NTR were generated. However, in several independent experiments no colonies were obtained with A26, G26, or C26 replicons (data not shown). In summary, these results conclusively show that a homopolymeric stretch of at least 26 uridine residues is required for HCV RNA replication and that other homopolymeric sequences cannot functionally replace homouridine.
FIG. 4.
(A) Only a homopolymeric uridine tract in the 3′ NTR supports RNA replication. Luciferase replicons carrying 26-nucleotide homopolymeric guanosine, adenosine, uridine, or cytidine tracts were transfected into Huh-7 cells, and RNA replication was determined after 24, 48, and 72 h. Bars, mean values of quadruplicate determinations and the error ranges. Values are normalized for transfection efficiency as determined from the luciferase activities measured 4 h after transfection (set at 100%). Wt, wild-type replicon; GND, replication-defective replicon that was used as a negative control. (B) Full-length 3′X is required for RNA replication. Luciferase replicons that lack SL1, SL2, or SL3, or the complete 3′X were transfected in parallel with the parental replicon (Wt) or the replication-deficient mutant (GND) into Huh-7 cells, and transient replication was determined as described above.
RNA replication requires full-length 3′X.
The most highly conserved region in the 3′ NTR of HCV is 3′X. Three stem-loop structures (SL1 to -3) were suggested by secondary structure prediction, and SL3 was confirmed by structure probing (7). To analyze their importance for RNA replication, we first constructed a replicon that lacked the complete 3′X region. This RNA terminated with a 50-nucleotide homouridine tract and the sequence AGU, which was added to preserve the restriction site for ScaI required to generate runoff in vitro transcripts. As shown in Fig. 4B, this construct, which was designated Δ3′X, did not replicate in the transient-replication assay, and we were also unable to generate Geneticin-resistant colonies after transfection of the corresponding neo replicon into Huh-7 cells (not shown). To analyze whether individual stem-loops in 3′X are dispensable for replication, three replicons that lacked either SL1, SL2, or SL3 were generated. To facilitate the cloning procedure, these deletions were introduced into constructs that carried either a 46- (ΔSL2 and ΔSL3) or a 50-nucleotide homopolymeric uridine stretch (ΔSL1 and Δ3′X). Moreover, while ΔSL1 and ΔSL2 corresponded to exact deletions of the respective stem-loops, ΔSL3 terminated with SL2 to which the dinucleotide sequence GU was added in order to preserve the ScaI restriction site. As shown in Fig. 4B, no replication was found with these three deletion mutants when they were tested in the transient-replication assay. In agreement with this observation, no Geneticin-resistant colonies were obtained when the mutations were introduced into selectable RNAs (not shown). These data demonstrate that a full-length 3′X is essential for efficient RNA replication.
Influence of mutations in the 3′ NTR on RNA stability and translation.
In principle the lack of RNA replication found with the mutants described above could also be explained by a destabilization of the RNA that was caused by the 3′ NTR alterations. For instance, Fang and coworkers suggested that, at least for some HCV isolates, 3′X increases RNA stability (15). Therefore, we measured the half-lives of the neo replicon RNAs carrying the various deletions and substitutions in the 3′ NTR after transfection into Huh-7 cells. Total RNA was prepared 2, 4, and 8 h after transfection, and the amount of replicon RNA was quantified by Northern blotting. Values were corrected for transfection efficiency as determined from the 2-h value and normalized for RNA amounts loaded onto the gel by using the beta-actin signal. As can be seen from the representative experiment shown in Fig. 5, within the accuracy of this assay none of the mutations introduced into the 3′ NTR affected RNA stability. In all cases replicon RNA was clearly detected at 4 and 8 h posttransfection, with only minor differences between the mutants. Although in this experiment the amounts of replicons ΔSL1 and ΔSL3 were lowest at 8 h posttransfection, the amount of the replicon lacking 3′X was as large as the amount of the one with parental RNA (wild type). Moreover, the amounts of wild-type and replication-deficient replicon (Δ5B) RNA were comparable at 8 h post transfection, showing that at this time point RNA replication did not affect our measurement. Thus, the inability of the mutants to replicate was not due to a decrease of RNA stability caused by mutations in the 3′ NTR.
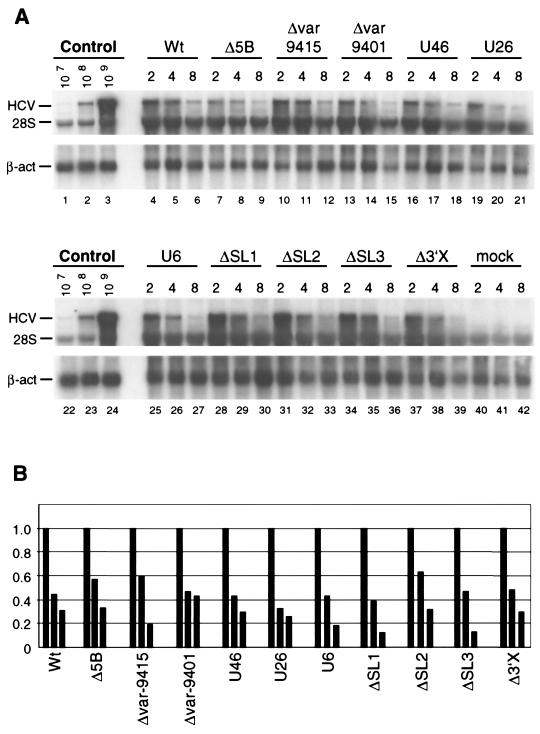
FIG. 5.
Stability of replicon RNAs is not affected by mutations in the 3′ NTR. (A) Huh-7 cells were transfected with neo replicons carrying given mutations in the 3′ NTR, and after 2, 4, and 8 h cells were lysed and total RNA was prepared and analyzed by Northern blotting. Given amounts of in vitro transcripts to which total RNA from naive Huh-7 cells was added as the carrier served as size markers (lanes 1 to 3 and 22 to 24). The positions of the replicon, beta-actin (β-act), and 28S rRNA are indicated at the left. (B) The amounts of replicon RNA were quantified by phospho imaging, and values were normalized by using the signals obtained with the beta-actin-specific probe. RNA amounts detected 2 h after transfection were set at 1. Transfections performed with the parental replicon (Wt) and the one carrying a deletion of the NS5B active site (Δ5B) served as controls. Note that, even 8 h after transfection, the amounts of these replicons are comparable, showing that up to this time point the measurement was not influenced by RNA replication.
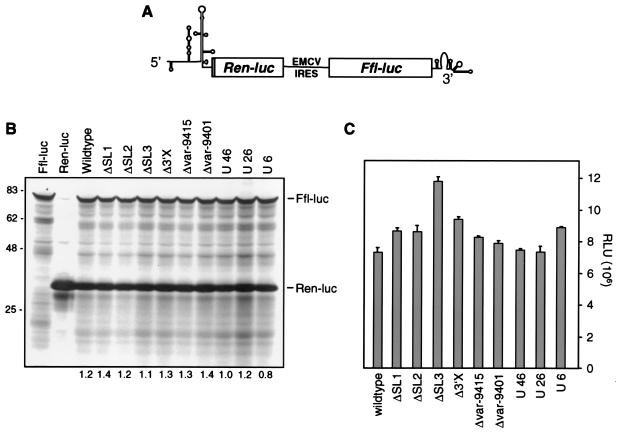
Apart from playing a role in RNA stability, the 3′ NTR may increase RNA translation, although this possibility is discussed controversially (24, 42). To examine whether the mutations introduced into the 3′ NTR affect RNA translation, we generated a bicistronic construct in which the Renilla luciferase reporter gene was fused to the HCV IRES whereas the firefly luciferase gene was translated under the control of the EMCV IRES (Fig. 6A). This design corresponded to a bicistronic subgenomic HCV replicon with the exception that the coding region for the replicase was replaced by a second reporter, which served as an internal control for the quality of the in vitro transcripts. Moreover, the series of constructs permitted the measurement of HCV IRES-mediated translation independent from RNA replication. Upon introduction of the various mutations described above into the 3′ NTR, in vitro transcripts were analyzed by in vitro translation using rabbit reticulocyte lysates. Proteins were radiolabeled with [35S]methionine-cysteine, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and quantified by phospho imaging. As shown in Fig. 6B, comparable amounts of Renilla luciferase translated from the HCV IRES were found with all RNAs irrespective of the particular mutation introduced into the 3′ NTR. In addition, the analogous result was found when the corresponding neo replicon RNAs were analyzed by in vitro translation in the same way and when NS3 was used as an internal standard (not shown). To test whether the mutations in the 3′ NTR also had no effect on RNA translation in cells, the bicistronic reporter constructs were transfected into Huh-7 cells and luciferase activities were measured after 3 h. The results in Fig. 6C show that only minor differences in the expression of Renilla luciferase could be observed. In this representative result, the ΔSL3 replicon was the most efficient, but this was not the case in several repetitions (not shown). Finally, the analogous result was found when the corresponding luciferase replicons were transfected and expression of both firefly luciferase and NS5B was determined (data not shown). In summary these data show that the mutations introduced into the 3′ NTR did not affect translation from the HCV IRES.
FIG. 6.
Sequences in the 3′ NTR do not affect RNA translation from the HCV IRES. (A) Structure of the bicistronic reporter construct carrying the luciferase genes from Renilla (Ren-luc) or the firefly (Ffl-luc) under control of the HCV or the EMCV IRES, respectively. Bars, reporter genes; solid line, EMCV IRES. The HCV NTRs are given with their secondary structures. (B) In vitro translation of bicistronic reporter constructs carrying the given mutations in the HCV 3′ NTR in rabbit reticulocyte lysate. Results obtained with RNAs encoding only the firefly or Renilla luciferase are shown in the two left lanes. Numbers below the lanes are the ratios between the amounts of the two reporters as determined by phospho imaging. The ratio found with the replicon carrying a homopolymeric uridine tract of 46 nucleotides was set at 1. Luciferases are identified at the right; the sizes of protein molecular mass standards are given at the left. (C) Translation efficiencies from the HCV IRES after transfection of given bicistronic RNAs into Huh-7 cells. Luciferase activities in lysates prepared from cells 3 h post transfection were measured. Values refer to relative light units (RLU) of Renilla luciferase after normalization for transfection efficiency by using the firefly luciferase activity.
Influence of additional 3′-terminal nucleotides on RNA replication.
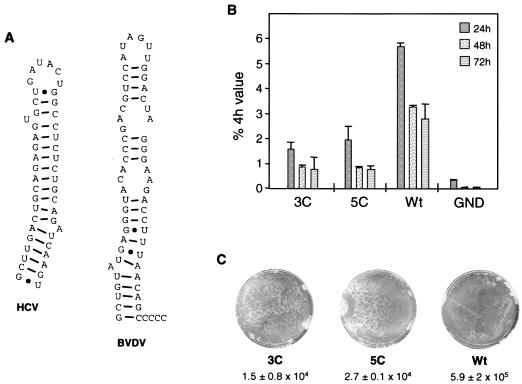
It has recently been shown that NS5B of HCV can initiate RNA synthesis de novo, and this mechanism is thought to operate also in vivo (38, 43, 66). The most efficient de novo initiation was found with GTP as the initiating nucleotide and templates that carry a 3′-terminal cytidylate (66). De novo initiation with a preference for GTP has also been described for NS5B of BVDV (26), underlining the close relationship between HCV and the pestiviruses. However, while the 3′ end of the BVDV genome carries three to five unpaired cytidylates, consistent with de novo initiation with GTP, the last base of the HCV genome is uracil base paired with a guanine (Fig. 7A). Although it has been shown that HCV NS5B can initiate RNA synthesis de novo also with ATP, this process is much less efficient than initiation with GTP (38). In light of this result and because of the higher replication efficiency of BVDV, we wanted to know whether the addition of a few cytidines to the 3′ end of the HCV genome would increase RNA replication. Therefore, we generated neo and luc replicons that carried three or five cytidines fused to the terminal uridine of SL3 and tested them for replication competence in Huh-7 cells. However, as shown in Fig. 7, both RNAs replicated with a much lower efficiency than the parental replicon in the transient-replication assay and they reduced the efficiency of colony formation by about 1 order of magnitude. This result shows that a 3′-terminal uridine is important for efficient RNA replication, and it highlights the fact that HCV and BVDV have different requirements for initiation of minus-strand RNA synthesis.
FIG. 7.
Importance of the correct sequence at the very 3′ end of the HCV replicon for RNA replication. (A) Secondary structures of the terminal stem-loop of HCV (7) and BVDV (64). (B) Transient replication of luciferase replicons carrying three or five extra cytidine residues at the 3′ end of the HCV 3′ NTR. (C) Colony formation of selectable replicons that carry 3′-terminal extensions. For further details, including the definitions of Wt and GND, see the legend to Fig. 2.
DISCUSSION
The study described here provides a detailed map of the sequences in the 3′ NTR crucial for RNA replication and corroborates and extends the results obtained in two different in vivo studies. In the first one, Yanagi and coworkers (62) tested several deletion mutants sequentially for infectivity in chimpanzees. They found that a deletion of 3′X or the poly(U/UC) tract rendered the genome nonviable whereas a partial deletion of the variable region was tolerated. Similar findings were made by Kolykhalov and coworkers (30) by showing that HCV genomes lacking 3′X or carrying a deletion of SL1 and SL2 are not infectious. Our studies described here are completely in line with these observations, but they disagree with two earlier reports suggesting that HCV genomes lacking 3′X and most of the poly(U/UC) region replicate in the human hepatoma cell lines HepG2 and Huh-7. While the reasons for this discrepancy are not clear, we note that even with selectable replicons lacking 3′X or the poly(U/UC) tract no colonies were obtained. Since this approach is highly sensitive and also allows the detection of pseudorevertants, our data provide strong evidence that both 3′X and a homouridine sequence are indispensable for HCV RNA replication. Moreover, for several other plus-strand RNA viruses, sequences at the 3′ end of the genome were shown to play crucial roles in RNA replication. For instance, the 3′ NTR of GB virus B, a virus that is very closely related to HCV, also has a tripartite structure composed of a variable region, a poly(U) tract of 9 to 30 nucleotides, and a conserved downstream sequence that can form a very stable 3′-terminal stem-loop involving the last 47 nucleotides (8, 50). Upon intrahepatic inoculation of tamarins (Saguinus spp.), only genomes with the complete 3′ NTR were viable, showing that this region is essential for infectivity (8, 51). For pestivirus BVDV, it was shown that mutations within the 3′-terminal stem-loop lead to the partial or complete inhibition of RNA replication of a subgenomic replicon (64). Finally, very similar findings were made for flaviviruses such as dengue virus (41) and tick-borne encephalitis virus (39). In summary, these data provide conclusive evidence that sequences in the 3′ termini of flavivirus genomes are absolutely essential for RNA replication in cell culture and in vivo.
In spite of a significant heterogeneity of the poly(U/UC) tract length, all HCV isolates studied thus far carry this sequence element in the 3′ NTR. Its conservation and the fact that a deletion of the poly(U/UC) tract completely blocked replication clearly demonstrate its importance for RNA replication. Interestingly, replicon RNAs carrying a hexauridine sequence replicated very inefficiently and only pseudorevertants with about 40-nucleotide uridine stretches could be recovered. This result appears to contradict the observation that a replicon with a homopolymeric uridine tract of 26 nucleotides replicated as efficiently as the wild type. However, the actual lengths of the poly(U) sequences of the pseudorevertants replicating in cells are difficult to determine due to errors that may be introduced during RT-PCR amplification and the cycle sequencing reaction. In contrast, the replicon with a 26-nucleotide homouridine tract was generated by using an oligonucleotide with a defined length (S9402StuI-30U-NheI; Table 1). Thus, our data show that a homouridine sequence of 26 residues can functionally replace the poly(U/UC) tract.
Currently, we can only speculate about the contribution of this polypyrimidine sequence for RNA replication, but it is interesting that both the NS5B RdRp and the NS3 helicase preferentially bind to uridine homopolymers (21, 25, 34). Therefore, this structurally flexible region may capture and position the replicase enzymes in close proximity of the site where minus-strand RNA synthesis is initiated. Moreover, efficient binding of NS3 and activation of the NTPase require homopolyuridine of at least 12 to 15 nucleotides (45), consistent with our results showing that a uridine homopolymer with a length between 6 and 26 nucleotides is required for RNA replication. Finally, cellular proteins, such as PTB, that bind to the poly(U/UC) tract most likely participate in the formation of a functional ribonucleoprotein complex, required for synthesis of minus-strand RNA (11, 18).
The observation that replicons lacking part or all of the variable region are replication competent, albeit with a low efficiency, fits well with the observations made with similar HCV mutants in vivo (62). In this respect it is interesting that variable sequences located in the 3′ NTR and upstream of a conserved region are also found in the genomes of pesti- and flaviviruses. Interestingly, comparable to what we describe here for HCV, for some flaviviruses such as Kunjin and tick-borne encephalitis viruses it was shown that deletions within these variable regions do not affect, or only moderately affect, RNA replication in cell culture and in vivo (27, 39, 41). Thus, the presence of a variable region in the 3′ NTR that is nonessential for RNA replication is a feature conserved at least among some members of the Flaviviridae family. The reduction of replication observed with HCV replicons suggests that the variable region plays some other, perhaps regulatory, role, but its precise function remains to be determined.
Essential stem-loop structures at the termini of many plus-strand RNA viral genomes represent binding sites for viral and cellular factors. These proteins are involved in several distinct steps of the viral life cycle, such as RNA replication (65) and genome packaging (48), or they facilitate interactions between sequences in the 5′ and 3′ termini of the viral genome (22). For HCV, several reports have demonstrated the binding of PTB to sequences in the 3′ NTR (11, 18, 23, 37, 57), but the importance of this interaction for RNA replication, if any, is not known. Our observation that the mutations introduced into SL1 that were required to create the BamHI recognition sequence completely blocked RNA replication suggests that PTB plays an important role. Interestingly, PTB can also bind to the 5′ NTR of HCV (1), and, based on the ability of this protein to dimerize, it may mediate a cross talk between the termini of the viral genome. Apart from PTB and other cellular factors (18, 37, 53), the binding of viral proteins NS3 and NS5B to sequences in the 3′ terminal region of the genome has been observed, but the functional relevance of these interactions is not known (3, 9, 43).
Several conflicting reports as to the role of the 3′ NTR in RNA stability and translation from the HCV IRES have been published. For instance, Murakami and coworkers (42) found that sequences in the 3′ NTR downregulate translation from the HCV IRES when tested in rabbit reticulocyte lysates. However, in the study by Ito et al. (24) it was suggested that RNAs carrying 3′X are translated three- to fivefold more efficiently in rabbit reticulocyte lysates and two- to threefold more efficiently in transfected Huh-7 cells. In the latter report monocistronic RNAs, in which a reporter gene was clamped between the HCV or the EMCV IRES and 3′X, were used. Since mutations affecting the PTB binding site in 3′X reduced the translation-enhancing effect, it was suggested that PTB is required for this effect, but it has not been shown whether the same is true in the context of the complete 3′ NTR. As shown in the present study, deletions and substitutions introduced into the 3′ NTR of either the replicon or a bicistronic reporter construct did not affect significantly the efficiency of translation from the HCV IRES. Ito et al. suggested that the 3′ NTR also enhances RNA translation from the EMCV IRES (24). Since we normalized our translation efficiencies by using firefly luciferase translated from the EMCV IRES (Fig. 6), one might argue that our approach would not reveal a difference in HCV IRES activity. However, even when the values obtained with the Renilla luciferase that was translated from the HCV IRES were not normalized, we observed at most a twofold difference among the various 3′ NTR mutants. For instance, in some experiments some of the mutants in which a single stem-loop in 3′X was deleted displayed a 1.5- to 2-fold-reduced translation efficiency of the HCV IRES, but this was not the case when 3′X was removed completely (data not shown). These results suggest that some minor experimental variations rather than the loss of PTB binding to 3′X were responsible for this effect. In summary, our data clearly show that the mutations introduced into the 3′ NTR primarily affected RNA replication.
As shown in this study, the HCV replicon system is a powerful tool that permits detailed studies of RNA replication in cell culture. In our opinion, the combination of transient-replication assays that are based on reporter-gene constructs with analyses of selectable RNAs are best suited. On one hand transient-replication assays allow a rapid test of a large number of mutants, which is important when using reverse genetics; on the other hand work with selectable replicons permits the isolation of pseudorevertants that can be even more informative than the mutant itself. By using these two approaches we established the first detailed map of sequences in the 3′ NTR required for RNA replication. It is obvious that the identification of cellular and viral proteins binding to these sequences and the definition of their roles in individual steps of RNA translation and replication will be the next important tasks.
Acknowledgments
We thank Ulrike Herian for excellent technical assistance and Nicole Krieger, Volker Lohmann, and Thomas Pietschmann for critical reading of the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB490, Teilprojekt A2) and the European Community (QLK2-1999-00356).
REFERENCES
- 1.Ali, N., and A. Siddiqui. 1995. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J. Virol. 69:6367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Banerjee, R., and A. Dasgupta. 2001. Specific interaction of hepatitis C virus protease/helicase NS3 with the 3′-terminal sequences of viral positive- and negative-strand RNA. J. Virol. 75:1708-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 5.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukh, J., C. L. Apgar, and M. Yanagi. 1999. Toward a surrogate model for hepatitis C virus: an infectious molecular clone of the GB virus-B hepatitis agent. Virology 262:470-478. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, J.-U., M.-F. Chang, and S. C. Chang. 1999. Specific interaction between the hepatitis C virus NS5B RNA polymerase and the 3′ end of the viral RNA. J. Virol. 73:7044-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Chung, R. T., and L. M. Kaplan. 1999. Heterogeneous nuclear ribonucleoprotein I (hnRNP-I/PTB) selectively binds the conserved 3′ terminus of hepatitis C viral RNA. Biochem. Biophys. Res. Commun. 254:351-362. [DOI] [PubMed] [Google Scholar]
- 12.Dash, S., A. B. Halim, H. Tsuji, N. Hiramatsu, and M. A. Gerber. 1997. Transfection of HepG2 cells with infectious hepatitis C virus genome. Am. J. Pathol. 151:363-373. [PMC free article] [PubMed] [Google Scholar]
- 13.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Failla, C., L. Tomei, and R. De Francesco. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang, J. W., and R. W. Moyer. 2000. The effects of the conserved extreme 3′ end sequence of hepatitis C virus (HCV) RNA on the in vitro stabilization and translation of the HCV RNA genome. J. Hepatol. 33:632-639. [DOI] [PubMed] [Google Scholar]
- 16.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gale, M. J., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 18.Gontarek, R. R., L. L. Gutshall, K. M. Herold, J. Tsai, G. M. Sathe, J. Mao, C. Prescott, and A. M. Del Vecchio. 1999. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3′NTR of the HCV RNA genome. Nucleic Acids Res. 27:1457-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67:2832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gwack, Y., D. W. Kim, J. H. Han, and J. Choe. 1996. Characterization of RNA binding activity and RNA helicase activity of the hepatitis C virus NS3 protein. Biochem. Biophys. Res. Commun. 225:654-659. [DOI] [PubMed] [Google Scholar]
- 22.Huang, P., and M. M. Lai. 2001. Heterogeneous nuclear ribonucleoprotein a1 binds to the 3′-untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 75:5009-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, T., and M. C. Lai. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J. Virol. 71:8698-8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ito, T., S. M. Tahara, and M. C. Lai. 1998. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 72:8789-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanai, A., K. Tanabe, and M. Kohara. 1995. Poly(U) binding activity of hepatitis C virus NS3 protein, a putative RNA helicase. FEBS Lett. 376:221-224. [DOI] [PubMed] [Google Scholar]
- 26.Kao, C. C., A. DelVecchio, and W. Zhong. 1999. De novo initiation of RNA synthesis by a recombinant Flaviviridae RNA-dependent RNA polymerase. Virology 253:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, D. W., Y. Gwack, J. H. Han, and J. Choe. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 215:160-166. [DOI] [PubMed] [Google Scholar]
- 29.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolykhalov, A. A., K. Mihalik, S. M. Feinstone, and C. M. Rice. 2000. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 33.Lin, C., B. M. Pragai, A. Grakoui, J. Xu, and C. M. Rice. 1994. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J. Virol. 68:8147-8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohmann, V., F. Körner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohmann, V., F. Körner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohmann, V., F. Körner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 37.Luo, G. 1999. Cellular proteins bind to the poly(U) tract of the 3′ untranslated region of hepatitis C virus RNA genome. Virology 256:105-118. [DOI] [PubMed] [Google Scholar]
- 38.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandl, C. W., H. Holzmann, T. Meixner, S. Rauscher, P. F. Stadler, S. L. Allison, and F. X. Heinz. 1998. Spontaneous and engineered deletions in the 3′ noncoding region of tick-borne encephalitis virus: construction of highly attenuated mutants of a flavivirus. J. Virol. 72:2132-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manns, M. P., M. Cornberg, and H. Wedemeyer. 2001. Current and future treatment of hepatitis C. Indian J. Gastroenterol. 20(Suppl. 1):C47-C51. [PubMed] [Google Scholar]
- 41.Men, R., M. Bray, D. Clark, R. M. Chanock, and C. J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami, K., M. Abe, T. Kageyama, N. Kamoshita, and A. Nomoto. 2001. Down-regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3′ untranslated region of RNA. Arch. Virol. 146:729-741. [DOI] [PubMed] [Google Scholar]
- 43.Oh, J. W., T. Ito, and M. C. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. T. Jackson, and C. T. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Preugschat, F., D. R. Averett, B. E. Clarke, and D. T. Porter. 1996. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J. Biol. Chem. 271:24449-24457. [DOI] [PubMed] [Google Scholar]
- 46.Pringle, C. R. 1999. Virus taxonomy-1999. The universal system of virus taxonomy, updated to include the new proposals ratified by the International Committee on Taxonomy of Viruses during 1998. Arch. Virol. 144:421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed, K. E., and C. M. Rice. 2000. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr. Top. Microbiol. Immunol. 242:55-84. [DOI] [PubMed] [Google Scholar]
- 48.Reusken, C. B., L. Neeleman, and J. F. Bol. 1994. The 3′-untranslated region of alfalfa mosaic virus RNA 3 contains at least two independent binding sites for viral coat protein. Nucleic Acids Res. 22:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rijnbrand, R., P. J. Bredenbeek, P. C. Haasnoot, J. S. Kieft, W. J. Spaan, and S. M. Lemon. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA. 7:585-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sbardellati, A., E. Scarselli, L. Tomei, A. S. Kekulé, and C. Traboni. 1999. Identification of a novel sequence at the 3′ end of the GB virus B genome. J. Virol. 73:10546-10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sbardellati, A., E. Scarselli, E. Verschoor, A. De Tomassi, D. Lazzaro, and C. Traboni. 2001. Generation of infectious and transmissible virions from a GB virus B full-length consensus clone in tamarins. J. Gen. Virol. 82:2437-2448. [DOI] [PubMed] [Google Scholar]
- 52.Spangberg, K., L. Goobar-Larsson, M. Wahren-Herlenius, and S. Schwartz. 1999. The La protein from human liver cells interacts specifically with the U-rich region in the hepatitis C virus 3′ untranslated region. J. Hum. Virol. 2:296-307. [PubMed] [Google Scholar]
- 53.Spangberg, K., L. Wiklund, and S. Schwartz. 2000. HuR, a protein implicated in oncogene and growth factor mRNA decay, binds to the 3′ ends of hepatitis C virus RNA of both polarities. Virology 274:378-390. [DOI] [PubMed] [Google Scholar]
- 54.Spangberg, K., L. Wiklund, and S. Schwartz. 2001. Binding of the La autoantigen to the hepatitis C virus 3′ untranslated region protects the RNA from rapid degradation in vitro. J. Gen. Virol. 82:113-120. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka, T., N. Kato, M. J. Cho, and K. Shimotohno. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215:744-749. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsuchihara, K., T. Tanaka, M. Hijikata, S. Kuge, H. Toyoda, A. Nomoto, N. Yamamoto, and K. Shimotohno. 1997. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J. Virol. 71:6720-6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood, J., R. M. Frederickson, S. Fields, and A. H. Patel. 2001. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J. Virol. 75:1348-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamada, N., K. Tanihara, A. Takada, T. Yorihuzi, M. Tsutsumi, H. Shimomura, T. Tsuji, and T. Date. 1996. Genetic organization and diversity of the 3′ noncoding region of the hepatitis C virus genome. Virology 223:255-261. [DOI] [PubMed] [Google Scholar]
- 62.Yanagi, M., M. St. Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoo, B. J., M. J. Selby, J. Choe, B. S. Suh, S. H. Choi, J. S. Joh, G. J. Nuovo, H. S. Lee, M. Houghton, and J. H. Han. 1995. Transfection of a differentiated human hepatoma cell line (Huh7) with in vitro-transcribed hepatitis C virus (HCV) RNA and establishment of a long-term culture persistently infected with HCV. J. Virol. 69:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, H., C. W. Grassmann, and S. E. Behrens. 1999. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J. Virol. 73:3638-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, W., and J. L. Leibowitz. 1995. A conserved motif at the 3′ end of mouse hepatitis virus genomic RNA required for host protein binding and viral RNA replication. Virology 214:128-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]