Abstract
Oligonucleotide directed misfolding of RNA (ODMiR) uses short oligonucleotides to inhibit RNA function by exploiting the ability of RNA to fold into different structures with similar free energies. It is shown that the 2′-O-methyl oligonucleotide, m(CAGCCUACCCGG), can trap Escherichia coli RNase P RNA (M1 RNA) in a nonfunctional structure in a transcription mixture containing RNase P protein (C5 protein). At about 200 nM, the 12-mer thus inhibits 50% of pre-tRNA processing by RNase P. Roughly 10-fold more 12-mer is required to inhibit RNase P containing full-length, renatured RNase P RNA. Diethyl pyrocarbonate modification in the presence of 12-mer reveals increased modification of sites in and interacting with P4, suggesting a structural rearrangement of a large pseudoknot important for catalytic activity. Thus, the ODMiR method can be applied to RNAs even when folding is facilitated by a cognate protein.
Keywords: RNase P, RNA folding, antisense oligonucleotides, misfolding
INTRODUCTION
RNA is becoming an attractive target for therapeutic intervention (Pearson and Prescott 1997; Hermann and Westhof 1998) as its roles in more processes are being discovered. For example, rRNA is the catalytic portion of the ribosome (Ban et al. 2000), and small interfering (si) RNAs regulate gene expression (Fire et al. 1998; Montgomery et al. 1998; Nykanen et al. 2001). Oligonucleotides are one promising class of therapeutics to target RNA. Oligonucleotides can be designed from simple base-pairing rules, analogs are synthetically accessible (Freier and Altmann 1997), and their pharmacokinetic properties are relatively independent of sequence (Crooke et al. 1996).
Currently, oligonucleotide-based therapeutics are being used clinically for treatment of cytomegalovirus (Vitravene; Galderisi et al. 1999) and hematological cancers (Genasense; Stein 2001). Often, oligonucleotide therapeutics use the antisense approach to target RNA and are typically 20 nt long. Even though antisense sequences of this length are effective in vivo, there are potential disadvantages. These include cost of synthesis and side effects due to binding with mismatches to bystander RNAs. Studies have shown, however, that RNA function can be inhibited in vitro with shorter oligonucleotides (Testa et al. 1999; Disney et al. 2001; Childs et al. 2002).
Previously (Childs et al. 2002), we described inhibition of the Candida albicans group I self-splicing intron by oligonucleotide directed misfolding of RNA (ODMiR). The ODMiR method uses a short oligonucleotide to trap RNA in a nonfunctional structure in a transcription mixture. Kinetic traps that cause inactivation of function have been detected during renaturation of other catalytic RNAs including the hepatitis delta virus (Been et al. 1992; Chadalavada et al. 2000, 2002), the Tetrahymena thermophila group I intron (Walstrum and Uhlenbeck 1990; Celander and Cech 1991; Pan and Woodson 1998; Russell et al. 2002), and the hammerhead ribozyme (Fedor and Uhlenbeck 1990). Inactivation is often the result of misfolding of secondary structure. In some cases, proteins are known to facilitate folding of an RNA into its active conformation in an RNA–protein complex (Lambowitz and Perlman 1990; Noller et al. 1992; Weeks and Cech 1995a,b; Zhang et al. 1995; Myers et al. 1996; Clodi et al. 1999; Webb et al. 2001). Here, we describe using short oligonucleotides to induce misfolding of Escherichia coli RNase P RNA (M1 RNA) in a transcription mixture containing RNase P protein (C5 protein; Kole and Altman 1979). Thus, the ODMiR method can be used to inhibit function of an RNA even in the presence of its cognate protein.
RNase P RNAs are responsible for the site-specific removal of the 5′ leader stem of all pre-tRNAs (Stark et al. 1978; Kole and Altman 1979) and are essential for cell survival. In E. coli, RNase P RNA is in a 1:1 complex with RNase P protein (Talbot and Altman 1994a,b), which is required for in vivo pre-tRNA processing (Altman et al. 1993; Pace and Brown 1995). In vitro, however, RNase P RNA can catalyze this reaction in the absence of protein (Guerrier-Takada et al. 1983). There is at least one slow step in the folding of E. coli RNase P RNA in which P7 undergoes a conformational change (Zarrinkar et al. 1996). More details of the folding pathway for Bacillus subtilis RNase P RNA are known, and are likely to be similar to E. coli RNase P RNA (Zarrinkar et al. 1996). They reveal that the major kinetic trap involves tertiary interactions between the catalytic (C) and specificity (S) domains (Pan and Sosnick 1997; Pan et al. 1999; Fang et al. 2002). This trap is eliminated when the domains are folded separately (Fang et al. 1999). The slow step in the folding of the catalytic domain involves consolidation of RNA structure around metal ions (Fang et al. 2002). These observations suggest that short oligonucleotides could direct RNase P RNA into an inactive structure.
RESULTS
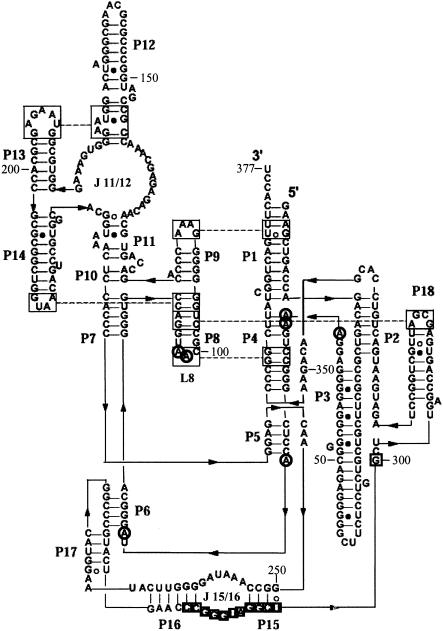
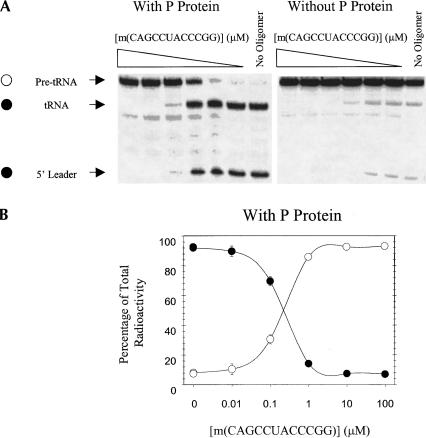
Figure 1▶ (Massire et al. 1998; Brown 1999) shows the functional secondary structure of E. coli RNase P RNA. Thirty-two DNA 12-mers complementary to consecutive regions of RNase P RNA were screened for inhibition of pre-tRNA processing in a transcription mixture containing E. coli RNase P protein (Table 1▶). Eight of these oligonucleotides inhibited at least 40% of pre-tRNA processing at 10 μM oligonucleotide concentration. Dose-response curves were measured for all eight of these sequences as their 2′-O-Me analogs (Table 1▶). The sequence m(CAGCCUACCCGG), which is complementary to nt 289–300, inhibited pre-tRNA processing most efficiently, with an IC50 of 200 nM (Fig. 2▶). As seen in Figure 2▶, the absence of RNase P protein reduces pre-tRNA processing to about 4% in a transcription mixture. Changing the oligonucleotide sequence to shift the binding site six residues in either direction slightly increases the IC50 to ~1 μM. Shortening the sequence to m(CAGCCUACCC) increases the IC50 to ~2 μM.
FIGURE 1.
Phylogenetic structure of RNase P RNA adapted from Massire et al. (1998) and http://www-ibmc.u-strasbg.fr/upr9002/westhof/. The oligonucleotide, m(CAGCCUACCCGG), is complementary to nt 289–300 (in boxes). Circles indicate sites of increased DEPC modification in the presence of m(CAGCCUACCCGG). Large boxes connected by dashed lines indicate tertiary interactions. (Reprinted from Journal of Molecular Biology, 279 773–793, Massire et al., © 1998 , with permission from Elsevier.)
TABLE 1.
Summary of ODMiR inhibition of pre-tRNA processing by E. coli RNase P and predicted ΔG°37’s of oligonucleotides binding to E. coli RNase P RNA by the OligoWalk program when RNA refolding is allowed
| Oligonucleotide | Region of complementarity | 10 μM DNA OligoWalk ΔG°37 | Pre-tRNA processing as percentage of control (10 μM DNA) | 1 μM RNA OligoWalk ΔG°37 | Pre-tRNA processing as percentage of control (1 μM 2′-O-Me RNA) | IC50 (μM) of 2′-O-Me oligo |
|---|---|---|---|---|---|---|
| Oligonucleotides that inhibita | ||||||
| CGGACTTTCCTC | 61–72 | −5.8 | 55 | −11.4 | 49 | ~1 |
| TCACCCTTACCT | 196–180 | −10.5 | 53 | −15.7 | 44 | ~1 |
| TACCGCACCCTT | 181–192 | −8.6 | 40 | −13.8 | 54 | ~1 |
| GTTACCAGCCGC | 205–216 | −6.4 | 33 | −11.8 | 71 | ~3 |
| GTGCCACGGACT | 217–228 | −4.7 | 54 | −11.4 | 86 | ~10 |
| AACCCCTATTTG | 253–264 | −4.8 | 32 | −9 | 66 | ~10 |
| CCGTACCTTATG | 265–276 | −4.2 | 41 | −8.9 | 44 | ~1 |
| ACCCGGGTTCAG | 283–294 | −11.7 | 49 | ~1 | ||
| CAGCCTACCCGG | 289–300 | −9.5 | 51 | −16.1 | 15 | ~0.2 |
| CAGCCTACCC | 291–302 | −14.4 | 67 | ~2 | ||
| CTCAAGCAGCCT | 295–306 | −19.9 | 48 | ~1 | ||
| TCAAGCAGCCT | 296–305 | −18.8 | 96 | ~10 | ||
| Oligonucleotides that do not inhibit | ||||||
| CTGGTCAGCTTC | 1–12 | −1.3 | 81 | −7.1 | N.D.b | N.D. |
| AGCGGCGACTGT | 13–24 | −5.8 | 95 | −13.4 | N.D.b | N.D. |
| GGACGACGACGA | 25–36 | 5.2 | 92 | −5.2 | N.D.b | N.D. |
| CTCCCCCGAAGA | 37–48 | −3.2 | 93 | −9.6 | N.D.b | N.D. |
| CCCTCCGCCCGT | 49–60 | −8.9 | 89 | −13.5 | N.D.b | N.D. |
| CCCTATGGAGCC | 73–84 | −9.1 | 85 | −15.7 | N.D.b | N.D. |
| CCTGGCACCCTG | 85–96 | −5.7 | 85 | −12.4 | N.D.b | N.D. |
| CCCCAGGCGTTA | 97–108 | −4.9 | 104 | −10.1 | N.D.b | N.D. |
| CGTGGGTTTCCC | 109–120 | −7.2 | 100 | −12.4 | N.D.b | N.D. |
| TGTTGCACTGGT | 121–132 | −7.8 | 104 | −14.3 | N.D.b | N.D. |
| CGGTTTGCTCTC | 133–144 | −12.1 | 87 | −17.6 | N.D.b | N.D. |
| GCGGGCCATCGG | 145–156 | −5.7 | 80 | −13.2 | N.D.b | N.D. |
| GATCCCGCTTGC | 157–168 | −6.2 | 87 | −11.7 | N.D.b | N.D. |
| GCGGTGCGCTCT | 193–204 | −5.9 | 99 | −12.5 | N.D.b | N.D. |
| GTGGAGTTTACC | 229–240 | −5.5 | 68 | −10.7 | N.D.b | N.D. |
| GCCTTGCTCCGG | 241–252 | −7.7 | 100 | −13.6 | N.D.b | N.D. |
| GTTCAGTACGGG | 277–288 | −4.7 | 98 | −11.5 | N.D.b | N.D. |
| CACTGGCTCAAG | 301–312 | −6.0 | 68 | −13.8 | N.D.b | N.D. |
| CCAGCAATCGCT | 313–324 | −7.0 | 94 | −13.0 | N.D.b | N.D. |
| CATTCATCTAGG | 325–336 | −9.3 | 81 | −15.2 | N.D.b | N.D. |
| GTCGTGGACAGT | 337–348 | −8.9 | 87 | −16.0 | N.D.b | N.D. |
| AAGCCGGGTTCT | 349–360 | −7.4 | 63 | −13.4 | N.D.b | N.D. |
| AAACTGACCGAT | 361–372 | −1.9 | 90 | −9.1 | N.D.b | N.D. |
| AGGTGAAACTGA | 366–377 | 1.2 | 99 | −8.1 | N.D.b | N.D. |
| GCACTGTCT | 11–19 | −11.4 | 101 | N.D. | ||
| TCCTCCC | 59–65 | −5.6 | 99 | N.D. | ||
| TATGGAGCCCG | 71–81 | −13.4 | 102 | N.D. | ||
| TATGGAG | 75–81 | −4.9 | 89 | N.D. | ||
| CCTGCCCTAT | 79–88 | −8.9 | 63 | N.D. | ||
| CCATCGGC | 144–151 | −10.5 | 101 | N.D. | ||
| CTTACCTGAT | 166–175 | −9.7 | 101 | N.D. | ||
| CTATTTGGCC | 250–259 | −13.6 | 98 | N.D. | ||
aT is replaced by U for 2′-O-Me sequences.
b(N.D.) not determined.
FIGURE 2.
Inhibition of processing of B. subtilis pre-tRNAAsp via ODMiR during transcription of E. coli RNase P RNA. (A) Autoradiogram of a gel for transcriptions in the presence or absence of m(CAGCCUACCCGG) with RNase P protein (left) and an autoradiogram of a gel for transcriptions in the presence or absence of m(CAGCCUACCCGG) without RNase P protein (right). (B) Plot of the percentage of pre-tRNA (open circles) and processed tRNA (filled circles) as a function of [m(CAGCCUACCCGG)]. All points have error bars, although in some instances they are smaller than the data points.
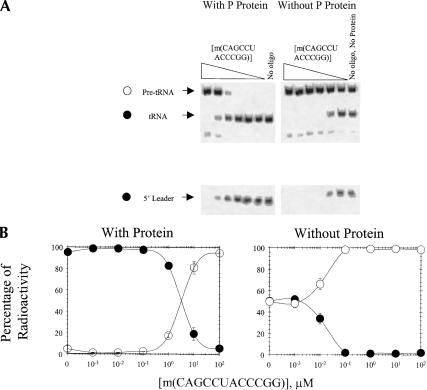
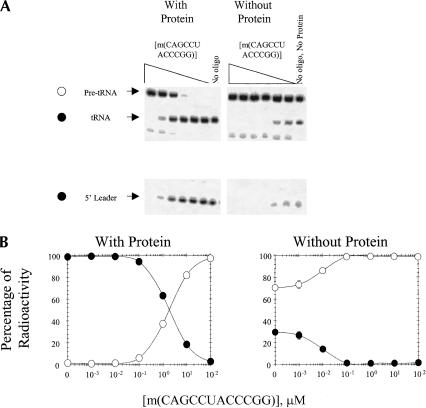
RNase P activity is also inhibited by m(CAGCCUACCCGG) when full-length RNase P RNA is renatured in the absence or presence of m(CAGCCUACCCGG). As shown in Figure 3▶, the IC50 for m(CAGCCUACCCGG) when RNase P RNA is renatured followed by addition of m(CAGCCUACCCGG) and RNase P protein is about 3 μM. If RNase P protein is not added to the reactions, however, the IC50 for m(CAGCCUACCCGG) is about 0.009 μM. When full-length RNase P RNA is renatured in the presence of m(CAGCCUACCCGG) followed by addition of RNase P protein, the IC50 is about 2 μM (Fig. 4▶). When RNase P protein is not added to the reactions in which full-length RNase P RNA is renatured in the presence of oligonucleotide, the IC50 for m(CAGCCUACCCGG) is about 0.013 μM (Fig. 4▶). Evidently, the IC50s are relatively independent of the point at which m(CAGCCUACCCGG) is added to full-length RNase P RNA, but are increased over 100-fold by the presence of protein.
FIGURE 3.
Inhibition of processing of B. subtilis pre-tRNA by addition of m(CAGCCUAC CCGG) to full-length, renatured RNase P RNA. (A) Autoradiogram of a gel in which m(CAGCCUACCCGG) was added after RNase P RNA was renatured. (Left) m(CAGCCUAC CCGG) and RNase P protein added after renaturation; (right) m(CAGCCUACCCGG) added after renaturation without RNase P protein. (B) Plot of the percentage of pre-tRNA (open circles) and processed tRNA (filled circles) as a function of [m(CAGCCUACCCGG)]. All points have error bars, although in some instances they are smaller than the data points.
FIGURE 4.
Inhibition of processing of B. subtilis pre-tRNA when full-length RNase P RNA is renatured in the presence of m(CAGCCUACCCGG). (A) Autoradiogram of a gel in which RNase P RNA was renatured in the presence of m(CAGCCUACCCGG). (Left) RNase P protein added after renaturation; (right) RNase P protein not added after renaturation. (B) Plot of the percentage of pre-tRNA (open circles) and processed tRNA (filled circles) as a function of [m(CAGCCUACCCGG)]. All points have error bars, although in some instances they are smaller than the data points.
The binding site for m(CAGCCUACCCGG) was determined experimentally by reverse transcription stops in the presence or absence of oligonucleotide. Stops are seen at the binding site because reverse transcriptase is unable to proceed through the oligonucleotide. As expected, m(CAGCCUACCCGG) binds to nucleotides 289–300.
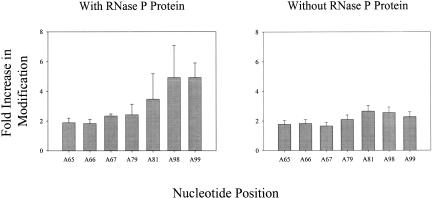
Differences in structure between the functional fold of RNase P RNA and the structure formed in the presence of m(CAGCCUACCCGG) were probed by diethyl pyrocarbonate (DEPC) modification. DEPC modifies purine N7s (Ehresmann et al. 1987; Doudna and Cech 1995), and therefore is a probe for changes in tertiary structure. As seen in Figures 1▶ and 5▶, m(CAGCCUACCCGG) enhances modification at A residues 65–67, 79, 81, and 98–99.
FIGURE 5.
Plots of the fold increase in DEPC modification at A residues 65–67, 79, 81, 98, and 99 in the presence (left) or absence (right) of RNase P protein when m(CAGCCUAC CCGG) is present.
DISCUSSION
RNA function has been inhibited by small molecules (Fourmy et al. 1996; Cho et al. 1998; Carter et al. 2000; Jin et al. 2000; Lynch and Puglisi 2001a,b; Kaul and Pilch 2002; Vicens and Westhof 2002; Lynch et al. 2003) and oligonucleotide-based therapeutics (Galderisi et al. 1999; Stein 2001; Disney et al. 2003). Oligonucleotide-based therapeutics can be rapidly designed and screened because much is known about molecular recognition between oligonucleotides and RNA, analogs are synthetically accessible (Freier and Altmann 1997), and their pharmacokinetic properties are likely to be similar (Crooke et al. 1996). In principle, insight into regions that can be targeted by oligonucleotide-directed misfolding can be gained from secondary structure prediction (Banerjee and Turner 1995; Pan and Woodson 1998; Mathews et al. 1999a; Chadalavada et al. 2000; Childs et al. 2002). Many RNAs function in RNA–protein complexes, however, and it is possible that the RNA–protein interaction could overcome an oligonucleotide strategy for inhibition. The results in Figure 2▶ show, however, that a 12-mer oligonucleotide can trap E. coli RNase P RNA during transcription in the presence of RNase P protein such that it is unable to process a pre-tRNA. Thus, the ODMiR strategy can be applied to inhibit the function of ribonucleoprotein complexes.
From an oligonucleotide screen of 32 DNA 12-mers, only 8 inhibited at least 40% of pre-tRNA processing at 10 μM. Thus, oligonucleotide complementarity to RNase P RNA is not sufficient for inhibition.
The most efficient oligonucleotide, m(CAGCCUACCCGG), inhibited E. coli RNase P function with an IC50 of about 200 nM (Fig. 2▶). This oligonucleotide (approximated as an RNA oligonucleotide) is predicted by the program OligoWalk (Mathews et al. 1999a) to have a total binding free energy of −16.1 kcal/mole, corresponding to a Kd of 5 pM, when the RNA is allowed to refold. Only one other 12-mer oligonucleotide tested is predicted to bind more tightly (Table 1▶). Of all 366 possible 12-mers, it ranks in the top 12% of oligonucleotides for favorable binding free energy. Thus, computer algorithms may be useful for identifying sequences worth screening even when the native structure has pseudoknots, which are not allowed by the algorithm (Zuker 1989). The more than 10,000-fold difference between measured IC50 and predicted Kd with refolding, however, indicates that there are many complexities not included in the algorithm. If refolding is not allowed, then the predicted Kd is about 1 nM.
The predicted binding free energy of −16.1 kcal/mole for m(CAGCCUACCCGG) can be compared with the Kd of 0.4 nM measured for RNase P protein binding to RNase P RNA under similar salt conditions (Talbot and Altman 1994b). This Kd corresponds to a ΔG°37 of −13.3 kcal/mole. If oligonucleotide inhibition of function required complete displacement of RNase P protein, then the expected free energy of binding would be −16.1 + 13.3 = −2.8 kcal/mole, corresponding to a Kd of ~10 mM. The least favorable IC50 for inhibition by m(CAGCCUACCCGG) is 3 μM. Evidently, the protein is still able to bind in the presence of m(CAGC CUACCCGG).
In a previous study of ODMiR inhibition of group I intron self-splicing (Childs et al. 2002), a successful 8-mer was designed on the basis of predicted suboptimal foldings of the intron. Several 2′-O-Me oligonucleotides were designed in a similar way for RNase P RNA, but only one inhibited activity at 10 μM. Evidently, our knowledge of RNA folding in the presence of pseudoknots and protein is insufficient for completely directed design of ODMiR oligonucleotides.
The m(CAGCCUACCCGG) oligonucleotide binds to nt 289–300 (3′ side of J15/16) as designed. It base pairs with a number of residues that have been implicated in pre-tRNA recognition, including G291, G292, and U294. In the current model, G291 forms a base triple with G259 in RNase P RNA and A76 in pre-tRNA; G292 forms a base triple with A258 in RNase P RNA and C75 in pre-tRNA; and U294 in RNase P RNA binds to R73 in pre-tRNA, where R is A or G (Kirsebom and Svard 1994; Heide et al. 2001). There is evidence that J15/16 is also a metal-ion-binding site (Kazakov and Altman 1991; Zito et al. 1993; Ciesiolka et al. 1994; Kufel and Kirsebom 1994). Part of the mode of inhibition may be competing with the pre-tRNA for this binding site or interfering with metal ion binding. Structure probing with DEPC, however, shows a significant structural rearrangement in L8 (A’s 98–99), J5/6 (A79 and A81), and the single-stranded region between P3 and P4 (A’s 65–67; Figs. 1▶, 5▶). Moreover, the IC50 for inhibition increases roughly 10-fold when m(CAGCCUACCCGG) is added to full-length RNase P RNA complexed with protein rather than to a transcription reaction. This suggests that oligonucleotide directed misfolding is the main mode of action.
A’s 65–67 are part of the pseudoknot containing P4 (Haas et al. 1991), and are important for catalytic activity (Kazantsev and Pace 1998). A’s 98 and 99 in L8 are predicted to be involved in a tertiary interaction with P4 (Massire et al. 1998). Thus, increased modification of A’s 65–67, 98, and 99 by DEPC suggests a structural rearrangement of part of the P4 pseudoknot. A’s 79 and 81 are in close proximity to the 4 bp of P6, which is also part of a pseudoknot (James et al. 1988). Deletion studies of RNase P RNA show that disruption of P6 increases the KM for pre-tRNA (Darr et al. 1992).
Improvements to secondary structure prediction and in understanding how large RNAs fold should eventually allow for selection of ODMiR targets from entire genomes. Such RNAs may include RNAs with secondary or tertiary structures important for catalysis, regulation of translation, localization, or interaction with other biomolecules.
MATERIALS AND METHODS
Buffers
HXMg is 50 mM HEPES (25 mM Na+), 135 mM KCl, and X mM MgCl2. Transcription buffer contains 1× H14Mg, 1 mM each NTP, 2 mM DTT, and 2 mM spermidine.
RNA and protein plasmids
Plasmids containing the E. coli RNase P RNA (M1) gene (pDW98) and RNase P protein (C5) gene (pECPE1), and the B. subtilis pre-tRNAAsp gene (pDW152; Beebe and Fierke 1994) were generously supplied by Norman R. Pace (University of Colorado, Boulder).
Oligonucleotides
Oligonucleotides were synthesized, deblocked, and purified by standard methods (Matteucci and Caruthers 1980; Caruthers et al. 1992; Xia et al. 1998). Concentrations were determined from predicted extinction coefficients and measured absorbances at 260 or 280 nm at 25°C (Puglisi and Tinoco 1989).
Preparation of E. coli RNase P protein
RNase P protein was expressed and purified by the method of A.W. Poole and N.R. Pace (pers. comm.). The RNase P protein plasmid was transformed into BL21 (DE3) pLysS cells for expression. Protein expression was induced by addition of 1 mM IPTG and incubation for 4 h at 37°C. Cells were lysed by sonication in 50 mM Tris-HCl (pH 8.0), 5 mM EDTA, 100 mM DTT, and 10% glycerol. Lysate was brought to final concentrations of 5 M urea, 50 mM NaCl, 50 mM Tris-HCl (pH 8.0), and 51 mM DTT and purified on an SP Sepharose column (Amersham) using a linear gradient from 50 mM to 2 M NaCl. Fractions containing RNase P protein were pooled and dialyzed into 5 M urea, 50 mM NaCl, 50 mM Tris-HCl (pH 7.5), and 1 mM DTT. The dialyzed fractions were loaded on a prepacked Mono-S column (Amersham) with a linear gradient from 50 mM to 2 M NaCl. Fractions containing RNase P protein were dialyzed into 1× TE (pH 7.5), 1 mM DTT, and 50% glycerol, and concentrated using a YM-10 cartridge (Centricon). RNase P protein was then passed through a 0.2-μm microfuge tube filter (Corning).
Preparation of pre-tRNA
B. subtilis pre-tRNAAsp was prepared by run-off transcription with T7 RNA polymerase (New England BioLabs) from its linearized plasmid. The pre-tRNA was purified on an 8% polyacrylamide denaturing gel, and electroeluted in 1× TBE. Purified pre-tRNA was desalted on a Sephadex column. Internally labeled material was prepared with [α-32P]ATP, and purified on an 8% polyacrylamide denaturing gel; the full-length product was excised from the gel and eluted by the crush-and-soak method. Internally labeled pre-tRNA was then concentrated with 2-butanol and ethanol precipitated. For each assay, pre-tRNA was annealed in 1× H0Mg buffer for 3 min at 90°C, followed by slow cooling to 37°C, at which point MgCl2 was added to a final concentration of 10 mM.
Oligonucleotide screen and dose-response curves
Transcription reactions were run in 20 μL containing 1× transcription buffer, 50 U T7 RNA polymerase, 100 ng linearized RNase P RNA plasmid, with or without 5 ng of RNase P protein, and trace internally labeled pre-tRNA annealed as above. Transcription mixtures with and without ODMiR oligonucleotides and with or without RNase P protein were incubated for 1 h at 37°C, and then were ethanol precipitated. Products were separated on an 8% polyacrylamide denaturing gel. Results were quantified with a phosphorimager and ImageQuant v5.2. IC50s were obtained by fitting dose-response curves to a four-parameter logistic curve (SigmaPlot 2001).
To determine the concentration of RNase P RNA transcribed in a 20-μL transcription reaction, 100 ng of linearized RNase P RNA plasmid was transcribed for 1 h at 37°C in 1× transcription buffer with 50 U T7 RNA polymerase, and 1 μCi (3000 Ci/mmole) [α-32P]ATP. The full-length product was compared to 1 μCi of [α-32P]ATP, and the concentration of full-length product was calculated to be about 2 nM.
Inhibition of full-length RNase P RNA by m(CAGCCUACCCGG)
The m(CAGCCUACCCGG) was also tested for inhibition of RNase P activity by renaturing RNase P RNA followed by addition of m(CAGCCUACCCGG) with and without RNase P protein. RNase P RNA was renatured in 1× H0Mg buffer at 90°C for 3 min, followed by slow cooling to 37°C. MgCl2 was added to 10 mM along with various concentrations of oligonucleotide. If included in the reactions, 5 ng of RNase P protein were added per 20 μL. Samples were incubated for 45 min at 37°C, and then renatured pre-tRNA was added. Samples were quenched with 2.5 volumes of ethanol after incubation for 1 h at 37°C. Experiments were also done in a similar manner but by renaturing RNase P RNA in the presence of m(CAGCCUACCCGG) with or without RNase P protein.
Diethyl pyrocarbonate probing
Structural changes due to ODMiR oligonucleotides were probed with DEPC by annealing 44 nM RNase P RNA with or without 10 μM m(CAGCCUACCCGG) for 3 min at 90°C, followed by slow cooling to 37°C. MgCl2 was added to 10 mM, and RNase P protein, if included, was added to 5 ng per 20 μL to give 1:1 stoichiometry. Samples were incubated for 45 min at 37°C, and then renatured pre-tRNA was added. The samples were equilibrated for 45 min at 37°C. DEPC was added to 650 mM and the samples were incubated for 20 min at 37°C. The reactions were quenched by addition of 2.5 volumes of ethanol (Banerjee and Turner 1995). RNase P RNA was ethanol precipitated and reverse transcribed to detect sites of modification. RNase P RNA was sequenced by the Sanger method with reverse transcriptase (Sanger et al. 1977). Structural changes for nucleotides were reported if there was at least a 1.9-fold increase in modification when normalized to full-length product.
OligoWalk predictions
The ΔG°37s of duplex formation between all possible 12-mer oligonucleotides and RNase P RNA were calculated by the OligoWalk program (Mathews et al. 1999a) with the following parameters: refold RNA, include suboptimal structures (predicted from the RNAstructure program; Mathews et al. 1999b), and DNA at 10 μM or RNA (approximation for 2′-O-Me RNA oligonucleotides) at 1 μM concentration.
Acknowledgments
The authors thank Norman Pace (University of Colorado, Boulder) for generously supplying all the plasmids used in this work and for helpful discussions, and Robert Bambara and Hui-I Kao for assistance with protein purification. This work was supported by NIH Grant GM22939 to D.H.T. J.L.C. was partially supported by NIH Grant T32 DE07202.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5780503.
REFERENCES
- Altman, S., Kirsebom, L., and Talbot, S. 1993. Recent studies of ribonuclease P. FASEB J. 7 7–14. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289 905–920. [DOI] [PubMed] [Google Scholar]
- Banerjee, A.R. and Turner, D.H. 1995. The time dependence of chemical modification reveals slow steps in the folding of a group I ribozyme. Biochemistry 34 6504–6512. [DOI] [PubMed] [Google Scholar]
- Beebe, J.A. and Fierke, C.A. 1994. A kinetic mechanism for cleavage of precursor tRNA(Asp) catalyzed by the RNA component of Bacillus subtilis ribonuclease P. Biochemistry 33 10294–10304. [DOI] [PubMed] [Google Scholar]
- Been, M.D., Perrotta, A.T., and Rosenstein, S.P. 1992. Secondary structure of the self-cleaving RNA of hepatitis delta-virus: Applications to catalytic RNA design. Biochemistry 31 11843–11852. [DOI] [PubMed] [Google Scholar]
- Brown, J.W. 1999. The ribonuclease P database. Nucleic Acids Res. 27 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., and Ramakrishnan, V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407 340–348. [DOI] [PubMed] [Google Scholar]
- Caruthers, M.H., Beaton, G., Wu, J.V., and Wiesler, W. 1992. Chemical synthesis of deoxyoligonucleotides and deoxyoligonucleotide analogs. Methods Enzymol. 211 3–20. [DOI] [PubMed] [Google Scholar]
- Celander, D.W. and Cech, T.R. 1991. Visualizing the higher-order folding of a catalytic RNA molecule. Science 251 401–407. [DOI] [PubMed] [Google Scholar]
- Chadalavada, D.M., Knudsen, S.M., Nakano, S., and Bevilacqua, P.C. 2000. A role for upstream RNA structure in facilitating the catalytic fold of the genomic hepatitis delta virus ribozyme. J. Mol. Biol. 301 349–367. [DOI] [PubMed] [Google Scholar]
- Chadalavada, D.M., Senchak, S.E., and Bevilacqua, P.C. 2002. The folding pathway of the genomic hepatitis delta virus ribozyme is dominated by slow folding of the pseudoknots. J. Mol. Biol. 317 559–575. [DOI] [PubMed] [Google Scholar]
- Childs, J.L., Disney, M.D., and Turner, D.H. 2002. Oligonucleotide directed misfolding of RNA inhibits Candida albicans group I intron splicing. Proc. Natl. Acad. Sci. 99 11091–11096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, J., Hamasaki, K., and Rando, R.R. 1998. The binding site of a specific aminoglycoside binding RNA molecule. Biochemistry 37 4985–4992. [DOI] [PubMed] [Google Scholar]
- Ciesiolka, J., Hardt, W.D., Schlegl, J., Erdmann, V.A., and Hartmann, R.K. 1994. Lead-ion-induced cleavage of RNase P RNA. Eur. J. Biochem. 219 49–56. [DOI] [PubMed] [Google Scholar]
- Clodi, E., Semrad, K., and Schroeder, R. 1999. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 18 3776–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke, S.T., Graham, M.J., Zuckerman, J.E., Brooks, D., Conklin, B.S., Cummins, L.L., Greig, M.J., Guinosso, C.J., Kornbrust, D., Manoharan, M., et al. 1996. Pharmacokinetic properties of several novel oligonucleotide analogs in mice. J. Pharmacol. Exp. Ther. 277 923–937. [PubMed] [Google Scholar]
- Darr, S.C., Zito, K., Smith, D., and Pace, N.R. 1992. Contributions of phylogenetically variable structural elements to the function of the ribozyme ribonuclease-P. Biochemistry 31 328–333. [DOI] [PubMed] [Google Scholar]
- Disney, M.D., Matray, T., Gryaznov, S.M., and Turner, D.H. 2001. Binding enhancement by tertiary interactions and suicide inhibition of a Candida albicans group I intron by phosphoramidate and 2′-O-Methyl hexanucleotides. Biochemistry 40 6520–6526. [DOI] [PubMed] [Google Scholar]
- Disney, M.D., Haidaris, C.G., and Turner, D.H. 2003. Uptake and antifungal activity of oligonucleotides in Candida albicans. Proc. Natl. Acad. Sci. 100 1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J.A. and Cech, T.R. 1995. Self-assembly of a group I intron active site from its component tertiary structural domains. RNA 1 36–45. [PMC free article] [PubMed] [Google Scholar]
- Ehresmann, C., Baudin, F., Mougel, M., Romby, P., Ebel, J.P., and Ehresmann, B. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, X.W., Pan, T., and Sosnick, T.R. 1999. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat. Struct. Biol. 6 1091–1095. [DOI] [PubMed] [Google Scholar]
- Fang, X.W., Thiyagarajan, P., Sosnick, T.R., and Pan, T. 2002. The rate-limiting step in the folding of a large ribozyme without kinetic traps. Proc. Natl. Acad. Sci. 99 8518–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor, M.J. and Uhlenbeck, O.C. 1990. Substrate sequence effects on hammerhead RNA catalytic efficiency. Proc. Natl. Acad. Sci. 87 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811. [DOI] [PubMed] [Google Scholar]
- Fourmy, D., Recht, M.I., Blanchard, S.C., and Puglisi, J.D. 1996. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science 274 1367–1371. [DOI] [PubMed] [Google Scholar]
- Freier, S.M. and Altmann, K.H. 1997. The ups and downs of nucleic acid duplex stability: Structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 25 4429–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galderisi, U., Cascino, A., and Giordano, A. 1999. Antisense oligonucleotides as therapeutic agents. J. Cell. Physiol. 181 251–257. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N., and Altman, S. 1983. The RNA moiety of ribonuclease-P is the catalytic subunit of the enzyme. Cell 35 849–857. [DOI] [PubMed] [Google Scholar]
- Haas, E.S., Morse, D.P., Brown, J.W., Schmidt, F.J., and Pace, N.R. 1991. Long-range structure in ribonuclease P RNA. Science 254 853–856. [DOI] [PubMed] [Google Scholar]
- Heide, C., Feltens, R., and Hartmann, R.K. 2001. Purine N7 groups that are crucial to the interaction of Escherichia coli RNase P RNA with tRNA. RNA 7 958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann, T. and Westhof, E. 1998. RNA as a drug target: Chemical, modelling, and evolutionary tools. Curr. Opin. Biotechnol. 9 66–73. [DOI] [PubMed] [Google Scholar]
- James, B.D., Olsen, G.J., Liu, J.S., and Pace, N.R. 1988. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell 52 19–26. [DOI] [PubMed] [Google Scholar]
- Jin, E., Katritch, V., Olson, W.K., Kharatisvili, M., Abagyan, R., and Pilch, D.S. 2000. Aminoglycoside binding in the major groove of duplex RNA: The thermodynamic and electrostatic forces that govern recognition. J. Mol. Biol. 298 95–110. [DOI] [PubMed] [Google Scholar]
- Kaul, M. and Pilch, D.S. 2002. Thermodynamics of aminoglycoside-rRNA recognition: The binding of neomycin-class aminoglycosides to the A site of 16S rRNA. Biochemistry 41 7695–7706. [DOI] [PubMed] [Google Scholar]
- Kazakov, S. and Altman, S. 1991. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli. Proc. Natl. Acad. Sci. 88 9193–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev, A.V. and Pace, N.R. 1998. Identification by modification-interference of purine N-7 and ribose 2′-OH groups critical for catalysis by bacterial ribonuclease P. RNA 4 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom, L.A. and Svard, S.G. 1994. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 13 4870–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole, R. and Altman, S. 1979. Reconstitution of RNase P activity from inactive RNA and protein. Proc. Natl. Acad. Sci. 76 3795–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J. and Kirsebom, L.A. 1994. Cleavage site selection by M1 RNA the catalytic subunit of Escherichia coli RNase P, is influenced by pH. J. Mol. Biol. 244 511–521. [DOI] [PubMed] [Google Scholar]
- Lambowitz, A.M. and Perlman, P.S. 1990. Involvement of aminoacyl-tRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem. Sci. 15 440–444. [DOI] [PubMed] [Google Scholar]
- Lynch, S.R. and Puglisi, J.D. 2001a. Structural origins of aminoglycoside specificity for prokaryotic ribosomes. J. Mol. Biol. 306 1037–1058. [DOI] [PubMed] [Google Scholar]
- ———. 2001b. Structure of a eukaryotic decoding region A-site RNA. J. Mol. Biol. 306 1023–1035. [DOI] [PubMed] [Google Scholar]
- Lynch, S.R., Gonzalez, R.L., and Puglisi, J.D. 2003. Comparison of X-ray crystal structure of the 30S subunit-antibiotic complex with NMR structure of decoding site oligonucleotide-paromomycin complex. Structure 11 43–53. [DOI] [PubMed] [Google Scholar]
- Massire, C., Jaeger, L., and Westhof, E. 1998. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 279 773–793. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Burkard, M.E., Freier, S.M., Wyatt, J.R., and Turner, D.H. 1999a. Predicting oligonucleotide affinity to nucleic acid targets. RNA 5 1458–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999b. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288 911–940. [DOI] [PubMed] [Google Scholar]
- Matteucci, M.D. and Caruthers, M.H. 1980. The synthesis of oligodeoxypyrimidines on a polymer support. Tetrahedron Lett. 21 719–722. [Google Scholar]
- Montgomery, M.K., Xu, S., and Fire, A. 1998. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 95 15502–15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, C.A., Wallweber, G.J., Rennard, R., Kemel, Y., Caprara, M.G., Mohr, G., and Lambowitz, A.M. 1996. A tyrosyl-tRNA synthetase suppresses structural defects in the two major helical domains of the group I intron catalytic core. J. Mol. Biol. 262 87–104. [DOI] [PubMed] [Google Scholar]
- Noller, H.F., Hoffarth, V., and Zimniak, L. 1992. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256 1416–1419. [DOI] [PubMed] [Google Scholar]
- Nykanen, A., Haley, B., and Zamore, P.D. 2001. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107 309–321. [DOI] [PubMed] [Google Scholar]
- Pace, N.R. and Brown, J.W. 1995. Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J. Bacteriol. 177 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, T. and Sosnick, T.R. 1997. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat. Struct. Biol. 4 931–938. [DOI] [PubMed] [Google Scholar]
- Pan, J. and Woodson, S.A. 1998. Folding intermediates of a self-splicing RNA: Mispairing of the catalytic core. J. Mol. Biol. 280 597–609. [DOI] [PubMed] [Google Scholar]
- Pan, T., Fang, X., and Sosnick, T.R. 1999. Pathway modulation, circular permutation and rapid RNA folding under kinetic control. J. Mol. Biol. 286 721–731. [DOI] [PubMed] [Google Scholar]
- Pearson, N.D. and Prescott, C.D. 1997. RNA as a drug target. Chem. Biol. 4 409–414. [DOI] [PubMed] [Google Scholar]
- Puglisi, J.D. and Tinoco Jr., I. 1989. Absorbance melting curves of RNA. Methods Enzymol. 180 304–325. [DOI] [PubMed] [Google Scholar]
- Russell, R., Zhuang, X., Babcock, H.P., Millett, I.S., Doniach, S., Chu, S., and Herschlag, D. 2002. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. 99 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger, F., Nicklen, S., and Coulson, A.R. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. 74 5436–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, B.C., Kole, R., Bowman, E.J., and Altman, S. 1978. Ribonuclease P: An enzyme with an essential RNA component. Proc. Natl. Acad. Sci. 75 3717–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, C.A. 2001. Antisense that comes naturally. Nat. Biotechnol. 19 737–738. [DOI] [PubMed] [Google Scholar]
- Talbot, S.J. and Altman, S. 1994a. Gel retardation analysis of the interaction between C5 protein and M1 RNA in the formation of the ribonuclease P holoenzyme from Escherichia coli. Biochemistry 33 1399–1405. [DOI] [PubMed] [Google Scholar]
- ———. 1994b. Kinetic and thermodynamic analysis of RNA–protein interactions in the RNase P holoenzyme from Escherichia coli. Biochemistry 33 1406–1411. [DOI] [PubMed] [Google Scholar]
- Testa, S.M., Gryaznov, S.M., and Turner, D.H. 1999. In vitro suicide inhibition of self-splicing of a group I intron from Pneumocystis carinii by an N3′ → P5′ phosphoramidate hexanucleotide. Proc. Natl. Acad. Sci. 96 2734–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens, Q. and Westhof, E. 2002. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding A site. Chem. Biol. 9 747–755. [DOI] [PubMed] [Google Scholar]
- Walstrum, S.A. and Uhlenbeck, O.C. 1990. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry 29 10573–10576. [DOI] [PubMed] [Google Scholar]
- Webb, A.E., Rose, M.A., Westhof, E., and Weeks, K.M. 2001. Protein-dependent transition states for ribonucleoprotein assembly. J. Mol. Biol. 309 1087–1100. [DOI] [PubMed] [Google Scholar]
- Weeks, K.M. and Cech, T.R. 1995a. Efficient protein-facilitated splicing of the yeast mitochondrial bI5 intron. Biochemistry 34 7728–7738. [DOI] [PubMed] [Google Scholar]
- ———. 1995b. Protein facilitation of group I intron splicing by assembly of the catalytic core and the 5′ splice site domain. Cell 82 221–230. [DOI] [PubMed] [Google Scholar]
- Xia, T., SantaLucia Jr., J., Burkard, M.E., Kierzek, R., Schroeder, S.J., Jiao, X., Cox, C., and Turner, D.H. 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry 37 14719–14735. [DOI] [PubMed] [Google Scholar]
- Zarrinkar, P.P., Wang, J., and Williamson, J.R. 1996. Slow folding kinetics of RNase P RNA. RNA 2 564–573. [PMC free article] [PubMed] [Google Scholar]
- Zhang, F., Ramsay, E.S., and Woodson, S.A. 1995. In vivo facilitation of Tetrahymena group I intron splicing in Escherichia coli pre-ribosomal RNA. RNA 1 284–292. [PMC free article] [PubMed] [Google Scholar]
- Zito, K., Huttenhofer, A., and Pace, N.R. 1993. Lead-catalyzed cleavage of ribonuclease-P RNA as a probe for integrity of tertiary structure. Nucleic Acids Res. 21 5916–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 1989. On finding all suboptimal foldings of an RNA molecule. Science 244 48–52. [DOI] [PubMed] [Google Scholar]