Abstract
Autosomal dominant dyskeratosis congenita (DKC) has been linked to mutations in the RNA component of telomerase, the ribonucleoprotein responsible for telomere maintenance. Recent studies have investigated the role of the GC (107–108) → AG mutation in the conserved P3 helix in the pseudoknot domain of human telomerase RNA. The mutation was found to significantly destabilize the pseudoknot conformation, resulting in a shift in the thermodynamic equilibrium to favor formation of a P2b hairpin intermediate. In the wild-type sequence, the hairpin intermediate was found to form a novel sequence of pyrimidine base pairs in a continuous stem capped by a structured pentaloop. The DKC mutant hairpin was observed to be slightly more stable than the wild-type hairpin, further shifting the pseudoknot-hairpin equilibrium to favor the mutant P2b hairpin. Here we examined the solution structure of the DKC mutant hairpin to identify the reason for this additional stability. We found that the mutant hairpin forms the same stem structure as wild-type and that the additional stabilization observed using optical melting can be explained by the formation of a YNMG-type tetraloop structure, with the last nucleotide of the pentaloop bulged out into the major groove. Our results provide a structural explanation for the increased stability of the mutant hairpin and further our understanding of the effect of this mutation on the structure and stability of the dominant conformation of the pseudoknot domain in this type of DKC.
Keywords: NMR, structure, pentaloop, pseudoknot, RDC, UUCG
INTRODUCTION
Telomerase is the ribonucleoprotein complex responsible for adding telomeric DNA repeats at the ends of linear chromosomes. Telomeric DNA repeats are eroded over successive rounds of DNA-templated DNA synthesis, and their presence prevents the loss of essential genetic information during genome replication. The telomerase complex consists of an RNA component (TR), a catalytic protein component (TERT), and a variety of accessory proteins (Kelleher et al. 2002; Harrington 2003). Although the length of the telomerase RNA varies significantly between vertebrates (400–500 nt), yeast (1000–1500 nt), and ciliates (150–200 nt; Chen et al. 2000), TR appears to contain a common core secondary structure for these evolutionarily distant organisms, including an RNA template for telomere addition and a hairpin (H)-type pseudoknot motif required for catalytic activity (Autexier et al. 1996; Gilley and Blackburn 1999; Tesmer et al. 1999; Tzfati et al. 2003).
Telomerase has been the focus of intense study due to its upregulation in many cancer cell lines and its role in preventing chromosomal instability (Blackburn 2001). In addition to links to certain human cancers, mutations in the RNA component of human telomerase (hTR), and thus telomerase activity, have also been linked to certain inherited human disorders such as autosomal dominant dyskeratosis congenita (DKC; Vulliamy et al. 2001a) and aplastic anemia (Vulliamy et al. 2002). DKC is a rare, inherited disorder characterized by abnormal skin pigmentation, leukoplakia, and nail dystrophy (Dokal 2000). The characteristic symptoms of DKC result from defects in highly proliferative tissues (such as bone marrow, skin, and nails) and could ensue from a telomere maintenance disorder (Mitchell et al. 1999b; Montanaro et al. 2002). Telomere lengths in patients with DKC (Vulliamy et al. 2001b) and some patients with ideopathic aplastic anemia (Ball et al. 1998) are found to be significantly reduced compared to age-matched controls. The earlier onset of disease symptoms in DKC patients in successive generations is consistent with the haploinsufficiency and phenotypic lag in consequences observed in mice heterozygous for TERT (mTERT+/−) or TR (mTR+/−; Hathcock et al. 2002; Liu et al. 2002).
DKC has both autosomal and X-linked inheritance patterns (Dokal 2000). The X-linked form of DKC results from a variety of mutations in the gene encoding dyskerin, a protein involved in pseudouridylation of ribosomal RNA, which has also been shown to bind to the H/ACA domain of telomerase RNA (Mitchell et al. 1999b). The autosomal dominant form of DKC is correlated with mutations in human telomerase RNA, not dyskerin, in three families with DKC inheritance (Vulliamy et al. 2001a). One mutation involves deletion of the terminal 78 nt of the hTR RNA (451 nt), eliminating the H/ACA and CR7 domains (Fig. 1A ▶) required for nucleolar localization, 3′-end processing, and RNA stability (Mitchell et al. 1999a; Dragon et al. 2000), whereas another is a point mutation (C408G) also involving the CR7 domain. Recent studies have shown that these mutations result in unstable hTR RNAs which do not accumulate in the cell (Fu and Collins 2003). The third mutation is a two-base substitution [GC (107–108) → AG] in the pseudoknot domain which is required for activity (Autexier et al. 1996; Tesmer et al. 1999) and involved in hTERT binding (Mitchell and Collins 2000). Some patients with aplastic anemia are also heterozygous for mutations in hTR, one of which occurs in the same region of the pseudoknot as the DKC mutation (Vulliamy et al. 2002). Fu and Collins (2003) also showed that the pseudoknot domain mutation does not affect accumulation of hTR RNA, but instead compromises the catalytic function of the mutant telomerase complex.
FIGURE 1.
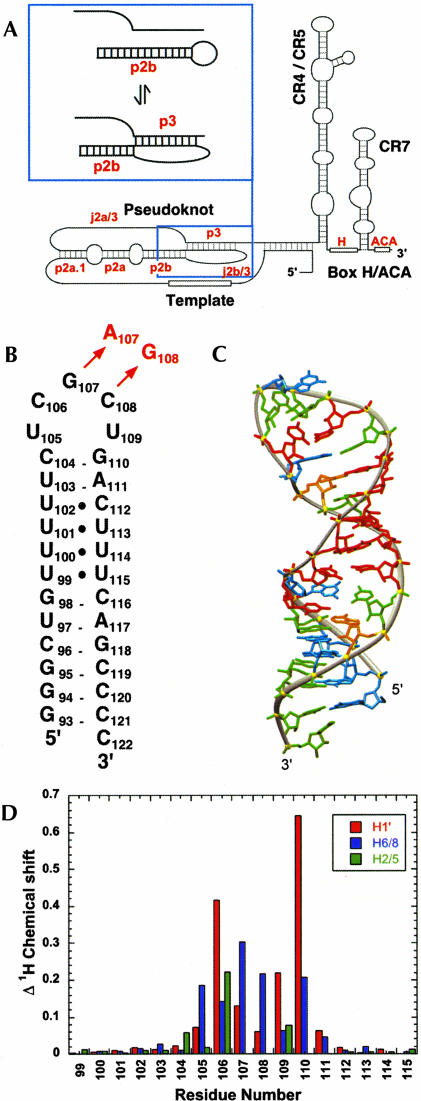
(A) Secondary structure of the human telomerase RNA, with the domains identified as described in Chen et al. (2000). Inset: Conformational equilibrium between the P2b-P3 pseudoknot and P2b hairpin conformations. (B) Secondary structure representation of the HPwt RNA representing nt 94–120 from the wild-type P2b hairpin. The DKC mutation in the P2b helix (HPdc) is indicated in red. (C) Solution structure of the HPwt hairpin (PDB accession code: 1NA2; Theimer et al. 2003). Nucleotides are colored red (U), green (C), orange (A), and cyan (G). (D) Observed changes in chemical shift (Δppm) between HPwt and HPdc are plotted by residue number and include H1′ (red), H6/8 (blue), and H5/2 (green) resonances.
The GC (107–108) → AG mutation occurs in the conserved P3 helix in the pseudoknot domain of human telomerase RNA. The P2b-P3 pseudoknot from human telomerase RNA, previously proposed to be in equilibrium with an alternate conformation in vitro and in vivo (Antal et al. 2002), was shown to exist in equilibrium with the P2b hairpin folding intermediate in vitro (Fig. 1A ▶, inset; Theimer et al. 2003). The hairpin stem has an unexpected and unique run of four consecutive pyrimidine base pairs. The terminal pentaloop of the wild-type hairpin forms a modestly stabilizing pentaloop structure in which the first three nucleotides of the loop stack on the 5′ side of the loop, with the last two nucleotides less ordered. The mutant P2b hairpin was found to be slightly more stable than the wild-type (Theimer et al. 2003). Although the structural basis for this increased stability was not identified, this small increase in P2b hairpin stability combined with the significant decrease in stability of the mutant P2b-P3 pseudoknot results in a shift in equilibrium to strongly favor the P2b hairpin conformation for the mutant sequence. A related NMR study, using a trans pseudoknot construct, also reported a structural equilibrium between pseudoknot and hairpin, but with the shift in equilibrium for the mutant pseudoknot attributed to hyperstablization of the mutant hairpin (Comolli et al. 2002). The shift in equilibrium is consistent with the observed decrease in telomerase activity for this mutation in vitro and in vivo (Comolli et al. 2002; Fu and Collins 2003) and with the shortened telomeres observed in patients with this mutation (Vulliamy et al. 2001b). These studies led to a proposal that the pseudoknot domain acts as a molecular switch in telomerase function (Comolli et al. 2002; Theimer et al. 2003).
To address the molecular basis for the observed increase in stability of the mutant P2b hairpin, we used UV melting and NMR spectroscopy to determine the thermodynamic properties and solution structure of a minimal RNA construct containing the DKC GC (107–108) → AG mutation, using heteronuclear multidimensional NMR spectroscopy. Here we identify the structural basis for the increased stability of the mutant hairpin observed in previous studies (Comolli et al. 2002; Theimer et al. 2003) as the formation of a pentaloop structure which mimics the YNMG family of tetraloops (Proctor et al. 2002) with no changes in the stem base-pairing compared to wild-type. We also show that the mutant hTR pentaloop is nearly identical in stability to a comparable tetraloop sequence. Finally, this pentaloop structure provides corroborating evidence of a structural basis for the recently proposed YNMG family of tetraloops (Proctor et al. 2002), and potentially extends the family to include YNMG(N) pentaloops.
RESULTS
Comparison of the wild-type and mutant P2b hairpins
The secondary structure of the wild-type P2b hairpin (HPwt) from human telomerase RNA (hTR) is given in Figure 1B ▶ with the GC (107–108) →AG mutation (HPdc) identified in red. The solution structure of the wild-type hairpin, which contains three consecutive U•U and one U•C base pairs toward the top of the 12-bp stem (Theimer et al. 2003), is shown in Figure 1C ▶. As described previously, NMR and thermodynamic evidence was consistent with formation of identical helical stems for the wild-type and mutant sequences, suggesting that the increased stability observed for the mutant hairpin derives from changes in the stacking of nucleotides in the pentaloop (Theimer et al. 2003). Consistent with this expectation, chemical shift mapping of the upper stem and pentaloop between the wild-type and mutant sequences shows significant chemical shift differences only for loop nucleotides and G110, the 3′ nucleotide of the closing C105-G110 base pair (Fig. 1D ▶). Chemical shift differences in the poly-pyrimidine tract (99–102 and 112–115) between the mutant and wild-type hairpins were less than 0.05 ppm for the H1′ or aromatic protons, consistent with identical structures. Because there was no additional information to be obtained from solving the poly-pyrimidine tract in the context of the mutant pentaloop, a minimal NMR construct, DCloop, was designed to contain nt 103–111 from the mutant hairpin closed by a sequence of three G•C bp for optimal hairpin stability and in vitro transcription efficiency (Fig. 2A ▶). Comparison of the pattern of H5-H6 TOCSY crosspeaks (Fig. 2B ▶) and NOESY spectra (data not shown) for HPwt, HPdc, and the DCloop RNAs show the same chemical shift differences for the loop and closing base pair of the DCloop and HPdc RNAs compared to HPwt. Additionally, this spectrum shows the overlap of pyrimidine tract aromatic proton chemical shifts with the mutant loop aromatic chemical shifts (Fig. 2B ▶) which was eliminated in the DCloop construct.
FIGURE 2.
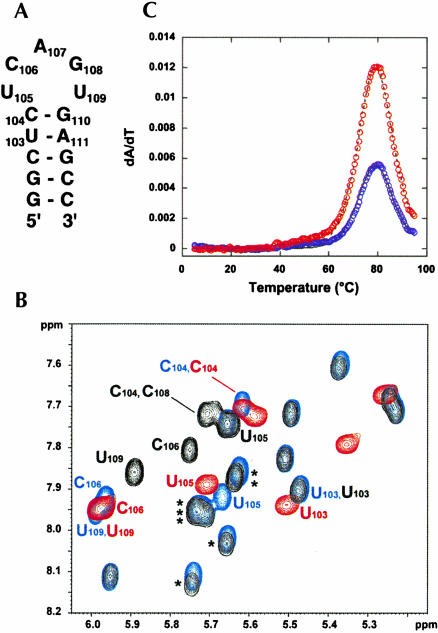
(A) Secondary structure of the minimal DCloop hairpin construct used for NMR studies using the numbering convention from HPwt. (B) 500 MHz 2D 1H-1H TOSCY spectra (50-msec mixing time) comparing H5-H6 correlations for HPwt (black), HPdc (blue), and DCloop (red) RNAs at 20°C. Asterisks (*) are used to indicate H5-H6 correlations from the poly-pyrimidine tract nucleotides. (C) First derivative UV melting profile of the DCloop hairpin (200 mM NaCl), with every 10th data point obtained at 260 and 280 nm shown as blue and red circles, respectively. The calculated fit to the data is indicated by solid (260 nm) and dashed (280 nm) lines.
Thermodynamic stability of the DCloop pentaloop
The thermodynamic stability derived from the mutant and wild-type hairpin pentaloop structures could not previously be calculated, because thermodynamic parameters for the poly-pyrimidine helix had not been explicitly determined. However, because the DCloop construct contains only Watson–Crick base pairs, the thermodynamics of which are extremely well characterized (Xia et al. 1998; Mathews et al. 1999), the thermodynamic contribution of the mutant pentaloop structure could be accurately determined. The optical melting profile of DCloop RNA at 200 mM NaCl reveals one intense transition (Fig. 2C ▶), with a melting temperature (tm) of 87.6°C and an enthalpy of −53.4 kcal/mole. The tm of the unfolding transition was unaffected by annealing conditions and independent of RNA concentration, consistent with its identification as the unfolding of a unimolecular hairpin. The observed stability of the DCloop hairpin was compared to thermodynamic predictions at 1 M NaCl for the nearest-neighbor stacking interactions of the Watson–Crick bp (Xia et al. 1998) and an entropic loop penalty of +5.6 kcal/mole for closing the pentaloop (Table 1 ▶; Mathews et al. 1999). The DCloop pentaloop structure therefore contributes −2.2 kcal/mole in enthalpy and −1.9 kcal/mole free energy [ΔG(37°C)] to the stability of the DCloop hairpin, which is comparable to the stability obtained from stabilizing tetraloop structures (Mathews et al. 1999; Proctor et al. 2002). Given the energetic contribution of the pentaloop structure to the stability of the DCloop hairpin and, by extension, the HPdc hairpin, it is possible to calculate that the pyrimidine tract contributes −23.1 kcal/mole in enthalpy and −1.0 kcal/mole in total free energy [ΔG(37°C)] to the stability of the P2b extended helical structure compared to the alternative, formation of a symmetric 8-nt internal loop (~ +3 kcal/mole; Mathews et al. 1999). The large ΔH contribution derives from the additional nearest-neighbor stacking interactions between the pyrimidine base pairs as well as stacking interactions with the Watson–Crick base pairs surrounding the poly-pyrimidine tract, although the total stacking energy is less than would be predicted for a similar number of Watson–Crick base pairs.
TABLE 1.
Thermodynamic parameters obtained at 1 M NaCl
| Observed | Predicteda | |||
| RNA | −ΔHtotal | −ΔGtotal | −ΔHtotal | −ΔGtotal |
| HPdc | 100.4 ± 1.1 | 10.8 ± 0.1 | 75.0 | 10.9 |
| DCloop | 53.4 ± 1.0 | 7.4 ± 0.3 | 51.2 | 5.5 |
| UCAG | 51.6 ± 1.5 | 7.2 ± 0.2 | 51.2 | 5.5 |
ΔH is model-dependent van’t Hoff enthalpy. ΔH and ΔG are reported in kcal/mole. ΔG is calculated at 37°C and compared to nearest-neighbor predictions for stacking (Xia et al. 1998), loop free energies, and tetraloop stabilization (Mathews et al. 1999). Parameters are the average of five independent measurements, and errors are standard deviations from repeated experiments.
aPrediction is based on the nearest-neighbor rules for stacking of the Watson-Crick bp in the stem and a +5.6 kcal/mole loop free-energy penalty for capping a helix with a 4- or 5-nt hairpin loop (Mathews et al. 1999).
Solution structure of the DCloop hairpin
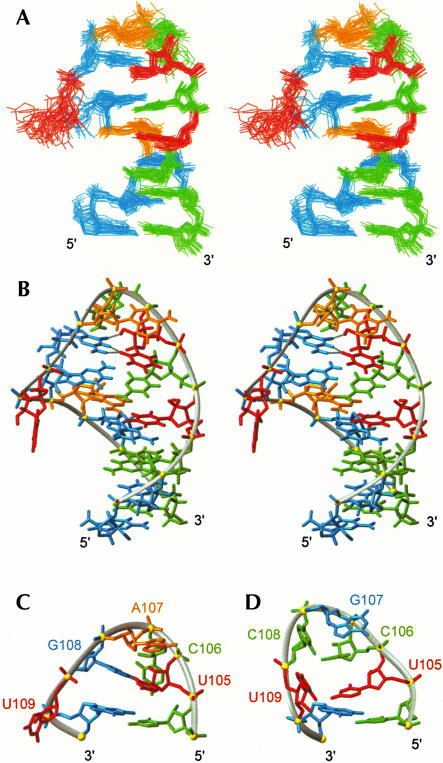
The structure determination of the DCloop hairpin used 283 NOE distance restraints, for an average of 18.8 NOE restraints per nucleotide, 86 dihedral angle restraints, and 14 residual dipolar couplings (Table 2 ▶). Superposition of the 20 lowest energy structures was performed over all heavy atoms except the bulge nucleotide, U109, and an RMSD to the mean of 0.63 ± 0.22 Å was observed for the ensemble (Fig. 3A ▶). The DCloop hairpin has a well defined structured loop on a 5-bp A-form helical stem, consistent with the experimentally observed thermodynamic stability. The anti-U105 and syn-G108 nucleotides form a non-Watson–Crick base pair that stacks on the closing C104•G110 base pair (Fig. 3B ▶). The second loop nucleotide, C106, is positioned in the minor groove of the loop with the H5-H6 edge facing the sugar moiety of U105 and the Watson–Crick face exposed to solution. The third loop nucleotide, A107, stacks on the G•U bp in the major groove with the Hoogsteen edge facing the C106 sugar and the Watson–Crick and sugar edges exposed to solution. The final loop nucleotide, U109, is bulged out into the major groove and is not well defined (Fig. 3A ▶).
TABLE 2.
Structural statistics for Dcloop
| Experimental data used for structure calculations | |
| NOE-derived distance restraints | |
| Intra-nucleotide NOEs | 157 |
| Inter-nucleotide NOEs | 125 |
| Hydrogen bond for paired residues | 16 |
| Dihedral restraints | 86 |
| Residual dipolar couplings | |
| 1DC-H (Hz) | 14 |
| Additional restraints | |
| Base pair planarity restraints | 6 |
| RMS deviation from experimental restraintsa | |
| Distance restraints (Å)b | 0.025 ± 0.001 |
| Dihedral restraints (°)c | 0.096 ± 0.027 |
| Dipolar couplings (Hz) | 0.308 ± 0.157 |
| Deviations from idealized geometrya | |
| Bonds (Å) | 0.004 ± 0.0001 |
| Angles (°) | 0.992 ± 0.004 |
| Impropers (°) | 0.399 ± 0.011 |
| Overall RMS deviation (Å) (residue: 1–9, 11–15)a | |
| From mean structure | 0.63 ± 0.22 |
| Mean pairwise | 0.95 ± 0.24 |
aAveraged over the accepted structures.
bNo violations > 0.2 Å.
cNo violations > 5°.
FIGURE 3.
(A) Stereoview of the solution structure ensemble of 20 lowest energy structures of DCloop RNA. Nucleotides are red (U), green (C), orange (A), and cyan (G). (B) Stereoview of the lowest energy structure of DCloop, with a major groove view of the pentaloop and hydrogen bonds for the U105-G108 base pair indicated by black dashed lines. (C) Major groove view of the DCloop pentaloop. (D) Major groove view of the HPwt pentaloop (PDB accession code: 1NA2; Theimer et al. 2003).
Initial structure calculations did not include hydrogen bonding restraints for the U105•G108 bp; however, all of the lowest energy structures consistently placed these nucleotides within hydrogen bonding distance and geometry. The observed orientation of the base pair in the calculated structures, extremely weak U105 and medium G108 imino proton intensities, partial protection of the G108 N2 amino protons, and undetectable U105-G108 imino-imino NOE crosspeak were inconsistent with a standard G•U reverse wobble base pair geometry (U105 and G108 imino protons hydrogen bonded to the O6 and O4 carbonyl of G108 and U105, respectively). Instead, the hydrogen bonding pattern for U105 and G108 could be inferred from their mutual orientation in structures calculated with no hydrogen bonds. These initial structures showed the bases positioned to form the U•G bp found in the UUCG tetraloop (discussed below). Inclusion of the hydrogen bonding restraints for this base pair (Allain and Varani 1995; Ennifar et al. 2000) did not cause any violation of the existing experimental restraints and were thus included in successive calculations.
The mutant DCloop hairpin has a completely different loop structure (Fig. 3C ▶) than that found for the wild-type loop sequence (Fig. 3D ▶; Theimer et al. 2003). Although the HPwt structure was solved with a similar quality and number of restraints (an average of 19.4 NOE restraints per nt) under identical solution conditions, the DCloop structure had 12 nonsequential loop NOE restraints, four times more than those observed for the HPwt loop, consistent with the mutant loop having a more compact structure. The placement of the first and last nucleotides in the loop is similar in the two structures, with the first U stacked over the closing base pair and the last U in the major groove. However, the first U in the wild-type hairpin is not in a base pair, and the other three nucleotides adopt completely different orientations and positions. Although overall the loop structures are different, the turn in the backbone occurs between the third and fourth nucleotides for both loops. It is clear from comparing the two structures that the better stacking and additional hydrogen bonds present in the mutant DCloop provide a structural explanation for the increased stability of the mutant compared to wild-type hairpin.
Comparison to the YNMG tetraloop family
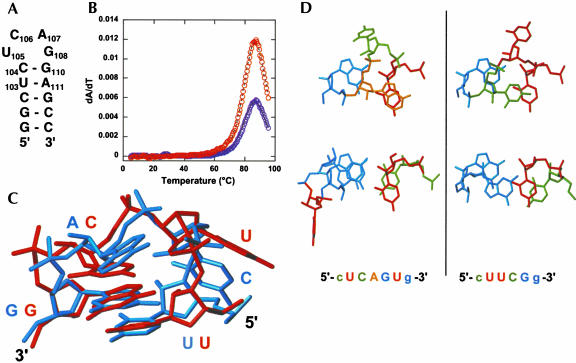
The structural results indicate that the increased thermodynamic stability of the mutant versus the wild-type P2b hairpin sequence derives from the formation of an additional base pair, hydrogen bonds, and stacking interactions in the pentaloop reminiscent of those observed in the YNMG family of tetraloops, of which UUCG is the most well characterized member (Proctor et al. 2002). A minimal RNA construct with the same Watson–Crick stem and loop nucleotide sequence as the DCloop RNA, except for deletion of U109, was designed to compare the thermodynamic properties of the DCloop pentaloop sequence with its UCAG tetraloop counterpart (Fig. 4A ▶). Because YNMG hairpins were previously described to form duplex at high concentration (Proctor et al. 2002), UCAG RNA samples were annealed prior to melting experiments. Melting profiles exhibited a single high-temperature transition similar to the DCloop RNA (Fig. 4B ▶), with thermodynamic parameters indicated in Table 1 ▶. The UCAG loop structure contributes −0.5 kcal/mole in enthalpy and −1.5 kcal/mole free energy [ΔG(37°C)] to the stability of the UCAG hairpin structure. This is consistent with the stability observed previously for the cUCAGg tetraloop in the context of a different helix (Proctor et al. 2002) and within experimental error of the total stability observed for the DCloop pentaloop structure. In fact, the stability difference between the DCloop and UCAG hairpins was found to be < 0.5 kcal/mole [ΔG(37°C)], under all solution conditions measured (data not shown). Therefore, although a single nucleotide bulge in the context of a helix contributes an entropic penalty of + 3.5 kcal/mole to the total stability of the structure (Mathews et al. 1999), a 3′ terminal bulge nucleotide appears to have an insignificant effect on the stability of this tetraloop.
FIGURE 4.
(A) Secondary structure of the UCAG tetraloop construct. Nucleotides are numbered using the numbering convention from DCloop. (B) UV melting profile of the UCAG hairpin measured at 200 mM NaCl. Data indicated as described in Figure 2C ▶. (C) Superposition of the DCloop hairpin structure with the UUCG tetraloop (PDB accession code: 1F7Y; Ennifar et al. 2000), superimposed over the UL1•GL4 base pair. The UUCG tetraloop is shown in red and the DCloop pentaloop in blue. (D) Comparison of the L1-L4 base pair and L3 nucleotide stacking interactions in the DCloop RNA (left) and UUCG tetraloop (right) structures. Nucleotides are colored as in Figure 1C ▶.
The UUCG tetraloop structure, which is the most stable and best characterized member of the YNMG family, was determined by both NMR and X-ray crystallography (Allain and Varani 1995; Ennifar et al. 2000), and has several distinctive features. Hydrogen bonding interactions occur between the first position (L1) nucleotide, UL1, O2 carbonyl and the fourth position (L4), GL4, H1 imino and H2 amino protons, as well as between the UL1 2′ hydroxyl proton and the O6 carbonyl of GL4. A hydrogen bond is also observed between the third position, CL3, H4 amino proton and the second position, UL2, pro-R phosphoryl oxygen. Nonhydrogen bonding features include a syn conformation for GL4, C2′-endo sugar puckers for UL2 and CL3, and nonstandard torsion angles for UL2 and the closing base pair G. Unusual chemical shifts for the loop and closing base pair include upfield chemical shifts for the closing base pair G H1′, CL3 H5′, and H5′′, and UL1 H2′ and downfield shifts for GL4 H2′ and H3′ (Allain and Varani 1995).
The DCloop has the same loop topology found in the UUCG tetraloop, with stacking of the L3 nucleotide over the basepair in the major groove and the minor groove position of the L2 nucleotide (Fig. 4C ▶). As described in the previous section, the hydrogen bonding pattern for the U•G basepair observed for DCloop is identical to the UUCG UL1•GL4 base pair (Fig. 3B ▶). The DCloop structure shows the same syn conformation for the GL4, C2′-endo sugar puckers for the CL2 and AL3, and similar nonstandard torsion angles for the loop nucleotides as described for UUCG (Allain and Varani 1995; Ennifar et al. 2000). In addition, unusual chemical shifts are also observed for the loop and closing base pair nucleotides in this structure, similar to the UUCG tetraloop (Allain and Varani 1995), although their relative magnitude is reduced.
Small differences between the UUCG tetraloop and the DCloop [UCAG(U)] are present, attributed primarily to the bulged U nucleotide. In the DCloop structure, the base of the GL4 nucleotide stacks over the base of the closing basepair G, rather than the sugar of the closing basepair G as seen in the UUCG structure (Fig. 4D ▶). This difference in stacking is reflected in the decreased upfield chemical shift of the closing base-pair G H1′ proton. The presence of the intervening bulge nucleotide allows better stacking of the G bases, most likely by decreasing torsional strain on the backbone. Additionally, the AL3 base stacks more over the UL1 base than CL3 in the UUCG structure (Fig. 4D ▶), resulting in smaller upfield shifts for the AL3 H5′ and H5′ protons, because the sugar is not stacked as well over the syn GL4. Overall, the bulge nucleotide can be easily accommodated by the UUCG tetraloop with only minor changes in the topology and characteristic features, and the DCloop pentaloop clearly displays the hallmark features of a YNMG tetraloop.
DISCUSSION
Telomere maintenance is particularly important in cells with high proliferation rates, such as the hemopoietic system, and mutations in the RNA component of telomerase have been linked to inherited human disorders with defects in highly proliferating cells (Ball et al. 1998; Vulliamy et al. 2001a, 2002). The DKC mutation, GC (107–108) → AG, compromises the catalytic function of the mutant telomerase complex (Fu and Collins 2003) consistent with the observed requirement of P2b-P3 pseudoknot formation for catalytic activity (Autexier et al. 1996; Tesmer et al. 1999). Previous studies have shown that this mutation destabilizes the P2b-P3 pseudoknot significantly and increases the stability of the mutant P2b hairpin conformation (Comolli et al. 2002; Theimer et al. 2003). We show here that the mutant P2b pentaloop forms a YNMG tetraloop structure with a single U nucleotide bulged into the major groove. The additional stability observed for the mutant P2b hairpin compared to wild-type results from significant changes in the stacking and hydrogen bonding interactions within the pentaloop alone, with no changes in the stem base-pairing, including the poly-pyrimidine tract. Further, we find that the total stability contributed by the mutant loop structure is similar to that observed for a related modestly stabilizing tetraloop. This slight stabilization of the mutant P2b hairpin was described previously as a small but contributing factor in shifting the pseudoknot-hairpin equilibrium to favor P2b hairpin formation by the DKC mutant sequence (Theimer et al. 2003). We see no evidence for the hyperstabilization of the mutant P2b structure (including the poly-pyrimidine tract) and different secondary structure compared to wild-type P2b predicted by Comolli et al. (2002). Although the occurrence of a stabilizing tetraloop structure in this particular case is due to random sequence mutation, and unlikely to be involved in tertiary structure or RNA–protein interactions, stabilizing the alternative conformation of the pseudoknot domain through formation of a more stable tetraloop structure contributes to the overall deleterious effect of the GC (107–108) → AG mutation.
Tetraloop structural motifs are a common structural feature of many ribosomal RNAs and other biologically important RNAs (Woese et al. 1990). Tetraloops have been found to be involved in RNA–RNA tertiary interactions (Tinoco 1996), RNA–protein interactions (Zell et al. 2002), and mediating correct folding of complicated RNA structures (Woese et al. 1990; Tinoco and Bustamante 1999). Phylogenetic studies of ribosomal RNAs have revealed several families of RNA tetraloops based on their frequent occurrence; the cUNCGg (where N is any nucleotide), cGNRAg (where R is a purine), and gCUUGc families (Woese et al. 1990) with lowercase letters surrounding the tetraloop sequence used to indicate the most common closing base pair. High-resolution structures have been determined for many tetraloops and have revealed that the increased thermodynamic stability of these tetraloop sequences compared to random loop sequences can be explained by extensive stacking and hydrogen bonding interactions in the loop regions (Heus and Pardi 1991; Allain and Varani 1995; Jucker and Pardi 1995; Cai et al. 1998; Ennifar et al. 2000).
A YNMG tetraloop family was recently proposed in which the UNCG tetraloop family is generalized to include tetraloops in which the first loop position may be either a U or a C (Y) and the third loop position may be either a C or an A (M) (Proctor et al. 2002). This extension is based on the observation of structural features, thermodynamic stability, and NMR and CD spectra characteristic of the UNCG family in the family of structures selected on the basis of stability using temperature-gradient gel electrophoresis (TGGE). In support of this proposal, the NMR structure of a uCACGg tetraloop from Coxsackievirus B3 which is involved in interactions with the viral 3C protease was solved and shown to contain the characteristic features of a UUCG tetraloop (Du et al. 2003). In the present study, we found that a disease-causing mutation in the P2b-P3 pseudoknot region of human telomerase RNA results in the formation of a pentaloop (cUCAGUg) which has the characteristic structural features of the YNMG family as well as similar stability, but utilizes other consensus nucleotides than the previously reported uCACGg structure (Du et al. 2003). Taken together, these structures provide strong evidence in favor of the proposal (Proctor et al. 2002) that tetraloops of the type (U/C)N(A/C)G form structures similar to the UUCG tetraloop and in combination with the UNCG family, comprise a larger YNMG structural family.
The solution structures of pentaloop forms of the GNRA tetraloop have previously been solved and contain the structural features of the standard GNRA tetraloop (Legault et al. 1998; Schärpf et al. 2000; Huppler et al. 2002). Therefore, a single nucleotide in the fourth position in the GNRA family (GNRNA) or the last position in the YNMG family (YNMGN) appears to be accommodated by the corresponding tetraloop structures without significantly compromising the consensus structure. These observations suggest that database searches for tetraloop occurrence may underrepresent the actual frequency of these structural motifs in biologically important RNAs. A database search of large and small subunit ribosomal RNA secondary structures (Proctor et al. 2002) shows that, after tetraloops (51%), pentaloops (21%) represent the next largest category of ribosomal RNA loops. As pentaloops occur with reasonably high frequency in important RNA structures, further occurrences of stabilizing tetraloop mimics are likely to be observed in a search of pentaloop structures.
MATERIALS AND METHODS
RNA synthesis and purification
Unlabeled and uniformly 13C,15N-labeled RNA oligonucleotides were prepared by in vitro transcription using purified His6-tagged T7 RNA polymerase and synthetic DNA oligonucleotides, and purified as described (Dieckmann and Feigon 1997). Unlabeled nucleoside triphosphates (NTPs) were purchased from Pharmacia, and individual 13C, 15N-labeled NTPs were purchased from Silantes. Purified RNAs were annealed at 65°C under dilute conditions (1–10 μM) in NMR buffer (10 mM sodium phosphate, pH 6.3, 200 mM KCl, 50 μM EDTA, 0.2 % NaN3) and concentrated by ultrafiltration to ~ 1.0 mM.
Thermal denaturation
Thermal melting experiments were collected on a Varian Cary 1 scanning spectrophotometer equipped with a Peltier heating accessory and temperature probe in double-beam mode. Percent transmittance was recorded as a function of probe temperature at 260 and 280 nm simultaneously. RNA samples (2–4 μM) were prepared, heated to 65°C for 10 min, unless otherwise indicated, slow-cooled to room temperature, and then equilibrated at 5.0°C for 30 min prior to data collection. High-concentration melting data were obtained using 0.1-cm pathlength cuvettes at 10- to 20-fold higher concentration than standard melting samples. The temperature was increased at a rate of 0.3°C/min from 5°–98°C with data collected at 0.3°C increments. Melting profiles were obtained by converting percent transmittance into absorbance and taking the first derivative of the absorbance with respect to temperature (∂A/∂T). Melting profiles were then subjected to nonlinear least squares parameter estimation of tm, i (melting temperature), ΔHi (van’t Hoff enthalpy), Ai260 and Ai280 (|g!A/|g!T amplitude at 260 and 280 nm) for each ith transition upon application of a sequential two-state unfolding model using the tmelt program (Theimer et al. 1998). Reported values are the average of five independent measurements.
The optical melting profiles of the DCloop hairpin RNA from stock solution revealed an additional unfolding transition with a tm of 42.4°C. Based on its high enthalpy (−113 kcal/mole) and observed concentration dependence of the tm, this transition was identified as duplex formation in the 1 mM stock solution. Annealing the DCloop RNA stock at 65°C for 10 min prior to melting experiments significantly reduced the duplex conformation. Comparison of the imino-imino region of a 2D 1H-1H NOESY, as well as 1D 1H spectra in 2H2O and 95% H2O/5% 2H2O, before and after annealing revealed that duplex formation was easily identifiable in NMR samples. Because duplex re-accumulated in the NMR sample over time, it was necessary to anneal the sample frequently during NMR data collection.
NMR spectroscopy
All NMR spectra were recorded on Bruker DRX 500 and 600 MHz spectrometers. Exchangeable proton spectra were measured in 95% H2O/ 5% 2H2O at 278K, and nonexchangeable proton spectra were measured in 99.999% 2H2O at 293K. The H2O resonance was suppressed in 95% H2O/5% 2H2O samples either by a 1–1 spin-echo or a WATERGATE water suppression scheme whereas low-power presaturation was used to suppress the HDO resonance in 99.999% 2H2O samples. Exchangeable protons and nitrogens in the Watson–Crick base pairs of the DCloop RNA were assigned using 2D NOESY (100 msec mixing time), 1H-15N HMQC, and 15N correlated CPMG-NOESY spectra (Cromsigt et al. 2001). Amino protons in the loop were assigned using a combination of 1H-15N HMQC and 15N correlated CPMG-NOESY spectra (Cromsigt et al. 2001). Nonexchangeable protons and carbons were assigned using 2D NOESY (50 and 250 msec), homonuclear TOCSY, 1H-13C CT-HSQC, 2D HCCH-COSY, 3D HCCH-TOCSY, and 3D NOESY-HMQC experiments (Cromsigt et al. 2001). Hydrogen bonding patterns were confirmed for the Watson–Crick base pairs using JNN-HNN-COSY (Luy and Marino 2000). 3JH3′P and 3JCP were measured using 31P spin echo difference CT-HSQCs to determine β and ɛ torsion angles (Wu et al. 2001). Residual dipolar couplings (RDCs) were measured for 1JHC in 5% C12E6/hexanol using CT-CE-HSQC (de Alba and Tjandra 2002). Spectra were processed and analyzed using Bruker XWINNMR 2.6 and Aurelia 3.108.
Structure calculation
Interproton distances from 2D NOESY and 3D NOESY-HMQC spectra were generated as described (Wu et al. 2001) except for the classification of semiquantitative NOEs, which were as follows: strong (1.8–3.5 Å), medium (2.5–4.5 Å), weak (3.5–5.5 Å), and very weak (4.5–6.5 Å). In total, 157 intranucleotide and 125 internucleotide distance restraints were generated. We obtained 86 dihedral angle constraints for α, β, δ, and ɛ backbone angles (Wu et al. 2001), and the γ dihedral angle for the nucleotides involved in base pairs in the helix except for the terminal base pair (C104–G110) were constrained to the A-form value (54 ± 30°; Cromsigt et al. 2001). Nucleotides with observable H1′–H2′ and H1′–H3′ correlations (C106, A107, and U109) in a 50-msec mixing time TOCSY, were constrained as C2′-endo (South-type; 145 ± 30°), and all other nucleotides which had no detectable H1′–H2′ correlations were constrained as C3′-endo (North-type; 82 ± 30°). Intranucleotide H1′ to aromatic NOEs from a 50-msec 2D NOESY indicated that all nucleotides fell into the anti range and could thus be constrained with a χ value of −160 ± 30°, except for G109 which was identified as a syn base and was restrained to a χ value of 60 ± 30°. Initial structure calculations included hydrogen-bonding distance restraints and weak planarity restraints (force constant of 3 kcal/Å2) for the five Watson–Crick base pairs only. Examination of the lowest energy structures consistently revealed distances and geometry consistent with a U6-G9 base pair, and therefore three hydrogen bond restraints (U6 O2 carbonyl to the G9 H1 imino and H2 amino protons and U6 2′ hydroxyl proton to G9 O6 carbonyl) were included for this base pair, as well as a weak planarity restraint consistent with the experimental data during successive rounds of calculation. As only one of the N6 amino protons for the AL3 nucleotide was detected, the presence of an AL3 exocyclic amino–CL2 pro-R phosphoryl oxygen hydrogen bond could not be confirmed for the DCloop structure. This hydrogen bond was therefore not included in the structure calculation presented here, although test calculations indicated that its inclusion does not violate the existing experimental restraints.
The structures of DCloop were calculated using XPLOR-NIH (Brünger 1992; Schwieters et al. 2003) starting from an extended, unfolded RNA conformation using NOE distances and dihedral angle restraints. The folding and refinement stages followed standard XPLOR protocols. Structures with no experimental restraint violations from the initial 200 calculated structures were then subjected to refinement against 14 residual dipolar couplings (RDCs). The protocol involved slow cooling from 1000K to 100K in 18 cycles of molecular dynamics corresponding to a total of 18 psec. During this stage, the force constant for dipolar couplings was slowly increased from 0.001 to 0.2 kcal mole−1Hz−2. The experimentally derived RDCs together with the lowest energy structures from the structure calculation employing NOE and dihedral restraints were used as input information. The grid search for the optimal values of the magnitude and asymmetry of the alignment tensor produced an optimal value of Da = −20 Hz and R = 0.575. The force constants used in the refinement stage of structure calculation were 50 kcal mole−1Å−2, 200 kcal mole−1deg−2, and 0.2 kcal mole−1Hz−2 for NOEs, dihedral angles, and RDCs, respectively. The correlation coefficients for the back-calculated RDCs increased from 0.85 to 0.98 after inclusion of the experimental RDCs in the structure calculation. Structural statistics for the 20 lowest energy structures are included in Table 2 ▶. All structures were viewed and analyzed using MOLMOL (Koradi et al. 1996).
Coordinate deposition
The coordinates for the 20 lowest energy structures of DCloop have been deposited in the Protein Data Bank (http://www.rcsb.org), accession code 1Q75. Chemical shift data for the DCloop RNA have been deposited in BioMagResBank (http://www.bmrb.wisc.org), accession code 5932.
Acknowledgments
We thank Robert Peterson for help with the NMR spectroscopy, Lukas Trantirek for assistance with the RDCs, Evan Feinstein for figure and manuscript preparation, and Craig Blois for sample preparation. This research was supported by the NSF and the NIH (J.F.) and an NIH NRSA postdoctoral fellowship (C.A.T.). The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5152303.
REFERENCES
- Allain, F.H.-T. and Varani, G. 1995. Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol. 250: 333–353. [DOI] [PubMed] [Google Scholar]
- Antal, M., Boros, E., Solymosy, F., and Kiss, T. 2002. Analysis of the structure of human telomerase RNA in vivo. Nucleic Acids Res. 30: 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier, C., Pruzan, R., Funk, W.D., and Greider, C.W. 1996. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 15: 5928–5935. [PMC free article] [PubMed] [Google Scholar]
- Ball, S.E., Gibson, F.M., Rizzo, S., Tooze, J.A., Marsh, J.C.W., and GordonSmith, E.C. 1998. Progressive telomere shortening in aplastic anemia. Blood 91: 3582–3592. [PubMed] [Google Scholar]
- Blackburn, E.H. 2001. Switching and signaling at the telomere. Cell 106: 661–673. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T. 1992. X-PLOR, version 3.1. A system for X-ray crystallography and NMR. Yale University Press, New Haven, CT.
- Cai, Z.P., Gorin, A., Frederick, R., Ye, X.M., Hu, W.D., Majumdar, A., Kettani, A., and Patel, D.J. 1998. Solution structure of p22 transcriptional antitermination N peptide box B RNA complex. Nat. Struct. Biol. 5: 203–212. [DOI] [PubMed] [Google Scholar]
- Chen, J.L., Blasco, M.A., and Greider, C.W. 2000. Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514. [DOI] [PubMed] [Google Scholar]
- Comolli, L.R., Smirnov, I., Xu, L.F., Blackburn, E.H., and James, T.L. 2002. A molecular switch underlies a human telomerase disease. Proc. Natl. Acad. Sci. 99: 16998–17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromsigt, J., van Buuren, B., Schleucher, J., and Wijmenga, S.S. 2001. Resonance assignment and structure determination for RNA. In Methods in enzymology (eds. T. James et al.), vol. 338, pp. 371–399. Academic Press, San Diego, CA. [DOI] [PubMed]
- de Alba, E. and Tjandra, N. 2002. NMR dipolar couplings for the structure determination of biopolymers in solution. Prog. Nucl. Magn. Reson. Spectros. 40: 175–197. [Google Scholar]
- Dieckmann, T. and Feigon, J. 1997. Assignment methodology for larger oligonucleotides: Application to an ATP-binding RNA aptamer. J. Biomol. NMR 9: 259–272. [DOI] [PubMed] [Google Scholar]
- Dokal, I. 2000. Dyskeratosis congenita in all its forms. Brit. J. Haematol. 110: 768–779. [DOI] [PubMed] [Google Scholar]
- Dragon, F., Pogacic, V., and Filipowicz, W. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20: 3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z.H., Yu, J.H., Andino, R., and James, T.L. 2003. Extending the family of UNCG-like tetraloop motifs: NMR structure of a CACG tetraloop from coxsackievirus B3. Biochemistry 42: 4373–4383. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Nikulin, A., Tishchenko, S., Serganov, A., Nevskaya, N., Garber, M., Ehresmann, B., Ehresmann, C., Nikonov, S., and Dumas, P. 2000. The crystal structure of UUCG tetraloop. J. Mol. Biol. 304: 35–42. [DOI] [PubMed] [Google Scholar]
- Fu, D. and Collins, K. 2003. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell 11: 1361–1372. [DOI] [PubMed] [Google Scholar]
- Gilley, D. and Blackburn, E.H. 1999. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc. Natl. Acad. Sci. 96: 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L. 2003. Biochemical aspects of telomerase function. Cancer Lett. 194: 139–154. [DOI] [PubMed] [Google Scholar]
- Hathcock, K.S., Hemann, M.T., Opperman, K.K., Strong, M.A., Greider, C.W., and Hodes, R.J. 2002. Haploinsufficiency of mTR results in defects in telomere elongation. Proc. Natl. Acad. Sci. 99: 3591–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus, H.A. and Pardi, A. 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253: 191–194. [DOI] [PubMed] [Google Scholar]
- Huppler, A., Nikstad, L.J., Allmann, A.M., Brow, D.A., and Butcher, S.E. 2002. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat. Struct. Biol. 9: 431–435. [DOI] [PubMed] [Google Scholar]
- Jucker, F.M. and Pardi, A. 1995. Solution structure of the CUUG hairpin loop: A novel RNA tetraloop motif. Biochemistry 34: 14416–14427. [DOI] [PubMed] [Google Scholar]
- Kelleher, C., Teixeira, M.T., Forstemann, K., and Lingner, J. 2002. Telomerase: Biochemical considerations for enzyme and substrate. Trends Biochem. Sci. 27: 572–579. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL—A program for display and analysis of macromolecular structures. J. Mol. Graphics 14: 51–55. [DOI] [PubMed] [Google Scholar]
- Legault, P., Li, J., Mogridge, J., Kay, L.E., and Greenblatt, J. 1998. NMR structure of the bacteriophage λ N peptide/BoxB RNA complex—Recognition of a gRNA gold by an arginine-rich motif. Cell 93: 289–299. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Kha, H., Ungrin, M., Robinson, M.O., and Harrington, L. 2002. Preferential maintenance of critically short telomeres in mammalian cells heterozygous for mTert. Proc. Natl. Acad. Sci. 99: 3597–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luy, B. and Marino, J.P. 2000. Direct evidence for Watson–Crick base pairs in a dynamic region of RNA structure. J. Am. Chem. Soc. 122: 8095–8096. [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Mitchell, J.R. and Collins, K. 2000. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell 6: 361–371. [DOI] [PubMed] [Google Scholar]
- Mitchell, J.R., Cheng, J., and Collins, K. 1999a. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 19: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, J.R., Wood, E., and Collins, K. 1999b. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402: 551–555. [DOI] [PubMed] [Google Scholar]
- Montanaro, L., Chilla, A., Trere, D., Pession, A., Govoni, M., Tazzari, P.L., and Derenzini, M. 2002. Increased mortality rate and not impaired ribosomal biogenesis is responsible for proliferative defect in dyskeratosis congenita cell lines. J. Invest. Dermatol. 118: 193–198. [DOI] [PubMed] [Google Scholar]
- Proctor, D.J., Schaak, J.E., Bevilacqua, J.M., Falzone, C.J., and Bevilacqua, P.C. 2002. Isolation and characterization of a family of stable RNA tetraloops with the motif YNMG that participate in tertiary interactions. Biochemistry 41: 12062–12075. [DOI] [PubMed] [Google Scholar]
- Schärpf, M., Sticht, H., Schweimer, K., Boehm, M., Hoffmann, S., and Rösch, P. 2000. Antitermination in bacteriophage λ: The structure of the N36 peptide-boxB RNA complex. Eur. J. Biochem. 267: 2397–2408. [DOI] [PubMed] [Google Scholar]
- Schwieters, C.D., Kuszewski, J.J., Tjandra, N., and Clore, G.M. 2003. The Xplor-NIH NMR molecular structure determination package. J. Mag. Reson. 160: 65–73. [DOI] [PubMed] [Google Scholar]
- Tesmer, V.M., Ford, L.P., Holt, S.E., Frank, B.C., Yi, X.M., Aisner, D.L., Ouellette, M., Shay, J.W., and Wright, W.E. 1999. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol. Cell. Biol. 19: 6207–6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, C.A., Wang, Y., Hoffman, D.W., Krisch, H.M., and Giedroc, D.P. 1998. Nonnearest neighbor effects on the thermodynamics of unfolding of a model mRNA pseudoknot. J. Mol. Biol. 279: 545–564. [DOI] [PubMed] [Google Scholar]
- Theimer, C.A., Finger, L.D., Trantirek, L., and Feigon, J. 2003. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. 100: 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco, I.J. 1996. RNA Enzymes: Putting together a large ribozyme. Curr. Biol. 6: 1374–1376. [DOI] [PubMed] [Google Scholar]
- Tinoco, I.J. and Bustamante, C. 1999. How RNA folds. J. Mol. Biol. 293: 271–281. [DOI] [PubMed] [Google Scholar]
- Tzfati, Y., Knight, Z., Roy, J., and Blackburn, E.H. 2003. A novel pseudoknot element is essential for the action of a yeast telomerase. Genes & Dev. 17: 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulliamy, T., Marrone, A., Goldman, F., Dearlove, A., Bessler, M., Mason, P.J., and Dokal, I. 2001a. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature 413: 432–435. [DOI] [PubMed] [Google Scholar]
- Vulliamy, T.J., Knight, S.W., Mason, P.J., and Dokal, I. 2001b. Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cells Mol. Dis. 27: 353–357. [DOI] [PubMed] [Google Scholar]
- Vulliamy, T., Marrone, A., Dokal, I., and Mason, P.J. 2002. Association between aplastic anaemia and mutations in telomerase RNA. Lancet 359: 2168–2170. [DOI] [PubMed] [Google Scholar]
- Woese, C.R., Winker, S., and Gutell, R.R. 1990. Architecture of ribosomal RNA: Constraints on the sequence of “tetra-loops.” Proc. Natl. Acad. Sci. 87: 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Pok, K., Butcher, S.E., Kang, S., Chanfreau, G., and Feigon, J. 2001. A novel family of RNA tetraloop structure forms the recognition site for S. cerevisiae RNase III. EMBO J. 20: 7240–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, T.B., SantaLucia, J., Burkard, M.E., Kierzek, R., Schroeder, S.J., Jiao, X.Q., Cox, C., and Turner, D.H. 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson–Crick base pairs. Biochemistry 37: 14719–14735. [DOI] [PubMed] [Google Scholar]
- Zell, R., Sidigi, K., Bucci, E., Stelzner, A., and Gorlach, M. 2002. Determinants of the recognition of enteroviral cloverleaf RNA by coxsackievirus B3 proteinase 3C. RNA 8: 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]